American Journal of Plant Sciences
Vol.4 No.1(2013), Article ID:27660,9 pages DOI:10.4236/ajps.2013.41021
Null Alleles in Gliadin Coding Loci and Wheat Allergenic Properties
![]()
1Cereals Department, Plant Breeding and Acclimatization Institute, Krakow, Poland; 2Jerzy Haber Institute of Catalysis and Surface Chemistry, Polish Academy of Sciences, Krakow, Poland; 3Department of Clinical and Environmental Allergology, Collegium Medicum Jagiellonian University, Krakow, Poland; 4The Franciszek Górski Institute of Plant Physiology, Polish Academy of Sciences, Krakow, Poland.
Email: *zhwaga@cyf-kr.edu.pl
Received October 20th, 2012; revised November 30th, 2012; accepted December 30th, 2012
Keywords: Wheat; Gliadins; Allergenicity; A-PAGE; RP-HPLC; ELISA; Plant Breeding
ABSTRACT
Wheat gliadin proteins-an important, nutritional component of many food products may also act as allergenic proteins causing various, clinical symptoms of IgE-mediated food allergies. Gliadins are coded by six complex loci on the chromosomes 1A, 1B, 1D, 6A, 6B and 6D of wheat genome. Each of the loci coding from a few to a dozen of polypeptides may spontaneously mutate to inactive gene variants called null alleles that do not code any proteins at all. The aim of the present work was to find out whether null alleles in some gliadin coding loci may decrease wheat allergenic properties. Six winter wheat genotypes: gliadin deletion lines (GDL) containing null alleles on 1D, 1B and 6B chromosomes and control lines (CL) containing active gene variants in all gliadin coding loci, were developed using plant breeding methods. Allergenic properties of the six analyzed hybrids were estimated by ELISA using polled sera of five patients allergic to gluten. Estimated immunoreactivity of GDLs was from 6% to 18% lower as compared with CLs. The obtained results evidenced that gliadin null alleles decrease wheat allergenic properties and may be used as parental forms for breeding of hypoallergenic wheat genotypes.
1. Introduction
Gliadins and glutenins are the main groups of wheat storage proteins and the main components of gluten matrix. As gluten proteins they are of essential importance for baking quality of wheat flour, food industry and human nutrition [1]. Gliadins are especially interesting biomolecules because of their complexity (they make a complex of monomeric proteins) and large polymorphism of individual monomeric fractions [2]. However, they are also highly allergenic proteins causing IgE mediated food allergies (e.g. asthma, urticaria, angioedema, WDEIA, atopic dermatitis) in people sensitized to wheat proteins [3,4].
Complexity, polymorphism and allergenic properties are determined genetically-genetics conditions of gliadins are specific and different from those of other non-gluten proteins of wheat kernels. Genes coding for gliadins are grouped as clusters and localized on the short arms of the chromosomes belonging to 1st and 6th homeological groups of wheat genome (1A, 1B, 1D, 6A6B and 6D; loci Gli A1, Gli B1, Gli D1, Gli A2, Gli B2 and Gli D2 respectively) [5]. Families of multiple alleles for each locus arose during evolution of Triticum aestivum species as results of non-lethal mutations hence a huge polymorphism among wheat cultivars and hybrid lines can be observed in separate protein complex by electrophoresis (A-PAGE and SDS-PAGE) or chromatography (RP-HPLC) [6,7]. Each individual allele code for one to dozen of protein fractions differed with regard to molecular weight ranging from 20 to 70 kDa, aminoacid composition with proline and glutamine as major amino acids grouped in short, repetitive sequences, and higher order structures with poly L-proline, β-turns and random coils observed most often [8]. A set of protein fractions expressed by individual gene cluster can be visualized as a group of electrophoretic bands or chromatographic peaks-they are called protein blocks [9]. Based on A-PAGE (the most efficiently electrophoretic method of gliadin separation) gliadins are divided into five groups: α, β, γ, ω-5 and ω-1.2, where α are the fastest and ω-1.2 the slowest moving protein fractions during separation on polyacrylamide gel.
Gliadin protein genes mutate spontaneously changing the number or electrophoretic mobility of individual protein bands. Some mutations consist of replacing individual nucleotides of DNA chain in regulatory regions of the genes. As a result codons responsible for incorporation of predicted amino acid into peptide during translation (e.g. glutamine codon CAA) may vary into stop codon (TAA) inhibiting translation of individual protein or groups of proteins [10]. Such inactive gene variants are called null alleles or pseudogenes. Phenotypic effects of gene inhibition are relatively easy to identify-electrophoregrams of null allelic genotypes lack a single band or groups of bands.
