 Advances in Microbiology, 2012, 2, 399-409 http://dx.doi.org/10.4236/aim.2012.23051 Published Online September 2012 (http://www.SciRP.org/journal/aim) Comparison of the Colonization Ability of Autochthonous and Allochthonous Strains of Lactobacilli in the Human Gastr ointestinal Tract Steven A. Frese, Robert W. Hutkins, Jens Walter* Department of Food Science and Technology, University of Nebraska, Lincoln, USA Email: *jwalter2@unl.edu Received July 3, 2012; revised August 8, 2012; accepted August 17, 2012 ABSTRACT Bacteria of the genus Lactobacillus are widely used as oral probio tics due to their putative health ben efits. In this study, we compared the colonization ability of two Lactobacillus strains that were identified as autochthonous to the human gastrointestinal tract (Lactobacillus reuteri ATCC PTA 6475 (MM4-1a) and Lactobacillus mucosae FSL-04) with that of an allochthonous strain (Lactoba cillus acidophilus DDS-1). Colonization ability was tested in a single-blinded, cross-over study, with twelve human subjects. The test strains were quantified in fecal samples by two independent methods, selective plating and molecular typing and quantitative real time PCR. The study revealed that the two autochthonous strains (L. reuteri ATCC PTA 6475 and L. mu co sae FSL-04) reached higher population levels in fecal samples and were recovered more frequently from subjects compared to the allochthonous strain (L. acidophilus DDS-1). All three strains became undetectable 8 days after the end of consumption with one exception, showing that persistence of all three strains remains short term in most individuals. In conclusion, th is study showed that au tochtho- nous Lactobacillus strains can be established more efficiently, albeit temporarily, in the human gastrointestinal tract, suggesting that evolutionary and ecological characteristics could be valuable criteria for the selection of probiotic strains. Keywords: Lactobacillus; Probiotic; Lactobacillus reuteri; Lactobacillus mucosae; Lactobacillus acidophilus; Gut Microbiology; Microbial Ecology; Colonization Resistance 1. Introduction Probiotic bacteria are defined by the FAO/WHO “as live organisms which when administered in adequate amounts confer a health benefit to the host.” Implied in this defi- nition is the expectation that these orally ingested organ- isms would reach the intestinal tract alive and at physio- logically relevant levels [1]. Probiotic microorganisms are hypothesized to be functionally active in the human gut, such that they can influence the host through a vari- ety of physiological mechanisms, including direct effects on the host immune system, in situ production of bioac- tive compounds, and competition with the resident mi- crobiota and pathogens [2]. Some probiotic functions, such as in vivo production of bioactive compounds and competition with pathogens, also require that probiotic bacteria are metabolically active in the human gut and competitive under the prevailing conditions. Accordingly, it is often considered an important prerequisite of probi- otic cultures that they originate from humans in order to ensure adaptation and persistence in the human gut [1, 3,4]. Although many of the commercially available probi- otic strains are of human fecal origin and are capable of surviving gastrointestinal passage, they are still rapidly eliminated after administration has ended [5-12]. The inability of probiotic bacteria to persist in the human in- testinal tract has been attributed to the phenomenon of colonization resistance, whereby the resident gut micro- biota restricts access of allochthonous organisms [13]. Moreover, even strains that are true autochthonous mem- bers of the microbiota of specific human subjects may not be able to colonize the gastrointestinal tract (GIT) of other humans due to individual differences [14]. However, it is also important to recognize that many strains currently used as probiotics belong to species which are considered allochthonous to the human intes- tinal tract [14,15]. Species often used as probiotics such as Lactobacillus acidophilus, Lactobacillus casei , Lac- tobacillus paracasei, Lactobacillus rhamnosus, Lactoba- cillus delbrueckii, Lactobacillus brevis, Lactobacillus *Corresponding a uthor. C opyright © 2012 SciRes. AiM  S. A. FRESE ET AL. 400 johnsonii, Lactobac illus plantarum, and Lactobacillus fermentum, although commonly found in fecal samples, have never been shown to form stable populations in the human gut and are likely to originate from food or the oral cavity [14,16]. In contrast, other species of Lactoba- cillus, in particular, Lactobacillus reuteri, Lactobacillus ruminis, Lactobacillus gasseri, and Lactobacillus sali- varius, have been reported to be autochthonous to the human gastrointestinal tract [15,16]. As autochthonous members of the gut microbiota, these species are likely to occupy specific niches that allow their replication and the establishment of stable population over long periods [17]. Recent comparative genome studies have begun to char- acterize the molecular basis of autochthony in the species L. gasseri, L. reuteri, and L. ruminis, and these studies identified adaptive traits that might contribute to the ecological success of lactobacilli in the human GIT [18- 20]. In this study, our goal was to compare survival and persistence rates of orally-consumed autochthonous and allochthonous Lactobacillus strains in human subjects. Three strains were included in this study, L. reuteri ATCC PTA 6475 (MM4-1a), L. mucosae FSL-04, and L. acidophilus DDS1. L. reuteri is a species long considered to be an auto chthonous member of the human gu t micro- biota [16]. Strain ATCC PTA 6475 is a member of the MLSA lineage II of L. reuteri, which is almost com- pletely composed of strains of human fecal origin, indi- cating that this subpopulation is adapted to the human GIT [21]. The second strain, L. mucosae FSL-04, was continuously isolated from fecal samples from a single healthy adult subject in high numbers during a 15-week period (see below). The third strain, L. acidophilus DDS-1, is a commonly-consumed probiotic [4,22] be- longing to the species L. acidophilus which is considered allochthonous to the human GIT [14-16,23,24]. To com- pare establishment and persistence of these three strains in the human gastrointestinal tract, we performed a hu- man cross-over study in which healthy individuals con- sumed the test strain and provided fecal samples that were then analyzed by cultural enumeration, molecular typing of isolates, and quant itati ve real-tim e PCR (qPCR ). 