American Journal of Plant Sciences
Vol. 4 No. 7 (2013) , Article ID: 34761 , 12 pages DOI:10.4236/ajps.2013.47184
Functional Aspects of Silencing and Transient Expression of psbS in Nicotiana benthamiana
![]()
1Department of Biochemistry and Genetics, The Connecticut Agricultural Experiment Station, New Haven, USA; 2Institute of Cell and Molecular Biology, Tartu University, Tartu, Estonia.
Email: Richard.Peterson@ct.gov
Copyright © 2013 Richard B. Peterson et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received May 9th, 2013; revised June 9th, 2013; accepted June 25th, 2013
Keywords: Fluorescence; Gas Exchange; Nonphotochemical Quenching; Quantum Yield; 810-nm Absorbance
ABSTRACT
MicroRNA-based gene silencing is a functional genomics tool for a wide range of eukaryotes. As a basis for broader application of virus-induced gene silencing (VIGS) to photosynthesis research, we employed a tobacco rattle virus (TRV) vector to silence expression of the nuclear psbS gene in Nicotiana benthamiana. The 22-kiloDalton psbS protein is essential for xanthophylland H+-dependent thermal dissipation of excitation in higher plants widely known as nonphotochemical quenching (NPQ). Controls treated with the TRV-VIGS vector containing a bacterial chloramphenicol resistance gene as the silencing target were included to test for non-silencing effects of the viral vector system. PsbS protein was undetectable and both psbS mRNA transcript levels and NPQ capacity were dramatically reduced in new leaf tissue of VIGS-psbS plants only. Photosynthetic performance in TRV-VIGS-treated and uninfiltrated plants was assessed by application of CO2 exchange, chlorophyll fluorescence, and in vivo absorbance changes at 810 nm. TRVVIGS caused a mild stress based on pigment content and light absorption characteristics in some cases. To assess transient complementation of NPQ, the endogenous psbS gene was silenced using only the transit sequence in the TRV vector followed by Agrobacterium-mediated transient expression of a modified gene consisting of an altered transit sequence fused to the native mature protein sequence. Nevertheless, NPQ in infused fully expanded leaves that expressed this re-introduced form was not fully restored indicating the possible importance of psbS incorporation prior to formation of grana stacks.
1. Introduction
The exponential growth in gene sequence information has led to use of reverse genetics to dissect complex functions by selective protein elimination. Most efforts have relied on permanent modification of the host genome by mutation or transformation. However, transgenic complementation studies are beset by limitations imposed by transformation frequency and silencing. Screening for deletion alleles is laborious and appropriate T-DNA or transposon insertion “knock-out” lines may be unavailable or non-viable as homozygotes.
Virus-induced gene silencing (VIGS) in plants can substantially improve the speed and convenience of the reverse genetics approach [1,2]. Post-transcriptional silencing of endogenous genes [3,4], induced by engineering genomic sequences into RNA genome viruses [5,6], circumvents the need to generate stable transgenic or mutant stocks. Silencing does not rely upon integration of transgenes into host genomes. Virus propagation plus dissemination and microRNA amplification plus transport combine to deliver VIGS at high levels in growing parts. Suppression of gene expression is often far higher than is achieved in stable antisense transgenic lines [7]. Gene silencing is highly sequence specific, requiring only a short section of perfect nucleic acid identity. Selective silencing of individual highly similar gene family members can be accomplished by targeting less conserved 5’ or 3’ UTR or promoter sequences. Alternatively, gene families can be co-silenced if shared sequences are targeted.
Photosynthesis is a dynamic and highly integrated process. Hence, ancillary effects of viral infection could confound interpretation of a specific silencing event [2]. The nuclear-encoded 22-kD psbS protein is essential for regulated dissipation of excess quanta in PSII referred to as nonphotochemical quenching (NPQ) [8]. Mutation of psbS in Arabidopsis thaliana does not impair growth nor dramatically alter Photosystem I/II (PSI/PSII) processes other than NPQ [8-12]. We sought to determine if a comparable psbS deletion phenocopy could be created in Nicotiana benthamiana by VIGS and assess possible adverse effects of the systemic infection. Hence, untreated plants were compared to plants infused with the tobacco rattle virus silencing system (TRV-VIGS) containing either of two silencing targets: the N. benthamiana psbS gene or an extraneous bacterial gene sequence as a control.
A practical extension of VIGS-psbS is reintroduction of psbS as a basis for structure/function and PSII assembly studies. We demonstrate Agrobacterium-mediated transient expression of high levels of native N. benthamiana psbS that partially restores NPQ capacity. The latter results are discussed in terms of developmental aspects of photosynthetic gene expression as well as protein complex formation and turnover in relation to grana stacking.
2. Materials and Methods
2.1. Plant Growth Conditions
Nicotiana benthamiana was grown in 4-L pots on a nutrient-supplemented peat substrate in a growth chamber. The irradiance was 250 mmol quanta m−2·s−1 with a light/dark cycle of 14/10 h, a temperature regime of 27˚C/20˚C, and relative humidity of 65%/90%.
2.2. Molecular Materials and Procedures
DNA constructs were prepared using standard procedures. Full-length N. benthamiana psbS cDNA (Gb EU645483) was generated by PCR using oligonucleotides NTABS1 and NTABS2 (Table 1), Qiagen Taq (Qiagen, Valencia, CA, USA), and DNA prepared from leaf RNA using M-MuLV reverse transcriptase (Roche, Indianapolis, IN, USA). The resulting DNA fragment was cloned into pCR2.1 (Invitrogen, Carlsbad, CA, USA) to generate plasmid pRH231. The tobacco rattle virus (TRV) system consists of two plasmids (pTRV1 and pTRV2-GATEWAY) with the engineered viral genome embedded in an Agrobacterium binary vector for efficient delivery in planta [13-15]. Target DNA for silencing RNA production was incorporated in the viral vector using the GATEWAY cloning system (Invitrogen). Five strains of Agrobacterium GV2260 were used containing the following plasmids: pTRV1 facilitates replication of the viral genome; pTRV2 with a chloramphenicol resistance marker (CmR) sequences; pTRV2 with the N. benthamiana phytoene desaturase gene (PDS); pDEST5 contained the pTRV2-GATEWAY vector with fulllength psbS cDNA from N. benthamiana; and pAV4 composed of pTRV2-GATEWAY containing the 184- base pair (bp) N. benthamiana psbS transit sequences [16]. Liquid cultures of pTRV2-containing Agrobacterium strains were each mixed 1:1 with strain YY192 containing pTRV1. Suspensions were injected into two fully expanded lower leaves of three-week-old N. benthamiana seedlings using a needleless hypodermic syringe. Measurements on newly emerged yet expanded leaves commenced two weeks after infusion.
Plasmid pRH231 was amplified with primers GATESPROF and GATESPROR or GATESPROF and GATESPROR2 and Qiagen Taq. The resulting DNA products were cloned into pDONOR207 as recommended with the GATEWAY cloning system (Invitrogen)

Table 1. Oligonucleotide
generating plasmids pENT5 and pAV3, respectively. These plasmids and pTRV2-GATEWAY were likewise treated as recommended to generate pDEST5 and pAV4, respectively.