Numerous examples of null alleles among gliadin genes, especially among α-gliadin genes localized on the chromosome 6A, were identified. Based on DNA sequencing and bioinformatics sequence alignment methods Anderson evaluated a total number of α-gliadin genes as more than 150—about one half of this number appears in the form of pseudogenes [11]. Null alleles were also observed among other gliadin groups. Some reports describe the effects of γ and ω-gliadin null alleles on baking quality [12,13]. However, null alleles are uniquely observed among common wheat cultivars. They appear spontaneously among hybrid genotypes developed by wheat breeders. It seems possible, that hybrids coming from cross combinations between commercial wheat cultivars and ancient types of wheat e.g. spelt wheat or some primitive, local biotypes grown in extreme environmental conditions such as on high altitudes (Andes, Himalaya) or high temperatures (south Mediterranean region) contain null alleles more often than hybrids coming from crossing between commercial wheat cultivars. Pseudogenes of gluten proteins are probably eliminated by artificial selection presumably because of their negative effects on some agricultural traits. Instead, the null allele of glutenin proteins localized in Glu A1 locus on the short arm of chromosome 1A is widespread among wheat cultivars and breeding lines investigated in Poland and other European countries in spite of its negative effects on wheat baking quality [14]. This is hypothetically because of linkage of Glu A1 locus with gene (genes) responsible for frost resistance and winter hardiness [15].
In our research on wheat allergenic properties we developed a set of hybrid genotypes containing pseudogenes in loci Gli B1, Gli D1 and Gli B2. Development of null alleles resulted in deletion of some A-PAGE bands coded by chromosomes 1B, 1D and 6B and belonging to ω-5, ω-1.2 and β-gliadins respectively. As food products made from wheat flour are daily consumed by millions of people all over the world allergenicity of wheat causes serious medical and nutritional problem. We suppose that decreased number of allergenic protein fractions may contribute to decreasing of the immunoreactivity of the whole gliadin complex. The present study is aimed to determine whether induction of pseudogenes in some Gli loci, present in null allelic lines developed by us, may contribute to decreasing of gliadin immunoreactive properties.
2. Material and Methods
Three hybrid lines of F4 generation coming from cross combination between spelt wheat (cultivar Oberkummler Rotkorn) and Polish high quality wheat line LAD 480 were reproduced on the plots (5 m2) in Plant Breeding and Acclimatization Institute in Krakow (Poland) in the years 2006, 2007 and 2008. Based on A-PAGE these lines were recognized as pure and homozygous lines with regard to genes coding for gliadin proteins. Electrophoretic separations of individual kernel harvested in 2008 displayed heterogeneity of gliadins spectra. Each of the three hybrids showed the presence of two closely related genotypes: one of them contained a protein pattern typical for reproduced hybrids and the second one lacking some protein bands as compared with the wild type pattern. Lacking bands were observed in ω-1.2, ω-5 and β-gliadin zones. Two hundred of half kernel for each of wild type and mutated genotypes were analyzed by A-PAGE while the other half kernels containing germs were reserved. Based on the obtained electrophoregrams germ populations were divided into two biotypes: one biotype contained full set of gliadin bands (wild type control line-CL) while the second one, lacking the predicted gliadin bands was called gliadin deletion line (GDL). Germs of selected biotypes were reproduced in fitotron and next on the 2 m2 plots in the years 2009, 2010 and 2011. Three sets of pairs of closely related genotypes were obtained. The purity of the obtained pairs of genotypes regarding gliadin protein composition was tested in each year of the experiment by A-PAGE. Kernels harvested in 2011 were used for further biochemical and immunological experiments.
A-PAGE, RP-HPLC and ELISA were performed as described in previous papers [16-18]. Shortly, A-PAGE were made using DESAGA (Germany) electrophoretic apparatus on 8% polyacrylamide gel in acidic conditions (pH = 3.1) of aluminum lactate buffer. Proteins were developed by three hours, next gels were stained in commasie brilliant blue solution and distained in distilled water.
RP-HPLC analyses were conducted using Agilent Technologies 1100 HPLC system with DAD detector on Zorbax 300SB-C18 classical column (4.5 × 150 mm, 5 μm, 300A pore diameter thermostated to 70˚C). The mobile phase composed of acetonitryle (ACN) and deionized water with 0.1% trifluoroacetic acid (TFA) additive. Separations were performed in a gradient program with ACN concentration varying between 25% - 50% and 1mL/min flow rate. The obtained RP-HPLC patterns for CLs and GDLs were compared with regard to two chromatographic parameters: peak high (PH) and area under the peak (AUP). The analysis considered the differences of individual peaks and groups of peaks corresponding to α, β, γ and ω gliadins.