2. Materials and Methods 2.1. Use of Human Subjects The human trial of this study was approved by the Insti- tutional Review Board of the University of Nebraska (IRB Approval Number: 2009079919FB), and written informed consent has been obtained from all subjects. 2.2. Bacterial Strains and Culture Conditions Strains used in this study are listed in Table 1. All strains were grown in Mann-Rogosa-Sharpe (MRS) (Difco) sup- plemented with 1.0% Maltose and 0.5% Fructose under anaerobic conditions at 37˚C. L. acidophilus DDS-1 was provided by Nebraska Cultures (Walnut Creek, CA USA). L. reuteri ATCC PTA 6475 (MM4-1a) was ob- tained from BioGaia (Stockholm, Sweden). L. mucosae FSL-04 was isolated from the feces of a healthy adult human during a previous human trial as described below [29]. 2.3. Molecular Typing and Identification of Isolates The molecular typing of strains was performed by Ran- dom Amplification of Polymorphic DNA (RAPD) using the primer M13V (Tab le 2) as described by Meroth et al. [32]. Isolates were assigned to species by 16S rRNA gene comparisons as described by Hammons et al. [33]. To obtain around 1 300 bp of the 16S rRNA gene for ex- Table 1. Strains used in this study. Strain Origin Reference or source L. reuteri ATCC PTA 6475 (MM4-1A) Human BioGaia AB (Stockholm, Sweden) L. reuteri 100- 2 3 Rat [25] L. reuteri mlc3 Mouse [21] L. reuteri lpuph Mouse [21] L. mucosae FSL-04 Human, fecal sample This study L. mucosae S5 (DSM 13346) Porcine, small intestine [26] L. mucosae 1028 Porcine, small intestine [27] L. mucosae 1031 Porcine, small intestine [27] L. acidophilus DDS-1 Milk Nebraska cultures (Walnut Creek, CA) L. acidophilus ATCC 4356 Human [28] Copyright © 2012 SciRes. AiM  S. A. FRESE ET AL. 401 Table 2. Primers used in this study. Primer name Sequence 5’ - > 3’ Target Reference 16S/p2 CTTGTACACACCGCCCGTC 1388-1406 16S rRNA [30] 23S/p10 CCTTTCCCTCACGGTACTG 546-474 23S rRNA [30] F_acidophilus_IS GAAAGAGCCCAAACCAAGTGATT 16S-23S rDNA spacer [31] R_acidophilus_IS CTTCCCAGATAATTCAACTATCGCTTA 16S-23S rDNA spacer [31] L. mucosae_For CACAATTAAACCGAGAACACC 16S-23S rDNA spacer This Study L. mucosae_Rev ATGATCTTACGATCACCTCAGTTA 16S-23S rDNA spacer This Study L. reuteri_For AACAATAAACCGAGAACACC 16S-23S rDNA spacer This Study L. reuteri_Rev CCTTCATAACTTAACCTAAACAA 16S-23S rDNA spacer This Study M13V GTTTCCCCAGTCACGAC -- act classification, PCR products were generated and se- quenced with primers 8F (5’-AGAGTTTGATCCTGG- CTCAG-3’) and 1391R (5’-GACGGGCGGTGWGTR- CA-3’). 2.4. Preparation of Lactobacillus Doses Food-grade freeze-dried powders of L. reuteri ATCC PTA 6475, L. mucosae FSL-04, and L. acidophilus DDS-1 were prepared by Culture Systems, Inc (Misha- waka, Indiana USA) and stored at –20˚C throughout the study. Upon receipt, viable cell counts and purity of all three preparations were determined and strain identity was verified by RAPD. Prior to each feeding period, vi- able cell counts were determined in the powders to adjust the daily dose of 109 cells, for each strain, where neces- sary. Subjects were provided with pre-dosed freeze-dried preparations of lactobacilli and instructed to store the preparations at 4˚C and reconstitute the powder in 40 mL cold or room-temperature milk and consume at their own convenience, with a meal. 2.5. Human Trial A blinded, crossover study was performed with twelve healthy adult humans (six male and six female, age 21 - 27). Subjects were selected to be tolerant of milk prod- ucts, free of chronic gastrointestinal disorders, non- vegetarians, and h ad not consumed antibio tics in the two months prior to the study. No dietary restrictions were placed on participants, except to avoid probiotic supple- ments, cultured dairy products, or products advertised as having “live and active cultures”. The study was con- ducted over three separate 8-week periods (each period separated by 3 to 4 weeks) where the individual strains were tested in succession. Each feeding period began with a two-week baseline period (no change in diet). The subjects then consumed a daily dose of 109 viable bacte- rial cells for 7 days, followed by a 5 week wash out pe- riod. Fecal samples were collected weekly, resulting in 2 fecal samples during the baseline period, and fecal sam- ples taken at day 1, 8, and 15 of the wash out periods. The human trial of this study was approved by the Insti- tutional Review Board of the University of Nebraska (IRB Approval Number: 20090-79919FB), and written informed consent was obtained from all subjects. Subjects completed a symptoms diary to assess the potential side effects of experimental strain administra- tion. The symptoms included were bowel movement, stool consistency, discomfort, flatulence, abdominal pain, and bloating, and subjects were asked to score them on a scale from 1 (none, normal, good well-being) to 5 (severe symptoms and discomfort). All twelve subjects com- pleted the trials, and self-reported compliance with the experimental treatments was 100%. 