Oligonucleotides NBTW1 and NBTW2 and separately NBTW3 and NBTW4 were amplified by PCR using the Expand High Fidelity system (Roche). The resulting products were joined using the same polymerase generating a 215-bp DNA fragment. Plasmid pRH231 was likewise amplified with oligonucleotides NBTW5 and NBEN20. This product was joined to the 215-bp DNA fragment to generate an 840-bp DNA fragment. The 840-bp fragment was cloned into pCR 2.1 (Invitrogen) generating plasmid pAV18. Plasmids pAV18 and pRH118 were cut with restriction endonuclease enzymes KpnI and BamHI and ligated. The resulting clone pNS451 includes the full-length N. benthamiana psbS cDNA under a CaMV35S promoter and nos termination sequences in the binary vector pCAMBIA1300 (CAMBIA, Canberra, Australia). DNA construct integrity was confirmed by sequence analysis.
Membrane protein fractions were prepared and probed for psbS by Western analysis as described previously [11]. The psbS content was related to a known mass of purified spinach psbS by densitometry. Levels of Lhca and Lhcb proteins were assessed immunologically (Agrisera). Chlorophyll (Chl) levels were measured in acetone extracts of leaf samples [17]. The density of Rubisco active sites was calculated after electrophoresis and densitometry of leaf extracts [18].
2.3. Gas Exchange and Optical Methods
The two-channel fast-response leaf gas exchange measurement system (Fast-Est, Tartu, Estonia) has been de scribed [17]. A portion of the attached test leaf was enclosed in the flow-through sandwich-type chamber (32 mm diameter, 3 mm height) and flushed with gas at a rate of 0.5 mmol·s−1. The upper epidermis of the leaf was pasted with starch to a window separating the chamber volume from a thermostatting water jacket. The water jacket was maintained at 22˚C and heat budget calculations indicated that leaf temperature never exceeded 23˚C. Uptake of CO2 was monitored with an infrared gas analyzer LI 6251 (LiCor, Lincoln, NE, USA) and a micro-psychrometer detected transpiration. Rates of CO2 and H2O exchange enabled calculation of dissolved CO2 concentration at the carboxylation site considering stomatal and mesophyll diffusion resistances. Linear electron transport rate (JC) associated with photosynthetic carbon metabolism was calculated as:
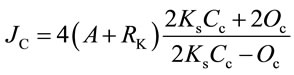 (1)
(1)
where A is the rate of net CO2 assimilation (mmol·m−2·s−1), RK is Krebs cycle respiration in the light, Ks is the Rubisco CO2/O2 specificity factor, and Cc and Oc are the dissolved CO2 and O2 concentrations (mM) at the carboxylation sites [19]. The gas phase O2 content was monitored using a calcia zirconia electrode Ametek S-3A (Thermox, Pittsburgh, PA, USA). Integration of the O2 pulse following a saturating single turnover Xe flash at low background O2 levels (10 to 50 mmol·mol−1) provided a measure of PSII reaction center (RC) density [20].
All light beams were directed to the leaf by a fiber optic guide (Fast-Est, Tartu, Estonia). Actinic white light (WL) and fluorescence saturation pulses (10,000 mmol·quanta·m−2·s−1 for 1.5 s) were provided by Schott KL 1500 sources. Measuring beams for fluorescence and 810-nm transmittance illuminated separate spots on the upper leaf surface. Far red light (FR, 50.6 mmol·quanta·m−2·s−1, 720 nm) was provided by a feedback-stabilized light-emitting diode source (Fast-Est, Tartu, Estonia).
Chl fluorescence yield was measured with a PAM-101 equipped with an ED-101 emitter-detector unit (H. Walz, Effeltrich, Germany). Corrections for leakage of the measuring beam to the detector, detector oversaturation, fluorescence undersaturation during pulses, and PSI fluorescence were applied [21]. The quantum yield of PSII electron transport based on fluorescence (YF) and corresponding electron transport rate (JF) are given by:
 (2)
(2)
and
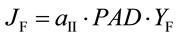 (3)
(3)
where Fs is the steady state and Fm is the pulse-saturated fluorescence yield and PAD (mmol·quanta·m−2·s−1) is the photon absorption density [22]. The PSII partitioning coefficient (aII) is estimated based on the quantum yields of electron flow to CO2 (YC) and PSII electron transport (YF) in limiting light (LL) such that:
 (4)
(4)
where
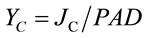 (5)
(5)
The corresponding partitioning coefficient for PSI (aI) was likewise based on the optically-measured rate of electron flow from plastoquinol to PSI (JI) and reduction level of P700 (P700red) as:
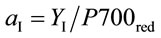 (6)
(6)
where
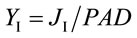 (7)
(7)
Light absorption by non-photosynthetic chromophores (anp) was calculated as 1 - aI - aII. Partitioning of FR quanta to PSI [aI(FR)] was based on O2 evolution (see above) assuming a PSII quantum efficiency of 0.8. NPQ is expressed in terms of the rate constant for regulated quenching of PSII excitation [23]:
 (8)
(8)
Leaf transmittance at 810 nm was monitored using a single-beam photometer FS810-A (Fast-Est, Tartu, Estonia). Signal deconvolution has been described [24].
2.4. Experimental Protocols
Test plants were pre-darkened for 12 h to ensure full relaxation of NPQ. After mounting and equilibrating the test leaf in the chamber, dark respiration rate and the dark-adapted minimal (Fod) and maximal (Fmd) fluorescence yields were recorded. Next, saturating single turnover flashes were applied to assess PSII RC density. The FR light titration procedure that followed enabled assessment of PSI donors and equilibrium constants [24]. Steady state levels of A, Fs, Fm, and the light-dark 810-nm absorption transient were recorded at incident irradiances of 2000, 1400, 760, 460, 260, 140, 75, 35, 13, and 0 mmol·quanta·m−2·s−1 (360 mmol CO2 mol−1, 21% O2). These quantities were then recorded at an irradiance of 2000 mmol·quanta·m−2·s−1 and a gas phase of 2000 mmol CO2 mol−1 (HCHL conditions). Measurements were recorded at an irradiance of 760 mmol·quanta·m−2·s−1 and CO2 levels of 360, 80, 40, 0, 200, and 520 mmol·mol−1. The O2 concentration was reduced to 2% and measurements were recorded at CO2 levels of 200, 100, 50, 0, and 200 mmol·mol−1. An integrating sphere and microspectrometer (Ocean Optics, Dunedin, FL, USA) apparatus was used to assess the spectral transmittance of a 2-cm2 disc from the chamber-enclosed area of the test leaf. Leaf absorption coefficients for WL and FR (aWL and aFR, respectively) were calculated considering the spectral emission profiles for these sources. The enclosed area of the test leaf (7.8 cm2) was excised, frozen in liquid N2, and stored at −80˚C pending membrane protein extraction and pigment analysis.
