American Journal of Plant Sciences
Vol.4 No.12(2013), Article ID:41295,4 pages DOI:10.4236/ajps.2013.412303
Genetic Relationships in Advanced Generation Hybrids Derived from Crosses between Texas (Poa arachnifera) and Kentucky (Poa pratensis) Bluegrass Using ISSR Markers
![]()
Southern Plains Range Research Station, United States Department of Agriculture, Agricultural Research Service, Woodward, USA.
Email: Jason.goldman@ars.usda.gov
Copyright © 2013 Jason Goldman. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received October 28th, 2013; revised November 30th, 2013; accepted December 11th, 2013
Keywords: Poa arachnifera; Poa pratensis; Interspecific Hybrids; Inter Simple Sequence Repeat; Genetic Similarity
ABSTRACT
Fertile, advanced generation hybrids derived from crosses between Texas (Poa arachnifera Torr.) and Kentucky (Poa pratensis L.) bluegrass have been selected. The hybrids are currently being evaluated for low-input turf potential. Since they are derived from hand-harvested seed from first-generation hybrids located in field nurseries their exact genetic origin is unknown. This experiment was conducted to determine if there are still genetic relationships among the advanced generation hybrids and some of the Texas and Kentucky parents in their pedigrees. Four Inter Simple Sequence Repeat (ISSR) primer combinations resolved on 6% nondenaturing polyacrylamide gels resulted in 179 polymorphic bands that were scored to create a genetic similarity matrix and dendrogram based on Jaccard’s coefficient. The clustering of the advanced generation hybrids was generally in agreement with what would be expected based on their pedigrees and indicated it was more likely to select a fertile hybrid from an advanced generation, rather than the F1 generation.
1. Introduction
Texas bluegrass (Poa arachnifera Torr.) is a highly rhizomatous, dioecious, perennial cool-season grass native to southern Kansas, Oklahoma, western Arkansas and most of Texas [1]. Interspecific hybrids with Kentucky bluegrass (Poa pratensis L.) have the potential to produce low-input turf type plants with greater heat and drought tolerance [2,3]. Assuming this grass will be propagated by seed, a Texas × Kentucky (TK) hybrid needs to inherit a Kentucky like inflorescence containing both male and female structures with the ability to self-pollinate and produce seed via apomixis. The cottony seed characteristic of the female Texas parent must also be drastically reduced or eliminated to enable combine harvesting and commercial-type seed cleaning for a hybrid to be accepted by the turf industry. Reveille [3] hybrid bluegrass is the only cultivar documented in the literature and is a first generation (F1) hybrid. Over the last 10 years, many Texas × Kentucky hybrids have been created and confirmed visually and by DNA fingerprinting with the same methods described by Goldman and Sims [4] and Goldman [5]. However, none of these F1 hybrids had a combination of all the essential traits required for further evaluation. Advanced generation hybrids in the current experiment are of two types. The Texas × Kentucky (TK) type involved evaluating first generation (F1) hybrids as spaced plants, hand harvesting seed from elite plants and then evaluating that seed in an additional spaced plant or small plot nursery where a single plant selection is the source of the fertile advanced generation hybrid. The (Texas × Kentucky) × Kentucky (TK) K type involved the same selection process as the TK type except an additional greenhouse cross in which a first generation TK hybrid is used as the female and another Kentucky source is used as the male. The advanced generation hybrids all have complete, fertile inflorescences and are being evaluated further for self-fertility, level of apomixis, and turf potential. The genetic origin of these seeds that gave rise to the selected advanced generation hybrids is unknown. It could have included self-pollination, cross-pollination with an adjacent full-sib hybrid, cross-pollination with other unrelated hybrids in the nursery, or with pure Texas or Kentucky bluegrass in the nursery, or some other genetic anomaly. The objective of the current experiment is to determine if there still exits a genetic relationship among the selected advanced generation hybrids and some of the Texas, Kentucky, and Texas × Kentucky parents in their pedigrees using intersimple sequence repeat (ISSR) DNA markers [5,6].
2. Materials and Methods
2.1. Plant Material
The advanced generation hybrid selections and breeding parents in Table 1, all single genotypes, were used for the DNA analysis. The hybrid derived from TK43 × Trenton (18:26) was selected after two rounds of field hand harvests and evaluation. An additional D4-10 ×

Table 1. Advanced generation hybrids and breeding parents used in the ISSR analysis.
Poland entry, not listed in Table 1, derived from seeded turf (multiple seedlings) was also included. Based on progeny spaced plant evaluation, D4-10 × Poland has been identified as highly apomictic. Trenton DNA was also derived from seeded turf.