2.6. Microbial Analysis of Fecal Samples Subjects provided fresh fecal samples in sterile fecal sample collection containers, and samples were proc- essed within four hours of defecation. A ten-fo ld dilution of each sample in sterile phosphate buffered saline (PBS) (pH 7.0) was immediately frozen at –80˚C for later DNA extraction (see below). Furthermore, a 10-fold dilution series was made with sterile saline (0.9% NaCl), and aliquots were plated on Rogosa SL Agar, which is selec- tive for lactobacilli. Plates were incubated anaerobically for 48 hours at 37˚C before enumeration. To analyze the total Lactoba- cillus population, 10 colonies were picked at random from a dilution agar plate containing about 100 colonies. Selection of colonies was randomized by drawing inter- secting lines across the plate, and picking colonies along the lines until ten had b een recov ered, in order to remove operator bias. The isolates were differentiated after sub- culture by RAPD analysis through direct comparison Copyright © 2012 SciRes. AiM  S. A. FRESE ET AL. 402 with the molecular fingerprint obtained with the test strains (Figure 1), a strategy which has been effectively used to differentiate Lactobacillus isolates from oral and fecal samples [33,34]. To determine the bacterial population of the test strains in fecal samples, total counts of lactobacilli (on Rogosa SL Agar) were multiplied by the percentage of the strain among the 10 typed colonies. Although this analysis would under-estimate the num- bers of the test strains in fecal samples from subjects with background Lactobacillus counts, the analysis still allowed the detection of a significant increase due to the administration of the strain during the test period. 2.7. Quantitative Real-Time PCR (qPCR) Lactobacillus strains were quantified in human fecal samples by qPCR. Strain specific PCR systems could not be designed for the human L. reuteri strains of the MLSA lineage II and for L. acidophilus strains, as these strains are clonal at all loci that were tested [20]. Therefore, species-specific primers that targeted the 16S-23S rDNA intergenic spacer region (Table 2) were used. Specific primers for L. acidophilus targeting this region were pre- viously described by Haarman and Knol [31]. Primers for L. reuteri and L. mucosae were designed to also target the same region within the 16S-23S rDNA spacer. For L. mucosae, the 16S-23S rDNA spacer region was ampli- fied from four strains (Table 1) by primers 16S/p2 and 23S/p10 [30] (Table 2) and sequenced (Table 3). Se- quences for L. reuteri were obtained from available ge- (a) (b) (c) (d) Figure 1. Identification of Lactobacillus strains by RAPD typing. RAPD patterns from L. reuteri MM4-1a (a); L. mu- cosae FSL-04 (b); and L. acidophilus DDS-1 (c). Each lane represents a RAPD-typed culture obtained from the origi- nal stock culture (lane 1), the freeze-dried powders used in the human trial (lane 2), and from colonies isolated from subject fecal samples after consumption of the strain (lane 3 - 5). Examples of isolates obtained by fecal culture during this study from multiple subjects indicating banding pat- terns are distinct between strains (d). Lane m: 1kb DNA ladder (New England Biolabs, Massachusetts USA). nome sequences (L. reuteri ATCC PTA 6475, 100-23, mlc3, and lpuph), and sequences were aligned using CLUSTALW to generate consensus sequences from which species-specific primers were constructed. Primers were validated for target specificity using DNA from lactoba- cilli-negative fecal samples (<102 CFU/g lactobacilli) and DNA isolated from strains of each of the species of Lactobacillus used (Table 1). Specificity for all primers was further validated in sili- cio using the NCBI database. Genomic DNA was extracted from fecal samples as described previously [24]. qPCR was performed in a Mastercycler Realplex2 (Eppendorf AG, Hamburg, Ger- many) using the Quanti-Fast SYBR Green PCR system (Qiagen, Düsseldorf, Germany) as directed by the manu- facturer. The PCR program consisted of a single 95˚C step for 5 mins, followed by 40 cycles of a two-step PCR reaction, beginning with a 10-second 95˚C denaturation step and a 30 second 60˚C annealing/extension step. Melting curves were also performed, consisting of a de- naturation step of 15 s at 95˚C, an increase from 60˚C - 95˚C over a 20-min period, and a final step of 15 s at 95˚C. Reactions were performed in 25 µL volumes con- taining 0.5 µm each primer and 1 µL of extracted DNA. Standard curves for absolute quantification were pre- pared from overnight cultures (14 h) of each organism (L. reuteri ATCC PTA 6475, L. mucosae FSL-04, and L. acidophilus DDS-1) that were plated in triplicate and enumerated, in parallel to DNA extraction from 1 ml of the culture. A tenfold dilution series was generated with this DNA in 10 mM Tris-Cl pH 8.0 to generate a range of DNA concentrations representing 109 cells to 104 cells, based on culture-based enumeration from the original culture. Reactions were performed in triplicate for the standard curves and in duplicate for fecal DNA samples. The correlation coefficient, r2, for the three standard Table 3. Sequences determ ine d during this study. Strain Sequence determined Accession number L. mucosae FSL-04 16S/23S intergenic spacer region JN368428 Partial 16S rRNA gene JN092131 L. mucosae S5 16S/23S intergenic spacer region JN592586 L. mucosae 1028 16S/23S intergenic spacer region JN592585 L. mucosae 1031 16S/23S intergenic spacer region JN592584 L. acidophilus DDS-116S/23S intergenic spacer region JN368427 L. acidophilus DDS-1Partial 16S rRNA gene JN368429 Copyright © 2012 SciRes. AiM  S. A. FRESE ET AL. Copyright © 2012 SciRes. AiM 403 curves were 0.99 and PCR efficiencies were 0.93 (L. mucosae) , 0. 98 (L. reuteri) and 1.00 (L. acidophilus). post-hoc test. 3. Results The qPCR systems were validated by spiking a lacto- bacilli-negative fecal sample (less than 102 CFU per gram lactobacilli as determined by selective culture on Rogosa SL Agar) with 10 to 108 cells per gram of each of the three test strains. DNA was extracted and qPCR was performed in duplicate as described above. Predicted numbers of lactobacilli demonstrated a linear dynamic range closely reflecting the number of known added cells (r2 > 0.98 for MM4-1a, 0.99 for FSL-04, and 0.93 for DDS-1) from 104 cells per g to 108 CFU per gram. Below 104 cells per gram fecal samples, background signal pre- vented accurate quan tification. Within the dynamic range, cell numbers determined by qPCR were in agreement with the cell numbers that were spiked to the samples as indicated by the slopes of the trend line in linear regres- sion (b = 0 .91 for ATCC PTA 64 75, b = 0.86 for FSL-04 and b = 1.01 for DDS-1) when predicted cell numbers were plotted against cells added. 