3. Results
3.1. Virus Induced Gene Silencing of psbS Expression
The primary effect of VIGS is reduction in mRNA transcript level for the targeted gene. Reverse Transcriptase (RT) PCR established that psbS transcript levels were specifically lowered in N. benthamiana plants treated with TRV-VIGS-psbS (pDEST5, full-length psbS cDNA) compared to uninfused controls (Figure 1). This reduc-
 (a)
(a)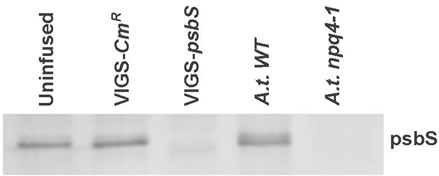 (b)
(b)
Figure 1. Assessment of psbS transcript levels. A. RT-PCR was performed using total RNA samples from leaves receiving no Agro-infiltration (uninfused 1-3) and from leaves inoculated with pTRV1 and pTRV2-NbpsbS (silenced 1-3). Samples from PCR cycles 18, 21, 24, and 27 amplified a 798-bp psbS-specific band and a 1.6-kb ubiquitin band (ubi) from cycles 27, 30, 33, and 36. B. An anti-psbS Western blot of leaf total membrane protein fractions compares psbS expression from uninfused N. benthamiana and from plants treated with a pTRV2 control construct (VIGS-CmR, see text) or pTRV2-NbpsbS (VIGS-psbS). We note that the response to VIGS-psbS was similar to the effect of deletion of psbS in Arabidopsis thaliana (A.t.).
tion in transcript level was associated with a loss of immunodetectable psbS (Figure 1, Table 2). As a positive control, we routinely confirmed the presence of VIGS in separate plants by targeting the phytoene desaturase gene (PDS), which causes bleaching of emerging leaves.
Application of VIGS typically results in a chimeric expression pattern for the targeted gene in plants [2]. We confirmed this with respect to psbS accumulation and expression of NPQ (Equation (8)). Figure 2 shows NPQ (panels A and B) and psbS levels (panels C and D) in leaves sampled 10 to 15 days after infusion with VIGSpsbS. Leaves already substantially developed at day zero exhibited the lowest level of additional expansion.
Younger leaves expanded relatively more (intermediate level) while leaves incipient at the time of inoculation were exposed to VIGS-psbS throughout their growth (100% expansion). Neither NPQ nor psbS levels in control (uninfused) leaves exhibited a clear effect of either sampling position along the long axis or leaf developmental stage for the low and intermediate expansion levels. However, NPQ was significantly elevated in the youngest leaves. In contrast, VIGS-psbS plants showed very low
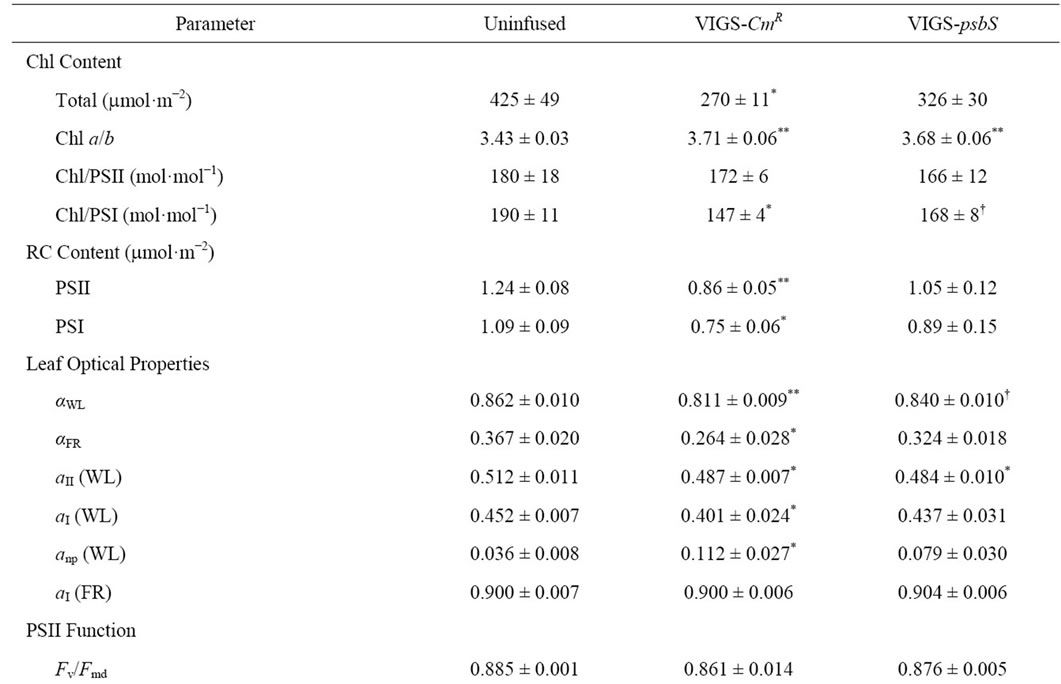

Table 2. Summary of leaf photosynthetic parameters for normal and TRV-VIGS-Treated N. benthamiana.
NPQ in newly developed leaves and in the basal and middle regions of leaves that underwent intermediate expansion. The same pattern was evident in the distribution of psbS. Cell division and chloroplast maturation is most active in the basal region of the developing dicotyledonous leaf. Silencing of psbS expression is most evident in leaf tissue formed post-inoculation.