Sandwich ELISA was performed using pooled sera of five patients of elevated concentration of sIgE to gluten and differentiated disease symptoms were used (Table 1).
Microtiter wells were coated with 70% ethanol gliadin extracts diluted 1:10,000 in four replications. Next, human sera diluted 1:2 in PBST were introduced to the wells followed by IgE antihuman antibodies coupled with horseradish peroxidase (Sigma Chemical Co., USA) diluted 1:1000. Optical Density (OD) values calculated by ELISA test reader Opsys MR (Thermo Lab Systems, USA) were approved as indicators of gliadins immunoreactive properties.
3. Results
3.1. A-PAGE
A-PAGE electrophoregrams of CL and GDL differing with regard to proteins controlled by the Gli D1 locus (1st set of analyzed hybrids) showed a complete lack of protein bands localized in ω-1.2 gliadin zone of GDL (Figure 1(a)). Moreover GDL lacks one, intensely stained protein band, localized in γ zone. Earlier studies on gliadin inheritance evidenced that all of lacking bands, localized in two distant electrophoretic zones, are controlled by genes clustered in complex locus on the chromosome 1D and make one gliadin block determined as Gli D1-1.2, based on the own gliadin classification used by the authors [19]. This is evidence that discovered mutation inhibited expression of the whole gliadin block Gli D1-1.2.
In the second set of compared hybrid lines, differentiated with regard to gliadins controlled by the locus Gli B1 on the chromosome 1B lack of individual, weakly stained protein band in ω-5 gliadin zone was observed for GDL. The lacking band is one of the components belonging to the block Gli B1-1 (Figure 1(b)).
Beta gliadins in the control line belonging to third set of hybrids, differentiated with regard to gliadins controlled by the locus Gli B2 on the chromosome 6B, are composed of four bands of similar staining intensity and equally distant from each other on A-PAGE electrophoregram (Figure 1(c)). Previous work confirmed all four bands are inherited together and belong to the Gli