3.1. Selection of Lactobacillus mucosae FSL-04 as an Autochthonous Member of the Human Intestinal Tract To identify human auto chthonou s lactobacilli, we screen- ed fecal samples from 11 human subjects that had par- ticipated in a previous study [29]. Fecal Lactobacillus populations were quantified over the duration of four months by plating serial dilutions on Rogosa SL Agar plates. As observed in previous studies [14,15,24,34], most of the subjects harbored low numbers of lacto bacilli in fecal samples (less than 106 CFU/g). However, one subject consistently shed high levels of lactobacilli in fecal samples ( Figure 2(a)) over the entire 4 month study. Two dominant colony types were identified, and 28 iso- lates were picked randomly during the 15 week duration of the trial representing both colony morphologies. Molecular typing of these isolates by RAPD revealed the presence of two unique strains. One strain was de- tected throughout the entire 15 week time period by RAPD-typing (Figure 2(b)) and classified as L. mucosae by 16S rRNA sequence analysis (>99.9% homology to 16S rRNA gene from L. mucosae CCUG 43179T). Given that this strain was found in the fecal samples of this hu- man subject in high numbers, w e concluded that it repre- sented an autochthonous member of the gut microbiota. 2.8. Statistical Analysis Results are presented as means ± standard deviations. Statistical tests for treatment effects of test strain admini- stration on the abundance of individual strains or abun- dance of Lactobacillus species were performed either by one-way analysis of variance (ANOVA) with repeated measure or by one-way ANOVA, followed by Tukey (a) (b) Figure 2. Identification of Lactobacillus mucosae FSL-04 as an autochthonous member of the human gut microbiota. (a) Total Lactobacilli (CFU/gram feces) enumerated from Rogosa SL agar over several weeks during isolation of FSL-04 from a single healthy adult human; (b) RAPD patterns of isolates identified as Lactobacillus mucosae obtained from these fecal samples. Lane m: 1kb DNA ladder (New England Biolabs, Massachusetts USA); Lane nc: Negative control. Numbered lanes corre- pond to the respective weeks isolates were obtained. s 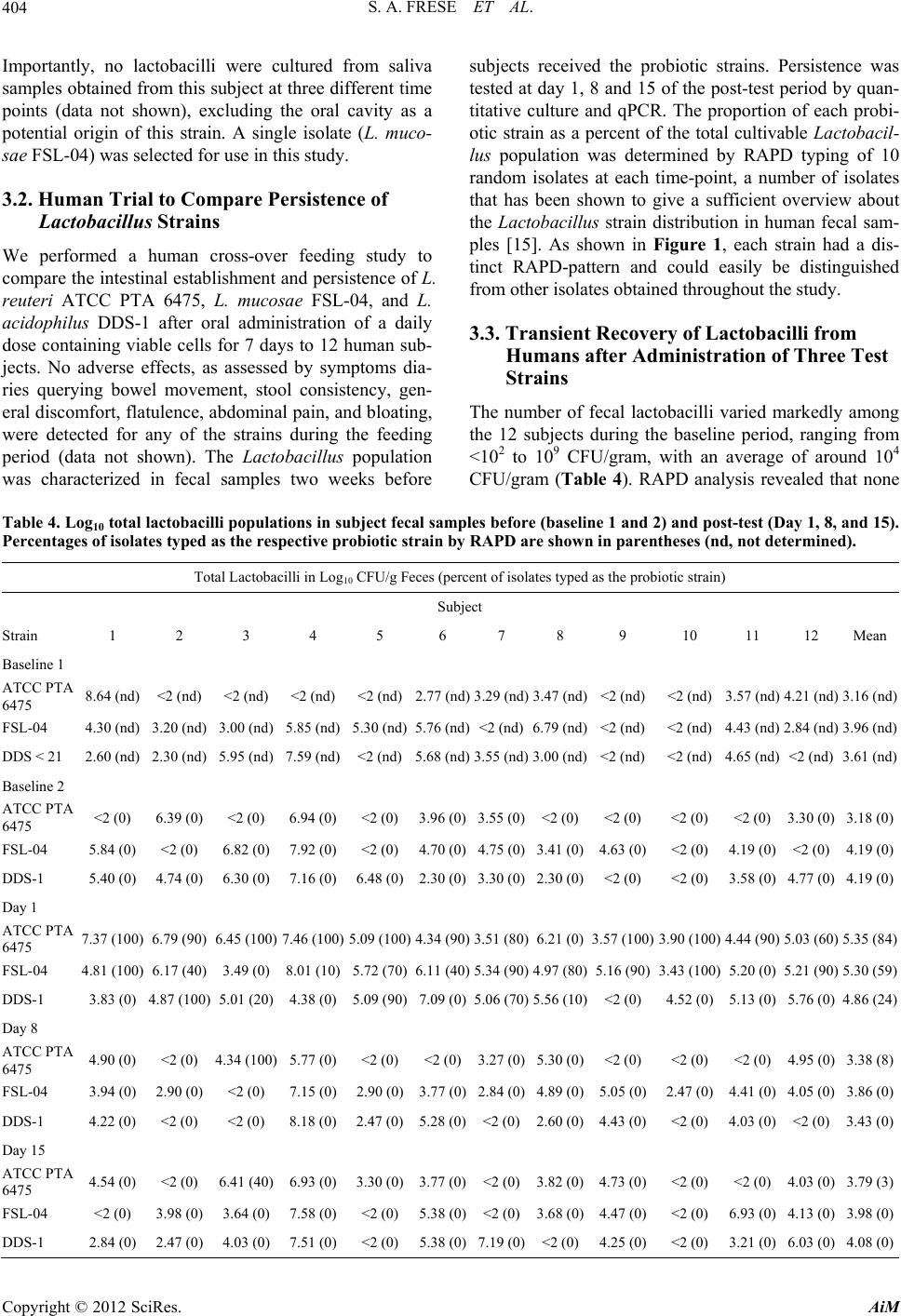 S. A. FRESE ET AL. 404 Importantly, no lactobacilli were cultured from saliva samples obtained from this subject at three different time points (data not shown), excluding the oral cavity as a potential origin of this strain. A single isolate (L. muco- sae FSL-04) was selected for use in this study. 3.2. Human Trial to Compare Persistence of Lactobacillus Strains We performed a human cross-over feeding study to compare the intestinal establishment and persistence of L. reuteri ATCC PTA 6475, L. mucosae FSL-04, and L. acidophilus DDS-1 after oral administration of a daily dose containing viable cells for 7 days to 12 human sub- jects. No adverse effects, as assessed by symptoms dia- ries querying bowel movement, stool consistency, gen- eral discomfort, flatulence, abdominal pain, and bloating, were detected for any of the strains during the feeding period (data not shown). The Lactobacillus population was characterized in fecal samples two weeks before subjects received the probiotic strains. Persistence was tested at day 1, 8 and 15 of the post-test period by quan- titative culture and qPCR. The proportion of each probi- otic strain as a percent of the total cultivable Lactobacil- lus population was determined by RAPD typing of 10 random isolates at each time-point, a number of isolates that has been shown to give a sufficient overview about the Lactobacillus strain distribution in human fecal sam- ples [15]. As shown in Figure 1, each strain had a dis- tinct RAPD-pattern and could easily be distinguished from other isolates obtained throughout the study. 3.3. Transient Recovery of Lactobacilli from Humans after Administration of Three Test Strains The number of fecal lactobacilli varied markedly among the 12 subjects during the baseline period, ranging from <102 to 109 CFU/gram, with an average of around 104 CFU/gram (Table 4). RAPD analysis revealed that none Table 4. Log10 total lactobacilli populations in subject fecal samples before (baseline 1 and 2) and post-test (Day 1, 8, and 15). Percentages of isolates typed as the respective probiotic strain by RAPD are shown in parentheses (nd, not determined). Total Lactobacilli in Log10 CFU/g Feces (percent of isolates typed as the probiotic strain) Subject Strain 1 2 3 4 5 6 7 8 9 10 11 12 Mean Baseline 1 ATCC PTA 6475 8.64 (nd) <2 (nd) <2 (nd) <2 (nd) <2 (nd)2.77 (nd)3.29 (nd)3.47 (nd)<2 (nd)<2 (nd) 3.57 (nd) 4.21 (nd)3.16 (nd) FSL-04 4.30 (nd) 3.20 (nd) 3.00 (nd) 5.85 (nd) 5.30 (nd)5.76 (nd)<2 (nd)6.79 (nd)<2 (nd)<2 (nd) 4.43 (nd) 2.84 (nd)3.96 (nd) DDS < 21 2.60 (nd) 2.30 (nd) 5.95 (nd) 7.59 (nd) <2 (nd)5.68 (nd)3.55 (nd)3.00 (nd)<2 (nd)<2 (nd) 4.65 (nd) <2 (nd)3.61 (nd) Baseline 2 ATCC PTA 6475 <2 (0) 6.39 (0) <2 (0) 6.94 (0) <2 (0) 3.96 (0)3.55 (0)<2 (0)<2 (0) <2 (0) <2 (0) 3.30 (0)3.18 (0) FSL-04 5.84 (0) <2 (0) 6.82 (0) 7.92 (0) <2 (0) 4.70 (0)4.75 (0)3.41 (0)4.63 (0)<2 (0) 4.19 ( 0) <2 (0)4.19 (0) DDS-1 5.40 (0) 4.74 (0) 6.30 (0) 7.16 (0) 6.48 (0)2.30 (0)3.30 (0)2.30 (0)<2 (0) < 2 (0) 3.58 (0) 4.77 (0)4.19 (0) Day 1 ATCC PTA 6475 7.37 (100) 6.79 (90) 6.45 (100) 7.46 (100) 5.09 (100)4.34 (90)3.51 (80)6.21 (0)3.57 (100)3.90 (100) 4.44 (90) 5.03 (60)5.35 (84) FSL-04 4.81 (100) 6.17 (40) 3.49 (0) 8.01 (10) 5.72 (70)6.11 (40)5.34 (90)4.97 (80)5.16 (90)3.43 (100) 5.20 (0) 5.21 (90)5.30 (59) DDS-1 3.83 (0) 4.87 (100) 5.01 (20) 4.38 (0) 5.09 (90)7.09 (0)5. 06 (70)5.56 (10)<2 (0) 4.52 (0) 5.13 (0) 5. 76 (0)4.86 (24) Day 8 ATCC PTA 6475 4.90 (0) <2 (0) 4.34 (100) 5.77 (0) <2 (0) <2 (0)3.27 (0)5.30 (0)<2 (0) <2 (0) <2 (0) 4.95 (0)3.38 (8) FSL-04 3.94 (0) 2.90 (0) <2 (0) 7.15 (0) 2.90 (0)3.77 (0)2.84 (0)4.89 (0)5.05 (0)2.47 (0) 4.41 (0) 4.05 (0)3.86 (0) DDS-1 4.22 (0) <2 (0) <2 (0) 8.18 (0) 2.47 (0)5.28 (0)<2 (0)2.60 (0)4.43 (0)<2 (0) 4.03 (0) <2 (0)3.43 (0) Day 15 ATCC PTA 6475 4.54 (0) <2 (0) 6.41 (40) 6.93 (0) 3.30 (0)3.77 (0)<2 (0)3.82 (0)4.73 (0)<2 (0) <2 ( 0) 4.03 (0)3.79 (3) FSL-04 <2 (0) 3.98 (0) 3.64 (0) 7.58 ( 0) <2 (0) 5.38 (0)<2 (0)3.68 (0)4.47 (0)<2 (0) 6.93 (0) 4.13 (0)3.98 (0) DDS-1 2.84 (0) 2.47 (0) 4.03 (0) 7.51 (0) <2 (0) 5. 