Table 2 confirms that co-suppression of psbS and NPQ was a specific result of TRV-VIGS treatment with psbS as the silencing target. Separate control plants (VIGS-CmR) were likewise treated with vector system containing a bacterial chloramphenicol resistance gene (kindly provided by S. P. Dinesh-Kumar) in the target domain to test for effects of proliferation of a non-silencing TRV-VIGS
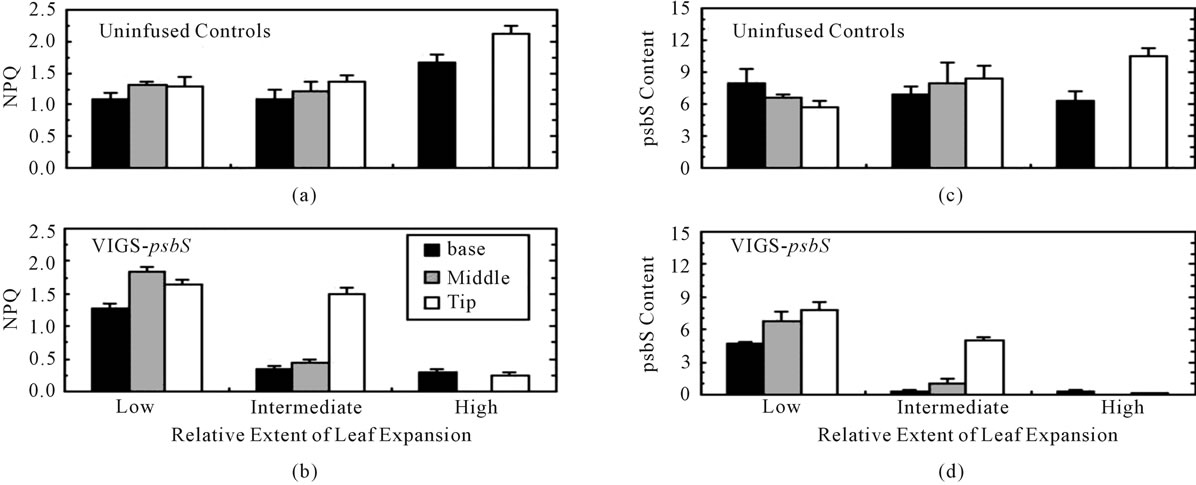
Figure 2. Developmental aspects of silencing of psbS expression in N. benthamiana. Three-week-old seedlings were inoculated with Agrobacterium/VIGS-psbS on day zero and then returned to the growth chamber. Beginning ten days later, 1.6-cm diameter discs were removed from selected leaves of fully dark-adapted plants in the basal, middle, and tip regions. Each disc was subjected to light stress treatment (500 mmol·quanta·m−2·s−1 in air) during which Fmd was estimated as the fluorescence peak emission and Fm was measured during a saturating multiple turnover flash 3 min later to calculate NPQ (Equation (8)). The leaf disc was then frozen in liquid N2 pending immunoassay of psbS (psbS content refers to mg per disc). The length of each leaf blade was measured on day zero and the day of sampling. Low, intermediate, and high extents of leaf expansion correspond to average increases of 48%, 71%, and 100%. Due to their small size, leaves in the highest expansion category were sampled at only two positions. Error bars represent ± SE.
system. We note that routine chromatographic analyses of photosynthetic pigments revealed normal function of the xanthophyll cycle in VIGS-psbS plants in response to changing irradiance.
3.2. VIGS in Relation to PSII and PSI Function
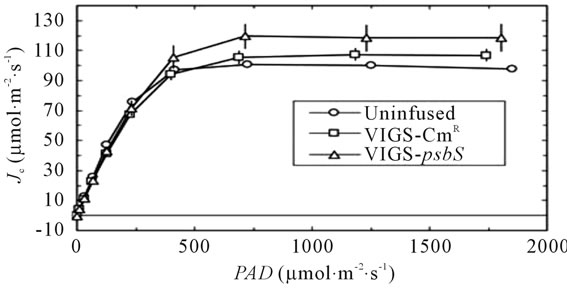
The sequential transport of electrons from H2O to CO2 involves cooperative energy transduction in PSII and PSI. Thus, rates of linear electron transport to CO2 (JC, Equation (1)) are informative in comparing uninfused, VIGSCmR, and VIGS-psbS plants. Values of JC were generally higher in TRV-VIGS-treated plants when irradiance was saturating, particularly for VIGS-psbS (Figure 3(a)). Thus, a reduction in H+-dependent photoprotective response can be compensated to a certain extent by higher rates of photochemistry. It is noteworthy that the maximal positive effect of TRV-VIGS on JC occurred at the higher O2 level (21%) although it was detectable at 2% O2 (Figure 3(b)). This enhancement could indicate a more prominent role for O2 as a terminal electron acceptor in TRV-VIGS-treated plants.
Previous reports have noted strongly positive relationships linking the quantum yields of PSII and PSI over a wide range of conditions [25,26]. Figure 4 compares the extent of reduction of P700 (PSI quantum yield, see Equations (6) and (7)) to the corresponding PSII quantum yield based on Chl fluorescence (YF, Equation (2)). We employed a redox equilibrium model [24] to deconvolute
 (a)
(a)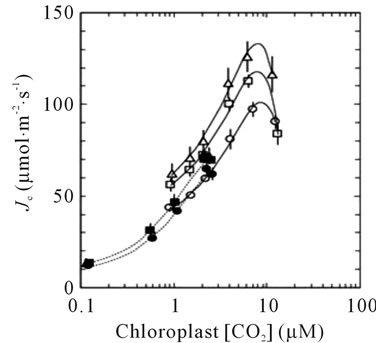 (b)
(b)
Figure 3. Responses of JC (Equation (1)) to irradiance (a) and Cc (b). Symbols are defined in (a). The open symbols in (b) indicate 21% O2 while the closed symbols indicate 2% O2 in the gas phase. The lines in (b) are second order polynomial fits.
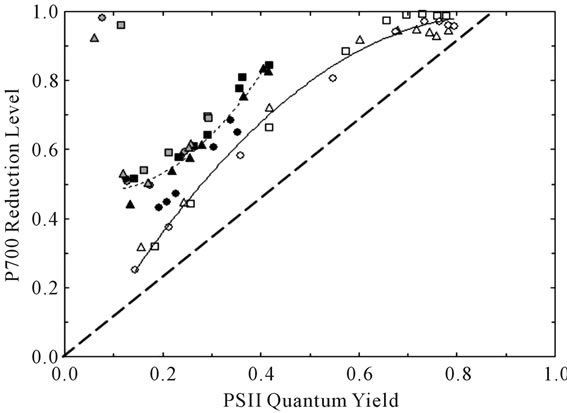
Figure 4. Relationship between the degree of reduction of the PSI reaction center P700 and PSII quantum yield. The symbol shapes are defined in Figure 3. The symbol shadings refer to the parameter varied: irradiance in 21% O2 (white), CO2 in 21% O2 (black), and CO2 in 2% O2 (gray). The solid and short-dashed lines are second order polynomial fits to the irradiance-dependent and CO2-dependent responses, respectively. Note that the symbols in the upper left of the figure were obtained under conditions of extremely limited acceptor availability (CO2-free 2% O2) wherein PSI acceptors were maximally reduced. The heavy-dashed line is the expected dependence assuming the complete absence of both PSI cyclic electron flow and blockage of P700 photooxidation due to reduced acceptors.
the 810-nm light-dark absorbance change associated with P700+ accumulation [27]. This model reports P700 reduction levels free of interference due to absorption by oxidized plastocyanin (PC+). It is well known that control of the rate of electron donation from plastoquinol profoundly affects the steady state reduction level of P700 [28]. In addition, PSI cyclic electron flow [29] and accumulation of reduced acceptors is associated with curvilinear plots like Figure 4 [30,31]. We point out the contrasting trends in extent of P700 reduction versus YF observed when CO2 (dashed line) or irradiance (solid line) was varied. Closer inspection of results obtained at 21% O2 and YF values < 0.42 revealed that the rate of P700 excitation was 28% and 36% faster than the rate of electron donation from plastoquinol when irradiance and CO2 were varied, respectively. Such imbalances in quantum and electron utilization are diagnostic for participation of the acceptor side in control of PSI energetics. We conclude that enhanced control by the PSI acceptor side contributed to the higher P700 reduction levels observed when CO2 was varied. These differences may involve latencies associated with the experimental protocol and illustrate plasticity in coordinate control of electron transport by the donor and acceptor sides. Compared to irradiance and CO2, TRV-VIGS imposed only minor effects on both P700 redox state and PSII quantum yield based on two-way analyses of variance.