Table 1. Characteristics of patients who donated the sera samples for experiments.
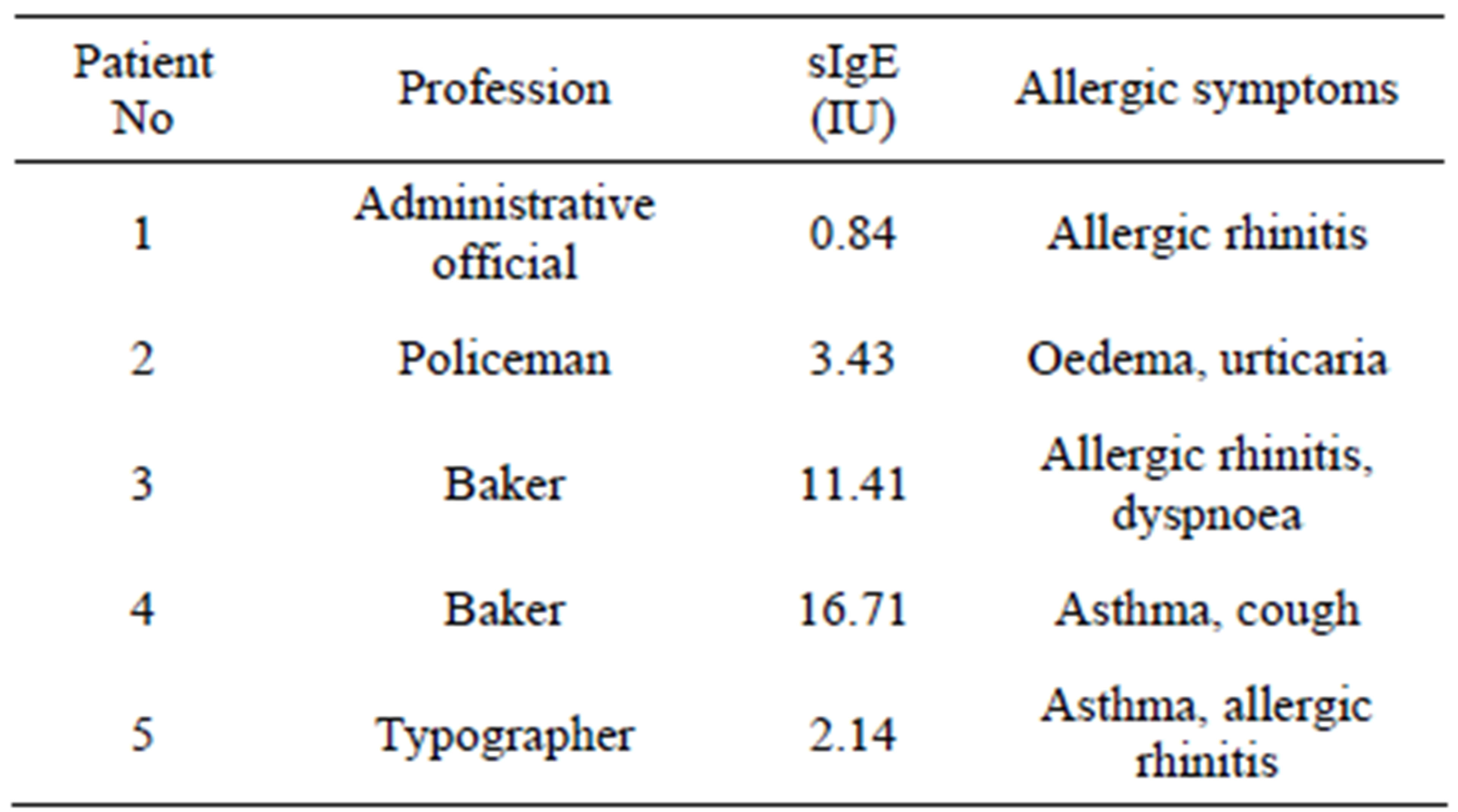

Figure 1. Gliadin protein of analyzed hybrid genotypes (CL and GDL) separated by A-PAGE. Gliadin bands present in CL but lacking in GDL are marked by the asterisk. Null alleles observed in GDL are localized on chromosomes: A-1D (locus Gli D1), B-1B (locus Gli B1) and C-6B (locus Gli B2). Gliadin classes (α, β, γ and ω) are marked on the left hand side of the Figure 2(a).
B2-2 block. In GDL lack of two from among four β-gliadin bands was observed-first of the lacking bands was of the slowest while the second one of the fastest electrophoretic mobility. The observed modification suggest that mutation in gliadin locus partly inhibited expression of genes coded for Gli B2-2 protein block.
3.2. RP-HPLC
Based on the comparison of A-PAGE and RP-HPLC for CLs and GDLs, gliadin chromatographic peaks can be classified into four zones in analyzed hybrid lines with retention times ranged between:
1) 20 - 26 min: ω-gliadins.
2) 26 - 31 min: β-gliadins.
3) 31 - 40 min: α-gliadins.
4) 40 - 60 min: γ-gliadins.
Significant differences between CLs and GDLs with regard to chromatographic parameters (PH and AUP) were observed in all analyzed pairs of hybrid genotypes
(Table 2).
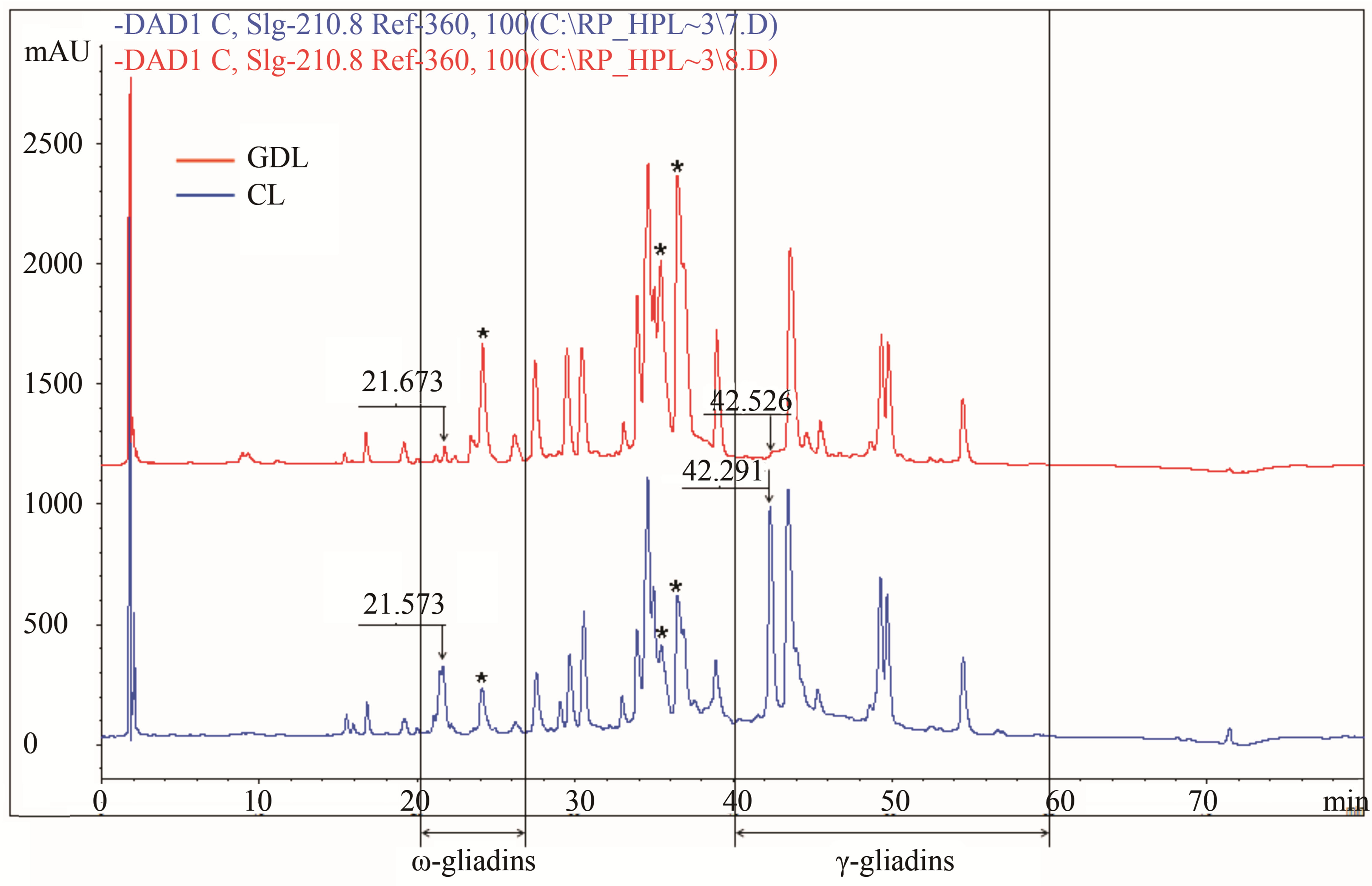
CLs and GDLs differing with regard to gliadins controlled by the chromosome 1D (1st group of analyzed hybrids) showed significant differences with regard to peaks RT = 21 and 42 min localized in ω and γ chromatographic zones respectively (Figure 2(a)). PH and AUP for both peaks were strongly reduced (about 70% - 90%) in GDL as compared with CL. At the same time hybrid belonging to the 1st group showed the effects of overexpression of genes coding for other fractions. PH and AUP values for peaks RT = 24 min (ω-gliadins), 35 min and 36 min (α-gliadins) in GDL were as much as twice higher than corresponding peaks in CL.