38 (0)7.19 (0)<2 (0)4.25 (0)<2 (0) 3.21 (0) 6.03 (0)4.08 ( 0) Copyright © 2012 SciRes. AiM  S. A. FRESE ET AL. 405 of the three test strains were detectable during the base- line. After subjects had consumed the test strains for 7 days, culture analysis of day 1 fecal samples showed an increase in total Lactobacillus numbers to about 105 CFU/gram. The RAPD analysis of random isolates re- vealed that the test strains could be detected in a majority of the subjects (Table 4), indicating that the increase in total lactobacilli detected soon after consumption was a result of administration of the respective experimental strain. We next estimated the fecal populations of the test strains by multiplying total lactobacilli counts by the relative proportions of each strain, as determined by RAPD analysis (see Materials and Methods). Admini- stration of L. reuteri ATCC PTA 6475 and L. mucosae FSL-04 led to a significant increase in the numbers of the respective strains at day 1 after consumption when com- pared to baseline, while administration of DDS-1 did not result in a significant increase (Figure 3(a) ). The popu la- tions of L. reuteri ATCC PTA 6475 (5.22 Log10 CFU/ gram) and L. mucosae (4.94 Log10 CFU/gram) were hi gher than that of L. acidophilus (4.26 Log10 CFU/gram), but differences did not reach statistical significance. The number of total lactobacilli returned to baseline at day 8 after consumption (Table 4 and Figure 3(a)), and the probiotic strains were not detectable anymore with one exception (L. reuteri ATCC PTA 6475 was detected at day 8 and 15 of the washout period in one subject). 3.4. L. reuteri ATCC PTA 6475 and L. mucosae FSL-04 Can be Established More Efficiently and in Higher Numbers than L. acidophilus DDS-1 All three test strains could be detected by the culture- RAPD method in a subset of subjects at day 1, but the rate of recovery differed between strains (Table 4). At day 1 after administration, 84% of the isolates were iden- tified as L. reuteri ATCC PTA 6475 at day 1, compared to 59% and 24% for L. mucosae FSL-04 and L. aci- dophilus DDS-1, respectively (Figure 3(b)). The differ- ence in recovery rate of L. reuteri ATCC PTA 6475 was significantly higher than that of L. acidophilus DDS-1 (Figure 3(b)). In addition, the autochthonous strains were detectable in fecal samples of more subjects. L. reuteri ATCC PTA 6475 was recovered from 11 out of the 12 subjects, L. mucosae FSL-04 and was present in 10 subjects, while L. acidophilus DDS-1 was present in 5 subjects. Although the back ground Lactobacillus population s in human fecal samples were low, they still confounded the culture-based analyses as they prohibited an exact quan- tification of the test strains. To determine the establish- ment of the test strains in th e human gastrointestinal tract, samples taken during baseline, day 1 post-test, and day 8 post-test were analyzed by species-specific qPCR. We used a species and not strain-specific primers for the qPCR, as human L. reuteri and L. acidophilus isolates are highly clonal, making the development of strain spe- cific primers impractical. Furthermore, we chose to target the same gene (the 16S-23S rRNA spacer region) in all three strains used during this study to avoid PCR bias. Population levels were determined by absolute quantifi- cation using standard curves obtained from bacterial cells. While strain-specific primers would be advantageous, the analysis of baseline fecal samples indicated that no con- founding Lactobacillus populations were present in any of the subjects. As shown in Figure 3(c), the leve ls of L. reuteri, L. mucosae, and L. acidophilus were generally below the detection limit (104 CFU/gram) in most of the samples during baseline and at day 8 of wash-out, indi- cating that background levels of the species did not con- found this technique. At day 1 of wash-out, there was a statistically signifi- cant increase in each of the three species when compared to the baseline and the day 8 wash-out, with L. reuteri ATCC PTA 6475 reaching 5.14 Log10 cells/gram, L. mucosae FSL-04 reaching 5.03 Log10 cells/gram, and L. acidophilus DDS-1 4.32 Log10 cells/gram feces. The qPCR analysis revealed that the two autochthonous strains L. reuteri ATCC PTA 6475 and L. mucosae FSL-04 reached significantly higher populations (p < 0.01) in fecal samples when compared to L. acidophilus DDS-1 (Figure 3(c)). 4. Discussion The objective of this study was to test whether autoch- thonous Lactobacillus strains (L. mucosae FSL-04 and L. reuteri ATCC PTA 6475) can be established more effi- ciently in the human gastroin testinal tract than an alloch- thonous strain (L. acidophilus DDS-1) fo llowing a 7-day feeding period. Accordingly, we characterized the Lac- tobacillus populations in fecal samples obtained from humans that had consumed standardized, freeze dried cell preparations of three test strains using both cul- ture-based and molecular (qPCR) methods. As observed in previous probiotic trials, all three Lactobacillus strains survived gastric passage and were temporarily detectable in the fecal samples [5,8,12,15,35]. Most importantly, our findings indicate that the two autochthonous strains (L. reuteri ATCC PTA 6475 and L. mucosa e FSL-04) reached higher populations and were generally more per- sistent than the allochthonous strain. In addition, the autochthonou s strains, and L. reuteri ATCC PTA 64 75 in particular, could be detected in a larger proportion of subjects and represented a larger percentage of the total actobacillus population (Table 4). L Copyright © 2012 SciRes. AiM 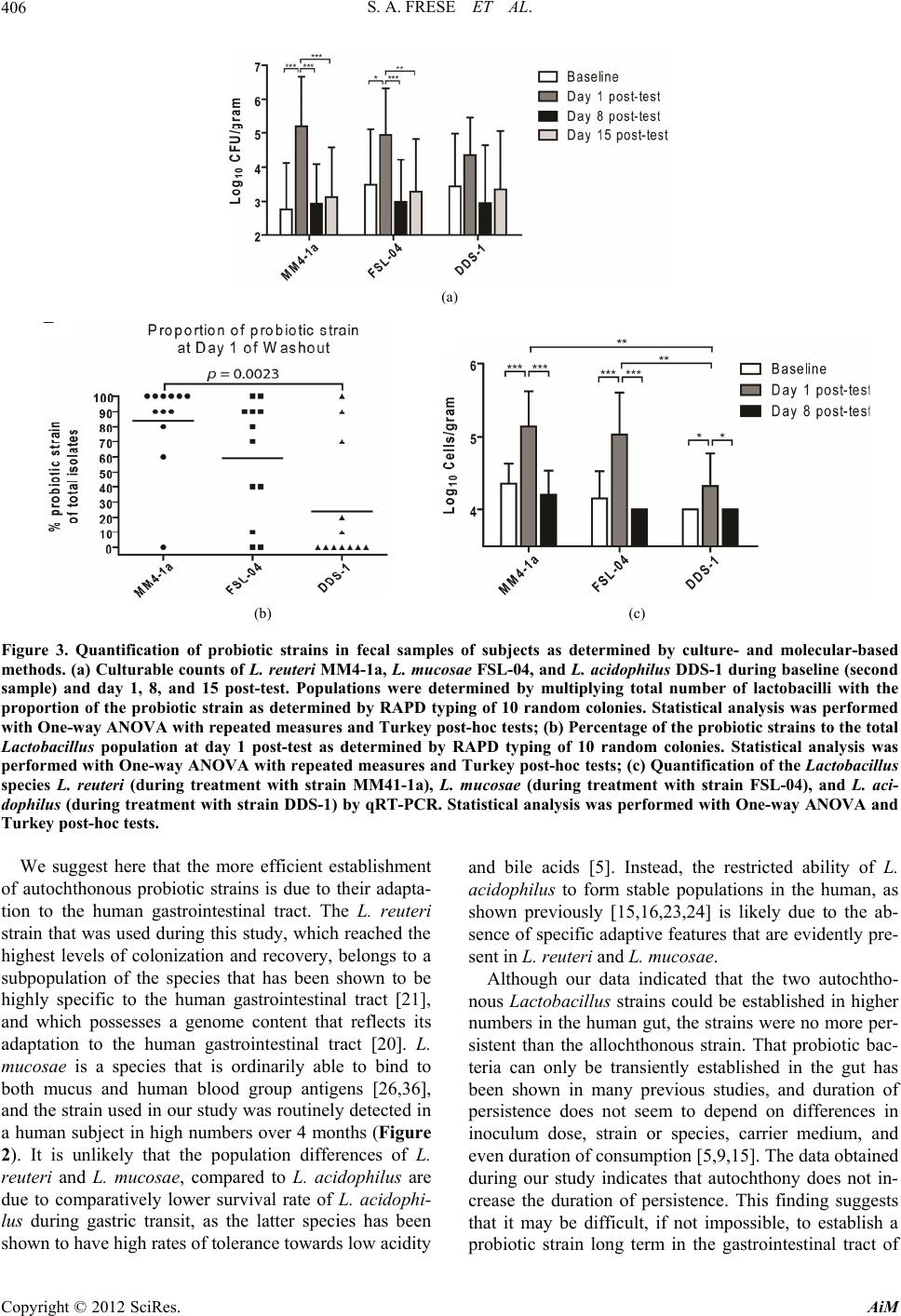 S. A. FRESE ET AL. 406 (a) (b) (c) Figure 3. Quantification of probiotic strains in fecal samples of subjects as determined by culture- and molecular-based methods. (a) Culturable counts of L. reute ri MM4-1a, L. mucosae FSL-04, and L. acidophilus DDS-1 during baseline (second sample) and day 1, 8, and 15 post-test. Populations were determined by multiplying total number of lactobacilli with the proportion of the probiotic strain as determined