Benchmark parameters reflecting composition and function of the photosynthetic apparatus for uninfused and VIGS-treated plants are shown in Table 2. TRV-VIGS was associated with a variable lowering of total Chl content. Consequently, absorption coefficients (aWL and aFR) for white light (WL) and far red (FR) light were lower. Coefficients for respective partitioning of absorbed WL quanta to PSII and PSI, aII (Equation (4)) and aI (Equation (6)), were marginally lower in virus-treated plants and this was ascribed to greater light interception by non-photosynthetic chromophores (i.e., cytochromes, anthocyanins, etc.) indicated by higher levels of anp. No effect of TRV-VIGS on partitioning of FR to PSI was detected.
We sporadically encountered reduced Chl levels in TRV-treated N. benthamiana consistent with reports of mild disease symptoms for this system [2,15]. This, in addition to TRV-dependent effects on RC density and Chl a/b (Table 2), led us to consider the possibility that some reorganization of PSI and PSII could occur. Figure 5 compares the relative abundances of light-harvesting proteins Lhcb1-6 and Lhca1-4 for the uninfused, CmR, and psbS-silenced plants of Table 2. Modest, yet significant, increases in Lhcb1, Lhcb3, Lhcb4, and Lhcb5 were noted for psbS-silenced N. benthamiana only. When the same comparisons were made for leaf tissue from wild type and the psbS deletion line of A. thaliana (Npq4-1) no significant differences in either Lhcb or Lhca protein levels were detected.
3.3. Transient Expression of psbS in N. benthamiana
Transient expression (TE) offers the possibility for facile assessment of gene transcription, translation, and protein function without the labor associated with stable incorporation into the nuclear genome [32]. Silencing of psbS in Figure 1 through 5 used the full-length N. benthamiana psbS cDNA in the target domain of the VIGS vector. In order to achieve transient expression of psbS we employed a 184-bp DNA construct containing most of the normal transit sequence in the silencing target domain. In order to transiently synthesize the wild type psbS in silenced tissue an altered psbS cDNA was generated in plasmid pNS451. This plasmid contained a full-length psbS cDNA clone in which 57 silent mismatches were engineered within the first 186 bp encoding the transit sequence [33]. This altered psbS cDNA therefore evaded silencing and encoded an otherwise wild type psbS protein which accumulated to levels that in some cases exceeded the control levels (Figure 6). Nevertheless, NPQ levels associated with the transiently expressed psbS were only about one-third that of the unsilenced controls. We note that the mobilities of SDS-treated control and transiently expressed psbS bands were identical on the immunoblots consistent with normal import of the latter into

Figure 5. Effects of either silencing in N. benthamiana ((a) and (b)) or deletion in A. thaliana of psbS ((c) and (d)) on levels of light-harvesting proteins of PSI (Lhca, (b) and (d)) and PSII (Lhcb, (a) and (c)). Quadruplicate aliquots of leaf membrane protein from each of four replicate leaves of Table 2 were examined by Western immunoblot. The sample aliquots within a given antibody test contained membrane protein corresponding to equal levels of leaf Chl. The sample sizes were adjusted to compensate for differences in antibody/antigen affinity among these Lhcs. Band intensities were determined by densitometry and the controls (uninfused and Col) were assigned a relative value of 1.00. Asterisks indicate a significant difference relative to the control (P < 0.05).
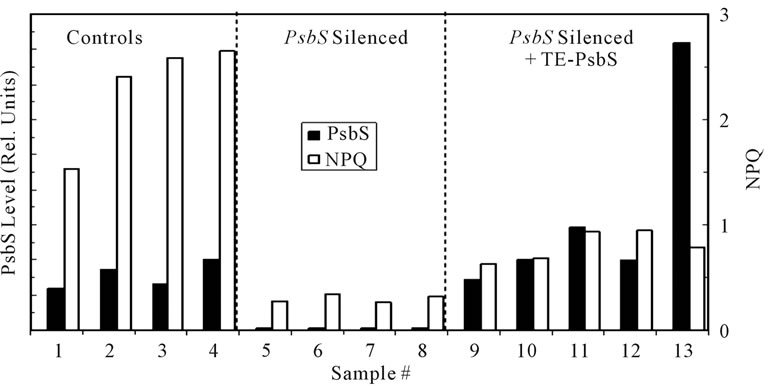
Figure 6. Transient expression (TE) of native psbS in N. benthamiana. Seedlings were silenced for expression of endogenous psbS using the DNA transit sequence as the silencing target. Newly emerged leaves were injected with Agrobacterium suspensions carrying psbS cDNA with a modified transit sequence (see text). Four days after injection 1.6-cm diameter leaf discs were tested for NPQ capacity following a 3-min exposure to high light (1000 mmol·quanta·m−2·s−1). Controls consisted of leaf samples from comparable unsilenced plants and samples from psbS-silenced leaves not inoculated with the Agrobacterium TE vector system. The levels of psbS in the discs were determined by quantitative Western immunoblot. Approximately one-half of the leaf injections produced significant levels of transiently expressed psbS.
the chloroplast. Furthermore, HPLC of pigment extracts indicated no appreciable differences (particularly zeaxanthin levels) between psbS-silenced and psbS-silenced + TE-psbS samples. In independent experiments, transcription of either full-length Arabidopsis psbS cDNA or a chimeric A. thaliana/N. benthamiana psbS cDNA was detected by RT-PCR with no change in expression of the silenced endogenous psbS (not shown). In both experiments psbS was detected by Western blot only in TE-psbS tissues. We conclude that psbS was derived from introduced transgenes and not from unsilencing of endogenous psbS.
4. Discussion
A comprehensive review of the mechanistic basis, implementation, and application of VIGS in plant research has recently appeared [2]. The most common use is gene identification in disease resistance mechanism studies [34]. Nevertheless, the range of processes accessible to genetic analysis using VIGS continues to widen. VIGS was used to characterize prospective drought tolerance gene homologs in N. benthamiana based on cDNAs identified in peanut [35]. Silencing successfully established the role of the ACO4 gene in flower senescence [36]. Although VIGS is efficient and reliable in solanaceous species, it has been reported for Arabidopsis as well [7]. Development of chloroplast thylakoid membranes in pea was impaired after suppression of Mg chelatase accumulation by VIGS [37]. We observed modestly higher JC values in psbS-silenced plants (Figure 3) confirming that leaves possess a limited capacity to increase energy use by noncyclic electron transport when energy dissipation by NPQ is reduced [12]. Dramatic differences were noted in the contributions of the donor and acceptor sides to control of electron flow through PSI over the wide range of irradiance and CO2 levels employed (Figure 4). Nevertheless, only weak and transient interactive effects of VIGS were observed on coordinate regulation of PSII and PSI. Our results support use of VIGS as a complementary, non-perturbing, tool for characterizing effects of multiple, yet selective, elimination of nuclear-encoded gene products such as light-harvesting (Lhc) proteins.