2.2. ISSR Analysis
Total genomic DNA from fresh leaf tissue was extracted and purified based on a 2% cetyltrimethylammonium bromide (CTAB) method [7]. DNA quality and quantity was determined after electrophoresis on an ethidium bromide stained 1% agarose gel before ISSR polymerase chain reaction (PCR). Four ISSR primer combinations (811 + 841, 811 + 826, 808 + 857, and 811 + 857) from the University of British Columbia Biotechnology Laboratory set nine (UBC9), previously described to produce robust fingerprints and identify interspecific hybrids with Texas bluegrass [5] were used for the analysis. PCR amplification conditions [5] were previously described. PCR products were resolved on a nondenaturing 6% polyacrylamide gel (17.1 w × 14.7 h cm) in 1X sodium boric acid buffer [8] for 2 h at 225 V. The gel was stained in ethidium bromide for 20 m before being digitally photographed and converted to a negative image. The PCR reaction and acrylamide gel analysis was repeated at least two times with all 18 entries included as a group. Only polymorphic, reproducible bands that appeared in direct alignment were scored. Bands were scored as “1” for presence and “0” for absence and used to calculate a pair-wise similarity matrix using Jaccard’s coefficient [9]. The similarity matrix was used to construct a dendrogram with the unweighted pair-group method by arithmetic averages (UPGMA) using the SAHN-clustering and TREE programs from the NTSYS-pc version 2.02 package [10].
3. Results and Discussion
The four ISSR primer combinations used for PCR resulted in 179 polymorphic bands that were scored over all entries. The number of polymorphic bands scored per primer combination ranged from six for 811 + 841 and 811 + 857 to 11 for 811 + 826. Nine polymorphic bands were scored from the 808 + 857 primer combination (Figure 1). The gels were scored conservatively because other reproducible polymorphic fragments that were extremely close in migration but may not have been in direct alignment were not scored. Nearly identical fingerprints were obtained for D4-10 × Poland single plant DNA and DNA derived from turf started from seedlings on all gels. This similarity was expected because D4-10 × Poland is highly apomictic.
The Jaccard’s similarity coefficient ranged from 0 to
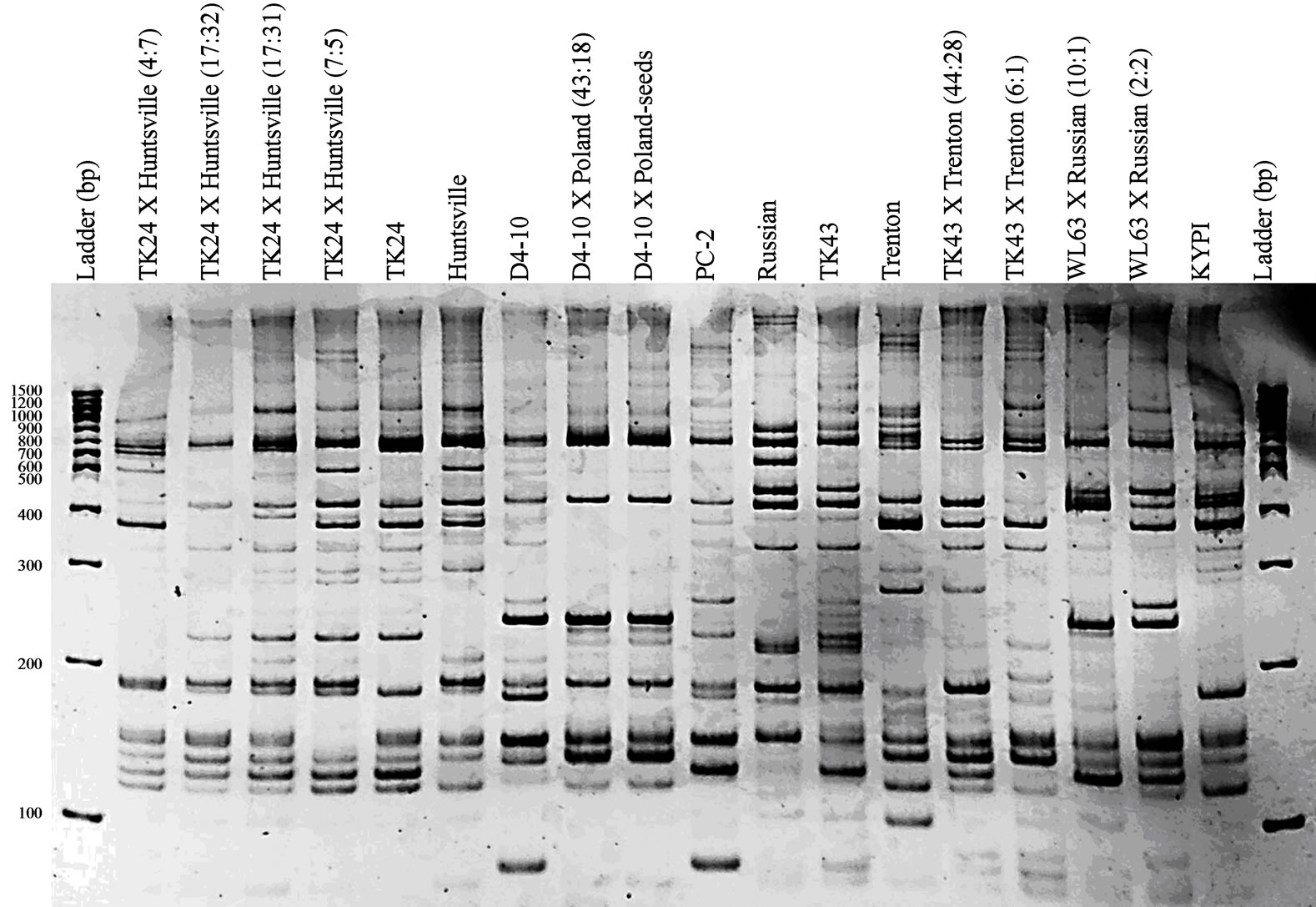
Figure 1. ISSR fingerprints of the advanced generation hybrids and breeding parents listed in Table 1 using the primer combination 808 + 857. D4-10 × Poland-seeds was DNA extracted from seeded turf, not a single genotype. The first and last lanes contain the same molecular weight ladder in base pairs (bp).