by RAPD typing of 10 random colonies. Statistical analysis was performed with One-way ANOVA with repeated measures and Turkey post-hoc tests; (b) Percentage of the probiotic strains to the total Lactobacillus population at day 1 post-test as determined by RAPD typing of 10 random colonies. Statistical analysis was performed with One-way ANOVA with repeated measures and Turkey post-hoc tests; (c) Quantification of the Lactobacillus species L. reuteri (during treatment with strain MM41-1a), L. mucosae (during treatment with strain FSL-04), and L. aci- dophilus (during treatment with strain DDS-1) by qRT-PCR. Statistical analysis was performed with One-way ANOVA and Turkey post-hoc tests. We suggest here that the more efficient establishment of autochthonous probiotic strains is due to their adapta- tion to the human gastrointestinal tract. The L. reuteri strain that was used during this study, which reached the highest levels of colonization and recovery, belongs to a subpopulation of the species that has been shown to be highly specific to the human gastrointestinal tract [21], and which possesses a genome content that reflects its adaptation to the human gastrointestinal tract [20]. L. mucosae is a species that is ordinarily able to bind to both mucus and human blood group antigens [26,36], and the strain used in our stud y was routinely detected in a human subject in high numbers over 4 months (Figure 2). It is unlikely that the population differences of L. reuteri and L. mucosae, compared to L. acidophilus are due to comparatively lower survival rate of L. acidophi- lus during gastric transit, as the latter species has been shown to have high rates of tolerance towards low acidity and bile acids [5]. Instead, the restricted ability of L. acidophilus to form stable populations in the human, as shown previously [15,16,23,24] is likely due to the ab- sence of specific adaptive features th at are eviden tly pre- sent in L. reuteri and L. mucosae. Although our data indicated that the two autochtho- nous Lactobacillus strains could be established in higher numbers in the human gut, the strains were no more per- sistent than the allochthonous strain. That probiotic bac- teria can only be transiently established in the gut has been shown in many previous studies, and duration of persistence does not seem to depend on differences in inoculum dose, strain or species, carrier medium, and even duration of consumption [5,9,15]. The data obtained during our study indicates that autochthony does not in- crease the duration of persistence. This finding suggests that it may be difficult, if not impossible, to establish a probiotic strain long term in the gastrointestinal tract of Copyright © 2012 SciRes. AiM  S. A. FRESE ET AL. 407 most human subjects, even if that microbe was derived from a stable component of one individual’s gut micro- biota. Therefore, the ability of the microbiota to prevent establishment of foreign organisms (colonization resis- tance), applies not only to pathogens, but to other gut microbes as well. The ecological principles that govern community as- sembly of the gut microbiota and determine which line- ages can become established are not completely under- stood. Modern concepts of community ecology suggest that a combination of niche-related and historic processes (e.g. in situ evolution of early colonizers) govern the process [37-39]. Historic factors such as colonization order, transmission, and niche construction are inherently stochastic, and therefore cause variation in individual microbiomes, while in situ evolution of members ensures that members are highly adapted to available niches [39]. These concepts have important implications if the goal is to establish a probiotic organism in the gut. First, the microbiome is individualized and composed of microbes that are adapted to occupy specific niches within each person’s particular community, and thus, members of mi- crobiomes are not necessarily interchangeable. Second, establishment of new microbes is only possible as long as niches are still open (early during assembly) or if niches become available (when community members are inten- tionally or accidentally removed from the community). If community structure becomes disrupted, for example through a high dose of antibiotics, microbes from another individual can be successfully and more permanently established [40]. Therefore, probiotic bacteria might be more permanently established when administered after antibiotic treatments or early in life. Although our findings indicate the autochthony of probiotic lactobacilli does not increase the duration of their persistence in the human gastrointestinal tract, the observation that autochthonous strains can be more effi- ciently established is clearly of practical importance. We not only detected L. reuteri ATCC PTA 6475 and L. mucosae FSL-04 in more subjects after administration, but these strains also reached about ten times higher cell numbers in fecal samples when compared to L. aci- dophilus DDS-1. These findings indicate that the effec- tive dose of a probiotic is likely to be higher when an autochthonous strain versus an allochthonous strain is used. If probiotic action is dependent on the metabolic functionality of the organisms in the gut, such as direct antagonism or the production of bioactive substances, then a higher dose of viable organisms present the gut is likely to increase the success of a probiotic strategy. In conclusion, th e results described in th is study support the notion that evolutionary and ecological characteristics are valuable criteria for the selection of probiotic strains [14]. Future studies should be aimed to test persistence of other autochthonous strains, especially L. ruminis, which appears to be the dominant Lactobacillus species in the human gut [15,16,41]. 