We selected the psbS gene for silencing for two reasons: 1) effects of mutational loss of psbS have been well characterized in Arabidopsis [8,10-12,38], and 2) the strong correlation between psbS expression and NPQ capacity enables a sensitive and convenient assessment of prerequisites for successful complementation by transient gene expression. The dominant effect of loss of psbS in Arabidopsis is abolishment of the dynamic H+-dependent qE phase of NPQ [39], although mild effects on the midpoint redox potential of QA [12] and grana stack formation [40] have been noted. The principal effects of TRV-VIGS-psbS treatment in this study were depletion of psbS mRNA transcript levels (Figure 1) leading to suppression of psbS accumulation and NPQ (Figures 2 and 6, Table 2). Treatment with TRV-VIGS produced some effects that contrasted with prior results with Arabidopsis. We observed significantly higher Chl a/b levels and modestly lower values of WL-partitioning to PSII (aII) and quantum efficiency (YC, LL) for TRV-VIGS-treated compared to uninfused plants (Table 2). Likewise, total Chl and RC contents were marginally lower for TRVVIGS-treated plants. Nevertheless, lower Chl levels were not associated with selective loss of any antenna pigment protein (Figure 5) indicating a probable effect of TRVVIGS on Chl biosynthesis. Interestingly, the intensity of these effects was greatest in the VIGS-CmR plants consistent with increased susceptibility to photodamage. The apparent easing by psbS-silencing of these symptoms may be due to increased expression of genes to detoxify reactive oxygen species [40] due, in turn, to the generally higher reduction state of the plastoquinone/plastoquinol pool associated with loss of psbS [12].
A potentially powerful methodological innovation for gene product structure/function studies is use of VIGS in combination with transient gene expression systems in N. benthamiana [32,42,43]. The psbS protein is a prime candidate for such an approach insofar as its role in the NPQ mechanism is unknown yet several specific functional domains have been defined [9,44]. We note that our repeated attempts to achieve Agrobacterium-mediated transient gene expression in Arabidopsis have been unsuccessful consistent with a previous report of a recalcitrant response in this species [45]. In contrast, transient expression of the psbS protein occurred at a relatively high frequency in N. benthamiana leaves (Figure 6). Transiently expressed psbS was indistinguishable from the normal protein based on molecular weight indicating proper translation and processing of the introduced gene. It is well established that the H+-dependent qE phase of NPQ is abolished when psbS is absent and intensified when psbS is overexpressed [11,38,40]. The low, but persistent, NPQ levels observed in psbS-silenced plants (Table 2, Figure 6) is due to an essentially irreversible type of NPQ distinct from qE [39]. Thus, the results of Figure 6 clearly indicate that transiently expressed psbS was less efficient in restoring qE.
The clear implication arising from the weak complementation of NPQ capacity by transiently expressed psbS is that proper incorporation of this protein does not occur when presented to a fully assembled PSII complex. This result can be interpreted in terms of current understanding of rules governing assembly and turnover of the constituents of PSII [46]. Biosynthesis of core proteins (D1, D2, cyt b559, CP43, CP47) occurs in a hierarchical manner implying that order of presentation of the constituents is important to efficient assembly and function. This program is furthermore affected by the degree of chloroplast maturity. Biosynthesis of some components is blocked in response to deletion of component genes encoding the peripheral low molecular weight PSII subunits (which include psbS) suggesting that a cascade of gene expression imposes an obligatory sequence of assembly [46]. Post-translational modification of proteins may also be linked to chloroplast developmental stage. It is thus pertinent to note that psbS is reportedly acetylated in spinach and pea thylakoids [47]. Nevertheless, the expression of limited psbS-dependent NPQ capacity following Agrobacterium-mediated re-introduction of psbS suggests that the timing of psbS incorporation into PSII may not be critical. Rather, the observations may be better explained in terms of accessibility of psbS to PSII as influenced by thylakoid membrane macrostructure.
A functional association may exist between qE and formation of grana stacks in the thylakoid system [40]. Specifically, full expression of qE may depend on concentration of PSII complexes into a granal macrostructure although formation of the latter does not require psbS. More importantly, with respect to this study, a granal macrostructure is unlikely to be compatible with turnover of constituent thylakoid membrane proteins. For example, although de novo synthesis of psbS was inhibited in plants systemically infected with TRV-VIGS-psbS (Figure 2) levels of this protein remained normal in tissue that was green at the time of Agro-infiltration implying low removal and replacement of psbS in the mature system. Indeed, the only PSII constituent specifically targeted for significant turnover is the photosensitive psbA gene product (D1 protein) which involves a complex repair cycle accompanied by migration of PSII to stroma-exposed margin regions of the grana stacks [46]. This interpretation implies that use of a systemic viral expression system could improve complementation of psbS-dependent NPQ capacity in leaf tissue by ensuring that the introduced psbS is available for insertion into the membrane prior to stacking. Nevertheless, the TE system described here is still useful as a rapid, perhaps preliminary, screening tool for surveys involving multiple psbS constructs and assessment of their capacity for transcription, translation, protein accumulation, targeting to the thylakoid membrane, and ability to support qE.
5. Acknowledgements
This work was supported by the US Department of Agriculture National Research Initiative Competitive Grants Program (grant No. 2004-35318-14124 to R.B.P.) and Estonian Science Foundation Grants 6607 and 6611. We thank members of the S. P. Dinesh-Kumar laboratory (Yale University, USA) for providing Agrobacterium strains, certain DNA constructs, and helpful advice. We also express gratitude to Claudia Buechel (Johann Wolfgang Goethe University, Germany) for providing purified spinach psbS and to Carol Clark, Regan Huntley, and Ada Vaill (CAES) for skillful technical assistance.
REFERENCES
- D. C. Baulcombe, “RNA Silencing in Plants,” Nature, Vol. 431, No. 7006, 2004, pp. 356-363. doi:10.1038/nature02874
- T. M. Burch-Smith, J. C. Anderson, G. B. Martin and S. P. Dinesh-Kumar, “Applications and Advantages of VirusInduced Gene Silencing for Gene Function Studies in Plants,” The Plant Journal, Vol. 39, No. 5, 2004, pp. 734- 746. doi:10.1111/j.1365-313X.2004.02158.x
- A. R. van der Krol, L. A. Mur, M. Beld, J. N. M. Mol and A. R. Stuitje, “Flavonoid Genes in Petunia: Addition of a Limited Number of Gene Copies May Lead to a Suppression of Gene Expression,” Plant Cell, Vol. 2, No. 4, 1990, pp. 219-299.
- C. Napoli, C. Lemieux and R. Jorgensen, “Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans,” Plant Cell, Vol. 2, No. 4, 1990, pp. 279-289.