Hybrids belonging to the 2nd set of analysed hybrids, differing with regard to presence/absence of individual, weakly stained ω-5 gliadin band showed stronger differentiation of chromatographic peaks than could be expected based on A-PAGE separations. GDL showed significant decreasing in PH and AUP for three peaks: RT = 32 min (β-gliadins), 43 min and 54 min (γ-gliadins) (Figure 2(b)). Peaks of RT = 32 min and 54 min were almost completely lost while the peak of RT = 43 min strongly reduced (about 60% of PH and 40% of AUP for GDL as compared with CL).
In turn genotypes belonging to the 3rd set of hybrids, differentiated with regard to β-gliadins composition showed significant differences for four peaks localized in the zone between RT = 26 and 38 min (Figure 2(c)). Two of the peaks (RT = 27,026 min and 28,935 min) present in CL were almost completely lost in GDL, while two other peaks (RT = 24,492 min and 29,946 min) in CL were considerably reduced in GDL (about 40% and 70% for PH and about 30% and 80% for AUP, respectively).
PH and AUP were also evaluated for groups of peaks corresponding to β, γ and ω-gliadins in the three sets of analyzed hybrids (Table 3). A decrease in total PH about 30% and 70% was observed for groups of peaks corresponding to β and γ gliadin classes respectively. No significant changes were stated however, for total PH calculated for peaks corresponding to ω-gliadin classes while comparing GDL and CL from the 1st set of analyzed hybrids. Total AUP decreased in GDLs just like total PH with the only exception for ω-gliadins where a considerable increase (about 18%) in GDL was observed.