0.92 among entries. The two TK24 × Huntsville selections derived from hybrid 17:14 (Table 1) had the highest genetic similarity (0.92), followed by D4-10 × Poland, parent, and seedlings (0.91). A single dendrogram (Figure 2) based on the similarity matrix was constructed with no ties. TK24, Huntsville, and the four TK24 × Huntsville advanced generation hybrids clustered together. Trenton and KYPI Kentucky bluegrass grouped together next to the cluster containing Huntsville. The next cluster contained the TK43 × Trenton and WL63 × Russian derived hybrids followed by D4-10 × Poland parent and progeny. Russian Kentucky bluegrass did not cluster with any of the other groups and was located next to the last cluster at the bottom of the tree that contained the Texas bluegrass females and TK43. The first generation TK hybrid breeding parents (TK24 and TK43) contained a similarity coefficient of 0.31 but clustered closer to their parent (TK43 and PC2; similarity 0.62) or advanced generation progeny (TK24 and TK24 × Huntsville hybrids; similarity 0.5 to 0.57).
The clustering of the advanced generation hybrids was generally in agreement with what would be expected based on their pedigrees. These results indicated that with the germplasm used in this experiment, it was more likely to select a fertile hybrid that merits further evalua-

Figure 2. UPGMA dendrogram based on Jaccard’s coefficient from 179 ISSR bands showing the relatedness among Texas, Kentucky, and hybrid bluegrass.
tion from an advanced generation, rather than the first generation (F1) and that these hybrids still contained Texas and Kentucky bluegrass DNA fragments from parents in their pedigrees.
REFERENCES
- A. S. Hitchcock, “Manual of the Grasses of the United States (revised by A. Chase),” US Government Printing Office, Washington DC, 1950, p. 1051.
- J. C. Read, “Utilization of Apomictic and Dioecious Methods of Reproduction in Breeding poa spp.,” International Turfgrass Society Research Journal, No. 9, 2001, pp. 202-205.
- J. C. Read, J. A. Reinert, P. F. Colbaugh and W. E. Knoop, “Registration of ‘Reveille’ Hybrid Bluegrass,” Crop Science, Vol. 39, No. 2, 1999, p. 590. http://dx.doi.org/10.2135/cropsci1999.0011183X003900020059x
- J. J. Goldman and P. L. Sims, “Production of an Interspecific Hybrid between Texas and Argentine Bluegrass,” Plant Breeding, Vol. 124, No. 4, 2005, pp. 419-420. http://dx.doi.org/10.1111/j.1439-0523.2005.01113.x
- J. J. Goldman, “The Use of ISSR Markers to Identify Texas Bluegrass Interspecific Hybrids,” Plant Breeding, Vol. 127, No. 6, 2008, pp. 644-646. http://dx.doi.org/10.1111/j.1439-0523.2008.01526.x
- E. Zietkiewicz, A. Rafalski and D. Labuda, “Genomic Fingerprinting by Simple Sequence Repeat (SSR)-Anchored Polymerase Chain Reaction Amplification,” Genomics, Vol. 20, No. 2, 1994, pp. 176-183. http://dx.doi.org/10.1006/geno.1994.1151
- S. O. Rogers and A. J. Bendich, “Extraction of DNA from Milligram Amounts of Fresh, Herbarium and Mummified Plant Tissues,” Plant Molecular Biology, Vol. 5, No. 2, 1985, pp. 69-76.
- J. R. Brody and S. E. Kern, “Sodium Boric Acid: A TrisFree, Cooler Conductive Medium for DNA Electrophoresis,” BioTechniques, Vol. 36, No. 2, 2004, pp. 214-216.
- P. Jaccard, “Nouvelles Recherches sur la Distribution Florale,” Bulletin de la Société Vaudoise des Sciences Naturelles, Vol. 44, No. 163, 1908, pp. 223-270.
- F. J. Rohlf, “NTSYS-PC: Numerical Taxonomy and Multivariate Analysis System, Version 2.02i,” Department of Ecology and Evolution, State University of New York, Setauket, 1998.