- M. H. Kumagai, J. Donson, G. Della-Cioppa, D. Harvey, K. Hanley and L. K. Grill, “Cytoplasmic Inhibition of Carotenoid Biosynthesis with Virus-Derived RNA,” Proceedings of the National Academy of Sciences, Vol. 92, No. 5, 1995, pp. 1679-1683. doi:10.1073/pnas.92.5.1679
- D. C. Baulcombe, “Fast Forward Genetics Based on Virus-Induced Gene Silencing,” Current Opinion in Plant Biology, Vol. 2, No. 2, 1999, pp. 109-113. doi:10.1016/S1369-5266(99)80022-3
- T. M. Burch-Smith, M. Schiff, Y. Liu and S. P. DineshKumar, “Efficient Virus-Induced Gene Silencing in Arabidopsis,” Plant Physiology, Vol. 142, No. 1, 2006, pp. 21-27. doi:10.1104/pp.106.084624
- X.-P. Li, O. Björkman, C. Shih, A. R. Grossman, M. Rosenquist, S. Jansson and K. K. Niyogi, “A PigmentBinding Protein Essential for Regulation of Photosynthetic Light Harvesting,” Nature, Vol. 403, No. 6768, 2000, pp. 391-395. doi:10.1038/35000131
- X.-P. Li, A. Phippard, J. Pasari and K. K. Niyogi, “Structure-Function Analysis of Photosystem II Subunit S (PsbS) in Vivo,” Functional Plant Biology, Vol. 29, No. 10, 2002, pp. 1131-1139. doi:10.1071/FP02065
- R. B. Peterson and E. A. Havir, “Photosynthetic Properties of an Arabidopsis thaliana Mutant Possessing a Defective psbS Gene,” Planta, Vol. 214, No. 1, 2001, pp. 142-152. doi:10.1007/s004250100601
- R. B. Peterson and E. A. Havir, “The Multiphasic Nature of Nonphotochemical Quenching: Implications for Assessment of Photosynthetic Electron Transport Based on Chlorophyll Fluorescence,” Photosynthesis Research, Vol. 82, No. 1, 2004, pp. 95-107. doi:10.1023/B:PRES.0000040477.43858.54
- R. B. Peterson, “PsbS Genotype in Relation to Coordinated Function of PS II and PS I in Arabidopsis Leaves,” Photosynthesis Research, Vol. 85, No. 2, 2005, pp. 205- 219. doi:10.1007/s11120-005-3106-7
- Y. Liu, M. Schiff and S. P. Dinesh-Kumar, “Virus-Induced Gene Silencing in Tomato,” The Plant Journal, Vol. 31, No. 6, 2002, pp. 777-786. doi:10.1046/j.1365-313X.2002.01394.x
- Y. Liu, M. Schiff, R. Marathe and S. P. Dinesh-Kumar, “Tobacco Rar1, EDS1 and NPR1/NIM1 Like Genes Are Required for N-Mediated Resistance to Tobacco Mosaic Virus,” The Plant Journal, Vol. 30, No. 4, 2002, pp. 415-429. doi:10.1046/j.1365-313X.2002.01297.x
- F. Ratcliff, A. M. Martin-Hernandez and D. C. Baulcombe, “Tobacco Rattle Virus as a Vector for Analysis of Gene Function by Silencing,” The Plant Journal, Vol. 25, No. 2, 2001, pp. 237-245. doi:10.1046/j.0960-7412.2000.00942.x
- S. P. Dinesh-Kumar, R. Anandalakshmi, R. Marathe, M. Schiff and Y. Liu, “Virus-Induced Gene Silencing,” Plant Functional Genomics, Vol. 236, 2003, pp. 287-294.
- A. Laisk, V. Oja, B. Rasulov, H. Rämma, H. Eichelmann, I. Kasparova, H. Pettai, E. Padu and E. Vapaavuori, “A Computer-Operated Routine of Gas Exchange and Optical Measurements to Diagnose Photosynthetic Apparatus in Leaves,” Plant, Cell & Environment, Vol. 25, No. 7, 2002, pp. 923-943. doi:10.1046/j.1365-3040.2002.00873.x
- W. Yamori, K. Noguchi and I. Terashima, “Temperature Acclimation of Photosynthesis in Spinach Leaves: Analyses of Photosynthetic Components and Temperature Dependencies of Photosynthetic Partial Reactions,” Plant, Cell & Environment, Vol. 28, No. 4, 2005, pp. 536-547. doi:10.1111/j.1365-3040.2004.01299.x
- A. Laisk and F. Loreto, “Determining Photosynthetic Parameters from Leaf CO2 Exchange and Chlorophyll Fluorescence: Rubisco Specificity Factor, Dark Respiration in the Light, Excitation Distribution Between Photosystems, Alternative Electron Transport and Mesophyll Diffusion Resistance,” Plant Physiology, Vol. 110, No. 3, 1996, pp. 903-912.
- V. Oja and A. Laisk, “Oxygen Yield from Single Turnover Flashes in Leaves: Non-Photochemical Excitation Quenching and the Number of Active PSII,” Biochimica et Biophysica Acta, Vol. 1460, No. 2-3, 2000, pp. 291- 301. doi:10.1016/S0005-2728(00)00155-9
- R. B. Peterson, V. Oja and A. Laisk, “Chlorophyll Fluorescence at 680 and 730 nm and Leaf Photosynthesis,” Photosynthesis Research, Vol. 70, No. 2, 2001, pp. 185- 196. doi:10.1023/A:1017952500015
- B. Genty, J. M. Briantais and N. R. Baker, “The Relationship between Quantum Yield of Photosynthetic Electron Transport and Quenching of Chlorophyll Fluorescence,” Biochimica et Biophysica Acta, Vol. 990, No. 1, 1989, pp. 87-92. doi:10.1016/S0304-4165(89)80016-9
- A. Laisk, V. Oja, B. Rasulov, H. Eichelmann and A. Sumberg, “Quantum Yields and Rate Constants of Photochemical and Nonphotochemical Excitation Quenching. Experiment and Model,” Plant Physiology, Vol. 115, No. 2, 1997, pp. 803-815.
- E. Talts, V. Oja, H. Rämma, B. Rasulov, A. Anijalg, A. Laisk, “Dark Inactivation of Ferredoxin-NADP Reductase and Cyclic Electron Flow under Far-Red Light in Sunflower Leaves,” Photosynthesis Research, Vol. 94, No. 1, 2007, pp. 109-120. doi:10.1007/s11120-007-9224-7
- J. Harbinson, B. Genty, N. R. Baker, “Relationship between the Quantum Efficiencies of Photosystems I and II in Pea Leaves,” Plant Physiology, Vol. 90, No. 3, 1989, pp. 1029-1034. doi:10.1104/pp.90.3.1029
- H. Eichelmann and A. Laisk, “Cooperation of Photosystems II and I in Leaves as Analyzed by Simultaneous Measurements of Chlorophyll Fluorescence and Transmittance at 800 nm,” Plant & Cell Physiology, Vol. 41, No. 2, 2000, pp. 138-147. doi:10.1093/pcp/41.2.138
- J. Harbinson and F. I. Woodward, “The Use of LightInduced Absorbance Changes at 820 nm to Monitor the Oxidation State of P-700 in Leaves,” Plant, Cell & Environment, Vol. 10, No. 2, 1987, pp. 131-140.