5. Acknowledgements The dedication of the participants in the human trial is gratefully acknowledged. We are thankful to Stefan Roos (Swedish University of Agricultural Sciences) for pro- viding L. mucosae S5, 1028, and 1031 and Dr. Todd R. Klaenhammer (North Carolina State University ) for providing L. acidophilus ATCC 4356. We thank Ne- braska Cultures (Walnut Creek, CA) for providing L. acidophilus DDS-1 and partially funding the study. REFERENCES [1] A. C. Ouwehand, S. Salminen and E. Isolauri, “Probiotics: An Overview of Beneficial Effects,” Antonie Van Leeu- wenhoek, Vol. 82, No. 1-4, 2002, pp. 279-289. doi:10.1023/A:1020620607611 [2] P. W. O’Toole and J. C. Cooney, “Probiotic Bacteria Influence the Composition and Function of the Intestinal Microbiota,” Interdisciplinary Perspectives on Infectious Diseases, Vol. 85, No. 1752, 2008, 9 Pages. [3] C. Dunne, L. Murphy, S. Flynn, L. O’Mahony, S. O’Hal- loran, M. Feeney, D. Morrisse y, G. Thornton, G. Fitzger- ald, C. Daly, B. Kiely, E. M. Quigley, G. C. O’Sullivan, F. Shanahan and J. K. Collins, “Probiotics: From Myth to Reality Demonstration of Functionality in Animal Models of Disease and in Human Clinical Trials,” Antonie Van Leeuwenhoek, Vol. 76, No. 1-4, 1999, pp. 279-292. doi:10.1023/A:1002065931997 [4] M. E. Sanders, “Probiotics: Considerations for Human Health,” Issue Nutrition Reviews, Vol. 61, No. 3, 2003, pp. 91-99. doi:10.1301/nr.2003.marr.91-99 [5] C. N. Jacobsen, N. Rosenfeldt, V. Hayford, A. E. Moller, P. L. Michaelsen, K. F. Paerregaard, A. Sandstrom, M. B. Tvede and M. Jakobsen, “Screening of Probiotic Activi- ties of Forty-Seven Strains of Lactobacillus spp. by in Vi- tro Techniques and Evaluation of the Colonization Abil- ity of Five Selected Strains in Humans,” Applied and En- vironmental Microbiology, Vol. 65, No. 11, 1999, pp. 4949-4956. [6] T. Vesa, P. Pochart and P. Marteau, “Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fer- mentum KLD, and Lactococcus lactis MG1363 in the Human Gastrointestinal Tract,” Alimentary Pharmacol- ogy & Therapeutics, Vol. 14, No. 6, 2000, pp. 823-828. doi:10.1046/j.1365-2036.2000.00763.x [7] C. de Champs, N. Maroncle, D. Balestri no, C. Ric h a nd C. Forestier, “Persistence of colonization of Intestinal Mu- cosa by a Probiotic Strain, Lactobacillus casei Subsp. rhamnosus LCR35, after Oral Consumption,” Journal of Clinical Microbiology, Vol. 41, No. 3, 2003, pp. 1270- 1273. doi:10.1128/JCM.41.3.1270-1273.2003 [8] N. Valeur, P. Engel, N. Carbajal, E. Connolly and K. Ladefoged, “Colonization and Immu nomodulation by Lac- Copyright © 2012 SciRes. AiM  S. A. FRESE ET AL. 408 tobacillus reuteri ATCC 55730 in the Human Gastroin- testinal Tract,” Applied and Environmental Microbiology, Vol. 70, No. 2, 2004, pp. 1176-1181. doi:10.1128/AEM.70.2.1176-1181.2004 [9] T. D. Klingberg and B. B. Budde, “The Survival and Persistence in the Human Gastrointestinal Tract of Five Potential Probiotic Lactobacilli Consumed as Freeze- Dried Cultures or as Probiotic Sausage,” International Journal of Food Microbiology, Vol. 109, No. 1-2, 2006, pp. 157-159. doi:10.1016/j.ijfoodmicro.2006.01.014 [10] R. Oozeer, A. Leplingard, D. D. Mater, A. Mogenet, R. Michelin, I. Seksek, P. Marteau, J. Dore, J. L. Bresson and G. Corthier, “Survival of Lactobacillus casei in the Human Digestive Tract after Consumption of Fermented Milk,” Applied and Environmental Microbiology, Vol. 72, No. 8, 2006, pp. 5615-5617. doi:10.1128/AEM.00722-06 [11] K. M. Tuohy, M. Pinart-Gilberga, M. Jones, L. Hoyles, A. L. McCartney and G. R. Gibson, “Survivability of a Pro- biotic Lactobacillus casei in the Gastrointestinal Tract of Healthy Human Volunteers and Its Impact on the Faecal Microflora,” Journal of Applied Microbiology, Vol. 102, No. 4, 2007, pp. 1026-1032. [12] Y. E. Dommels, R. A. Kemperman, Y. E. Zebregs, R. B. Draaisma, A. Jol, D. A. Wolvers, E. E. Vaughan and R. Albers, “Survival of Lactobacillus reuteri DSM 17938 and Lactobacillus rhamnosus GG in the Human Gastroin- testinal Tract with Daily Consumption of a Low-Fat Pro- biotic Spread,” Applied and Environmental Microbiology, Vol. 75, No. 19, 2009, pp. 6198-6204. doi:10.1128/AEM.01054-09 [13] B. Stecher and W. D. Hardt, “Mechanisms Controlling Pathogen Colonization of the Gut,” Current Opinion in Microbiology, Vol. 14, No. 1, 2011, pp. 82-91. doi:10.1016/j.mib.2010.10.003 [14] J. Walter, “Ecological Role of Lactobacilli in the Gastro- intestinal Tract: Implications for Fundamental and Bio- medical Research,” Applied and Environmental Microbi- ology, Vol. 74, 2008, pp. 4985-4996. doi:10.1128/AEM.00753-08 [15] G. W. Tannock, K. Munro, H. J. Harmsen, G. W. Welling, J. Smart and P. K. Gopal, “Analysis of the Fecal Micro- flora of Human Subjects Consuming a Probiotic Product Containing Lactobacillus rhamnosus DR20,” Applied and Environmental Microbiology, Vol. 66, No. 6, 2000, pp. 2578-2588. doi:10.1128/AEM.66.6.2578-2588.2000 [16] G. Reuter, “The Lactobacillus and Bifidobacterium Mi- croflora of the Human Intestine: Composition and Suc- cession,” Current Issues in Intestinal Microbiology, Vol. 2, No. 2, 2001, pp. 43-53. [17] D. C. Savage, “Microbial Ecology of the Gastrointestinal Tract,” Annual Review of Microbiology, Vol. 31, 1977, pp. 107-133. doi:10.1146/annurev.mi.31.100177.000543 [18] M. A. Azcarate-Peril, E. Altermann, Y. J. Goh, R. Tallon, R. B. Sanozky-Dawes, E. A. Pfeiler, S. O’Flaherty, B. L. Buck, A. Dobson, T. Duong, M. J. Miller, R. Barrangou and T. R. Klaenhammer, “Analysis of the Genome Se- quence of Lactobacillus gasseri ATCC 33323 Reveals the Molecular Basis of an Autochthonous Intestinal Organ- ism,” Applied and Environmental Microbiology, Vol. 74, No. 15, 2008, pp. 4610-4625. doi:10.1128/AEM.00054-08 [19] B. M. Forde, B. A. Neville, M. M. O’Donnell, E. Ribou- let-Bisson, M. J. Claesson, A. Coghlan, R. P. Ross and P. W. O’Toole, “Genome Sequences and Comparative Ge- nomics of Two Lactobacillus ruminis Strains from the Bovine and Human Intestinal Tracts,” Microbial Cell Factories, Vol. 10, Suppl. 