- A. Laisk, H. Eichelmann, V. Oja and R. B. Peterson, “Control of Cytochrome b6f at Low and High Light Intensity and Cyclic Electron Transport in Leaves,” Biochimica et Biophysica Acta, Vol. 1708, No. 1, 2005, pp. 79-90. doi:10.1016/j.bbabio.2005.01.007
- A. Laisk, H. Eichelmann, V. Oja, E. Talts and R. Scheibe, “Rates and Roles of Cyclic and Alternative Electron Flow in Potato Leaves,” Plant & Cell Physiology, Vol. 48, No. 11, 2007, pp. 1575-1588. doi:10.1093/pcp/pcm129
- J. Harbinson, B. Genty and N. R. Baker, “The Relationship between CO2 Assimilation and Electron Transport in Leaves,” Photosynthesis Research, Vol. 25, No. 3, 1990, pp. 213-224. doi:10.1007/BF00033162
- A. Laisk and V. Oja, “Range of Photosynthetic Control of Postillumination P700+ Reduction Rate in Sunflower Leaves,” Photosynthesis Research, Vol. 39, No. 1, 1994, pp. 39-50. doi:10.1007/BF00027141
- B. B. H. Wulff, C. M. Thomas, M. Smoker, M. Grant and J. D. G. Jones, “Domain Swapping and Gene Shuffling Identify Sequences Required for Induction of an AvrDependent Hypersensitive Response by the Tomato Cf-4 and Cf-9 Proteins,” Plant Cell, Vol. 13, No. 2, 2001, pp. 255-272.
- S. M. Gómez, K. Y. Bil’, R. Aguilera, J. N. Nishio, K. F. Faull, J. P. Whitelegge, “Transit Peptide Cleavage Sites of Integral Thylakoid Membrane Proteins,” Molecular & Cellular Proteomics, Vol. 2, 2003, pp. 1068-1085. doi:10.1074/mcp.M300062-MCP200
- I. Hein, M. Barciszewska-Pacak, K. Hrudikova, S. Williamson, M. Dinesen, I. E. Soenderby, S. Sundar, A. Jarmolowski, K. Shirasu and C. Lacomme, “Virus-Induced Gene Silencing-Based Functional Characterization of Genes Associated with Powdery Mildew Resistance in Barley,” Plant Physiology, Vol. 138, No. 4, 2005, pp. 2155-2164. doi:10.1104/pp.105.062810
- M. Senthil-Kumar, G. Govind, K. Li, K. S. Mysore and M. Udayakumar “Functional Characterization of Nicotiana benthamiana Homologs of Peanut Water DeficitInduced Genes by Virus-Induced Gene Silencing,” Planta, Vol. 225, No. 3, 2007, pp. 523-539. doi:10.1007/s00425-006-0367-0
- J.-C. Chen, C.-Z. Jiang, T. E. Gookin, D. A. Hunter, D. G. Clark and M. S. Reid, “Chalcone Synthase as a Reporter in Virus-Induced Gene Silencing Studies of Flower Senescence,” Plant Molecular Biology, Vol. 55, No. 4, 2004, pp. 521-530. doi:10.1007/s11103-004-0590-7
- T. Luo, S. Luo, W. L. Araújo, H. Schlicke, M. Rothbart, J. Yu, T. Fan, A. R. Fernie, B. Grimm and M. Luo, “Virus-Induced Gene Silencing of Pea CHLI and CHLD Affects Tetrapyrrole Biosynthesis, Chloroplast Development and the Primary Metabolic Network,” Plant Physiology and Biochemistry, Vol. 65, 2013, pp. 17-26. doi:10.1016/j.plaphy.2013.01.006
- X.-P. Li, P. Müller-Moulé, A. M. Gilmore and K. K. Niyogi, “PsbS-Dependent Enhancement of Feedback DeExcitation Protects Photosystem II from Photoinhibition,” Proceedings of the National Academy of Sciences, Vol. 99, No. 23, 2002, pp. 15222-15227. doi:10.1073/pnas.232447699
- R. B. Peterson and E. A. Havir, “Contrasting Modes of Regulation of PSII Light Utilization with Changing Irradiance in Normal and PsbS Mutant Leaves of Arabidopsis thaliana,” Photosynthesis Research, Vol. 75, No. 1, 2003, pp. 57-70. doi:10.1023/A:1022458719949
- A. Z. Kiss, A. V. Ruban and P. Horton, “The PsbS Protein Controls the Organization of the Photosystem II Antenna in Higher Plant Thylakoid Membranes,” The Journal of Biological Chemistry, Vol. 283, 2008, pp. 3972- 3978. doi:10.1074/jbc.M707410200
- T. Pfannschmidt, J. F. Allen and R. Oelmüller, “Principles of Redox Control in Photosynthesis Gene Expression,” Physiologia Plantarum, Vol. 112, No. 1, 2001, pp. 1-9. doi:10.1034/j.1399-3054.2001.1120101.x
- A. Bendahmane, G. Farnham, P. Moffett and D. C. Baulcombe, “Constitutive Gain-of-Function Mutants in a Nucleotide Binding Site-Leucine Rich Repeat Protein Encoded at the Rx Locus of Potato,” The Plant Journal, Vol. 32, No. 2, 2002, pp. 195-204. doi:10.1046/j.1365-313X.2002.01413.x
- A. J. Bernal, Q. Pan, J. Pollack, L. Rose, A. Kozik, N. Willits, Y. Luo, M. Guittet, E. Kochetkova and R. W. Michelmore, “Functional Analysis of the Plant Disease Resistance Gene Pto Using DNA Shuffling,” The Journal of Biological Chemistry, Vol. 280, 2005, pp. 23073- 23083. doi:10.1074/jbc.M500992200
- N. P. Schultes and R. B. Peterson, “Phylogeny-Directed Structural Analysis of the Arabidopsis PsbS Protein,” Biochemical and Biophysical Research Communications, Vol. 355, No. 1426, 2007, pp. 464-470. doi:10.1016/j.bbrc.2007.01.173
- T. Wroblewski, A. Tomczak and R. Michelmore, “Optimization of Agrobacterium-Mediated Transient Assays of Gene Expression in Lettuce, Tomato, and Arabidopsis,” Plant Biotechnology Journal, Vol. 3, No. 2, 2005, pp. 259-273. doi:10.1111/j.1467-7652.2005.00123.x
- E. Baena-González and E.-M. Aro, “Biogenesis, Assembly and Turnover of Photosystem II Units,” Philosophical Transactions of the Royal Society B, Vol. 357, No. 1426, 2002, pp. 1451-1460. doi:10.1098/rstb.2002.1141
- S. M. Gómez, J. N. Nishio, K. F. Faull and J. P. Whitelegge, “The Chloroplast Grana Proteome Defined by Intact Mass Measurements from Liquid Chromatography Mass Spectrometry,” Molecular & Cellular Proteomics, Vol. 1, 2002, pp. 46-59. doi:10.1074/mcp.M100007-MCP200.