1, 2011, p. S13. doi:10.1186/1475-2859-10-S1-S13 [20] S. A. Frese, A. K. Benson, G. W. Tannock, D. M. Loach, J. Kim, M. Zhang, P. L. Oh, N. C. Heng, P. B. Patil, N. Juge, D. A. Mackenzie, B. M. Pearson, A. Lapidus, E. Dalin, H. Tice, E. Goltsman, M. Land, L. Hauser, N. Ivanova, N. C. Kyrpides and J. Walter, “The Evolution of Host Specialization in the Vertebrate Gut Symbiont Lac- tobacillus reuteri,” PLoS Genet, Vol. 7, No. 2, 2011, Ar- ticle ID: e1001314. doi:10.1371/journal.pgen.1001314 [21] P. L. Oh, A. K. Benson, D. A. Peterson, P. B. Patil, E. N. Moriyama, S. Roos and J. Walter, “Diversification of the Gut Symbiont Lactobacillus reuteri as a Result of Host- Driven Evolution,” The ISME Journal, Vol. 4, No. 3, 2010, pp. 377-387. doi:10.1038/ismej.2009.123 [22] S. Rabot, J. Rafter, G. T. Rijkers, B. Watzl and J. M. An- toine, “Guidance for Substantiating the Evidence for Beneficial Effects of Probiotics: Impact of Probiotics on Digestive System Metabolism,” Journal of Nutrition, Vol. 140, No. 3, 2010, pp. 677S-689S. doi:10.3945/jn.109.113738 [23] M. E. Sanders and T. R. Klaenhammer, “Invited Review: The Scientific Basis of Lactobacillus acidophilus NCFM Functionality as a Probiotic,” Journal of Dairy Science, Vol. 84, No. 2, 2001, pp. 319-331. doi:10.3168/jds.S0022-0302(01)74481-5 [24] J. Walte r, C. He rte l, G. W. Tannock, C. M. Lis, K. Munro and W. P. Hammes, “Detection of Lactobacillus, Pedio- coccus, Leuconostoc, and Weissella Species in Human Feces by Using Group-Specific PCR Primers and Dena- turing Gradient Gel Electrophoresis,” Applied and Envi- ronmental Microbiology, Vol. 67, No. 6, 2001, pp. 2578- 2585. doi:10.1128/AEM.67.6.2578-2585.2001 [25] E. Wesney and G. W. Tannock, “Association of Rat, Pig, and Fowl Biotypes of Lactobacilli with the Stomach of Gnotobiotic Mice,” Microbial Ecology, Vol. 5, No . 1, 1979, pp. 35-42. doi:10.1007/BF02010576 [26] S. Roos, F. Karner, L. Axelsson and H. Jonsson, “Lacto- bacillus Mucosae sp. nov., a New Species with in Vitro Mucus-Binding Activity Isolated from Pig Intestine,” In- ternational Journal of Systematic and Evolutionary Mi- crobiology, Vol. 50, No. 1, 2000, pp. 251-258. doi:10.1099/00207713-50-1-251 [27] L. Axelsson and S. Lindgren, “Characterization and DNA Homology of Lactobacillus Strains Isolated from Pig In- testine,” Journal of Applied Bacteriology, Vol. 62, No. 5, 1987, pp. 433-440. doi:10.1111/j.1365-2672.1987.tb02673.x [28] C. Efthymiou and P. A. Hansen, “An Antigenic Analysis of Lactobacillus Acidophilus,” The Journal of Infectious Diseases, Vol. 110, No. 3, 1962, pp. 258-267. doi:10.1093/infdis/110.3.258 Copyright © 2012 SciRes. AiM  S. A. FRESE ET AL. Copyright © 2012 SciRes. AiM 409 [29] I. Martinez, J. Kim, P. R. Duffy, V. L. Schlegel and J. Walter, “Resistant Starches Types 2 and 4 Have Differen- tial Effects on the Composition of the Fecal Microbiota in Human Subjects,” PLoS One, Vol. 5, No. 11, 2010, p. e15046. doi:10.1371/journal.pone.0015046 [30] P. Kabadjova, X. Dousset, V. Le Cam and H. Prevost, “Differentiation of Closely Related Carnobacterium Food Isolates Based on 16s-23s Ribosomal DNA Intergenic Spacer Region Polymorphism,” Applied and Environ- mental Microbiology, Vol. 68, No. 11, 2002, pp. 5358- 5366. doi:10.1128/AEM.68.11.5358-5366.2002 [31] M. Haarman and J. Knol, “Quantitative Real-Time PCR Analysis of Fecal Lactobacillus Species in Infants Re- ceiving a Prebiotic Infant Formula,” Applied and Envi- ronmental Microbiology, Vol. 72, No. 4, 2006, pp. 2359- 2365. doi:10.1128/AEM.72.4.2359-2365.2006 [32] C. B. Meroth, J. Walter, C. Hertel, M. J. Brandt and W. P. Hammes, “Monitoring the Bacterial Population Dynamics in Sourdough Fermentation Processes by Using PCR-De- naturing Gradient Gel Electrophoresis,” Applied and En- vironmental Microbiology, Vol. 69, No. 1, 2003, pp. 475- 482. doi:10.1128/AEM.69.1.475-482.2003 [33] S. Hammons, P. L. Oh, I. Martinez, K. Clark, V. L. Schlegel, E. Sitorius, S. E. Scheideler and J. Walter, “A Small Variation in Diet Influences the Lactobacillus Strain Composition in the Crop of Broiler Chickens,” Systematic and Applie d Microbi ology, Vol. 3 3, No. 5, 2010, pp. 275-281. doi:10.1016/j.syapm.2010.04.003 [34] B. Dal, F. Walter, W. J. Hammes and C. Hertel, “In- creased Complexity of the Species Composition of Lactic Acid Bacteria in Human Feces Revealed by Alternative Incubation Condition,” Microbial Ecology, Vol. 45, No. 4, 2003, pp. 455-463. doi:10.1007/s00248-003-2001-z [35] J. Sui, S. Leighton, F. Busta and L. Brady, “16s Ribo- somal DNA Analysis of the Faecal Lactobacilli Composi- tion of Human Subjects Consuming a Probiotic Strain Lactobacillus acidophilus NCFM,” Journal of Applied Microbiology, Vol. 93, No. 5, 2002, pp. 907-912. doi:10.1046/j.1365-2672.2002.01767.x [36] M. Watanabe, H. Kinoshita, M. Nitta, R. Yukishita, Y. Kawai, K. Kimura, N. Taketomo, Y. Yamazaki, Y. Tat- eno, K. Miura, A. Horii, H. Kitazawa and T. Saito, “Iden- tification of a New Adhesin-Like Protein from Lactoba- cillus mucosae ME-340 with Specific Affinity to the Hu- man Blood Group a and b Antigens,” Journal of Applied Microbiology, Vol. 109, No. 3, 2010, pp. 927-935. doi:10.1111/j.1365-2672.2010.04719.x [37] L. Dethlefsen, P. B. Eckburg, E. M. Bik and D. A. Rel- man, “Assembly of the Human Intestinal Microbiota,” Trends in Ecology & Evolution, Vol. 21, No. 9, 2006, pp. 517-523. doi:10.1016/j.tree.2006.06.013 [38] J. Cavender-Bares, K. H. Kozak, P. V. Fine and S. W. Kembel, “The Merging of Community Ecology and Phy- logenetic Biology,” Ecology Letters, Vol. 12, No. 7, 2009, pp. 693-715. doi:10.1111/j.1461-0248.2009.01314.x [39] J. Walter and R. Ley, “The Human Gut Microbiome: Ecology and Recent Evolutionary Changes,” Annual Re- view of Microbiology, Vol. 65, No. 10, 2011, pp. 411- 429. doi:10.1146/annurev-micro-090110-102830 [40] A. Khoruts, J. Dicksved, J. K. Jansson and M. J. Sadowsky, “Changes in the Composition of the Human Fecal Micro- biome after Bacteriotherapy for Recurrent Clostridium difficile-Associated Diarrhea,” Journal of Clinical Gast ro- enterology, Vol. 44, No. 5, 2010, pp. 354-360. [41] J. O’Callaghan and P. W. O’Toole, “Lactobacillus: Host- Microbe Relationships,” Current Topics in Microbiology and Immunology, in Press, 2012. Abbreviations and Acronyms GI: gastrointestinal; RAPD: random amplification of polymorphic DNA; qPCR: quantitative real-time PCR; PCR: polymerase chain reaction; MRS: mann-rogosa-sharpe media; NCBI: national center for biotechnology information; MLSA: multi-locus sequencing an alysis; IRB: institutional review bo ard; CFU: colony-forming units.
|