Table 2. Decreasing of individual peaks high and area under the peaks observed in GDL as compared with CL based on RP-HPLC separations.
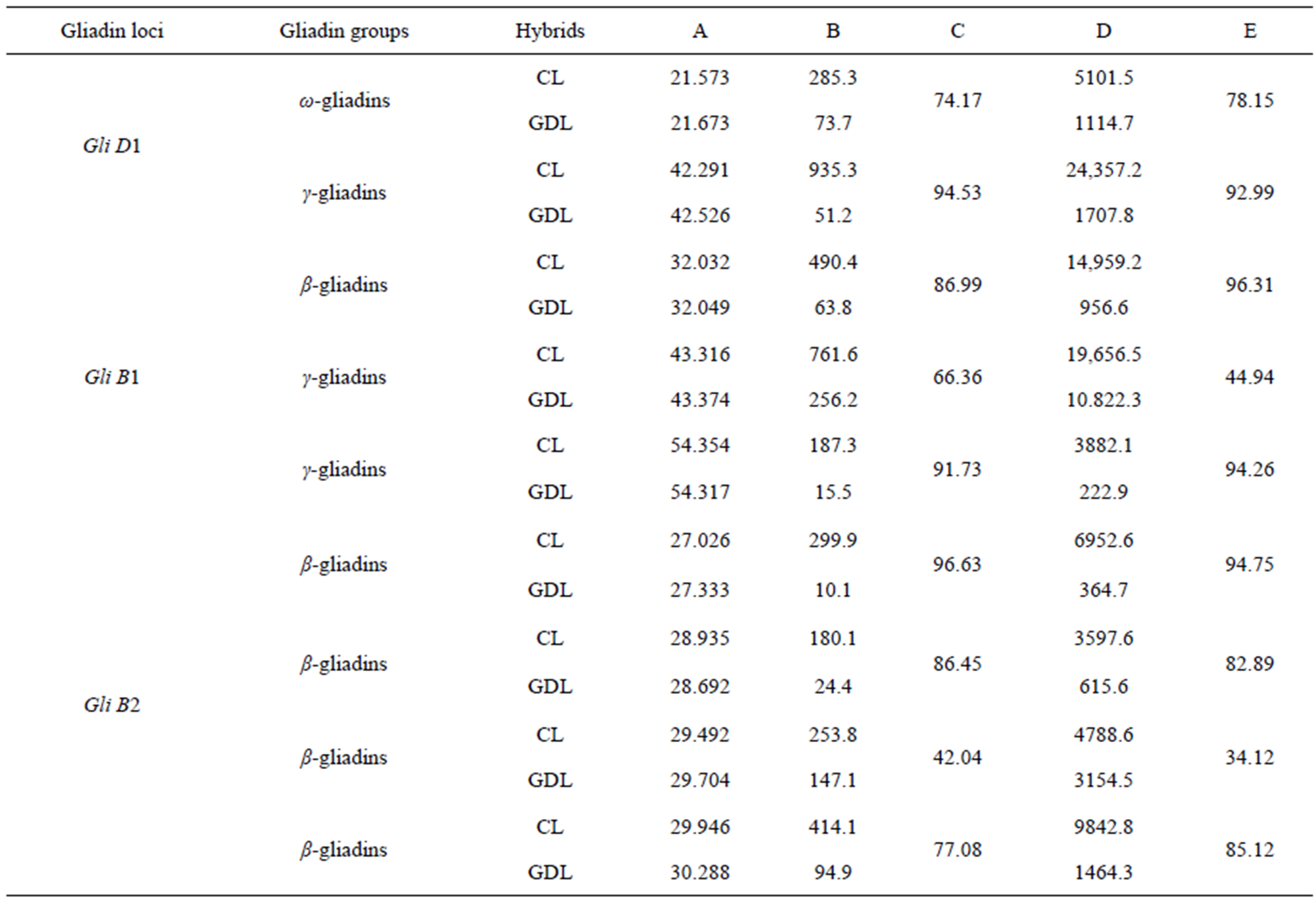
CL: Control lines; GDL: Gliadin deletion lines; A: Retention times (min); B: Peaks high (mAU); C: Proportional decreasing of peak high observed for GDL in relation to CL; D: Area under the peaks (mAUxs) ; E: Proportional decreasing of area under the peaks observed for GDL in relation to CL (%).
 (a)
(a)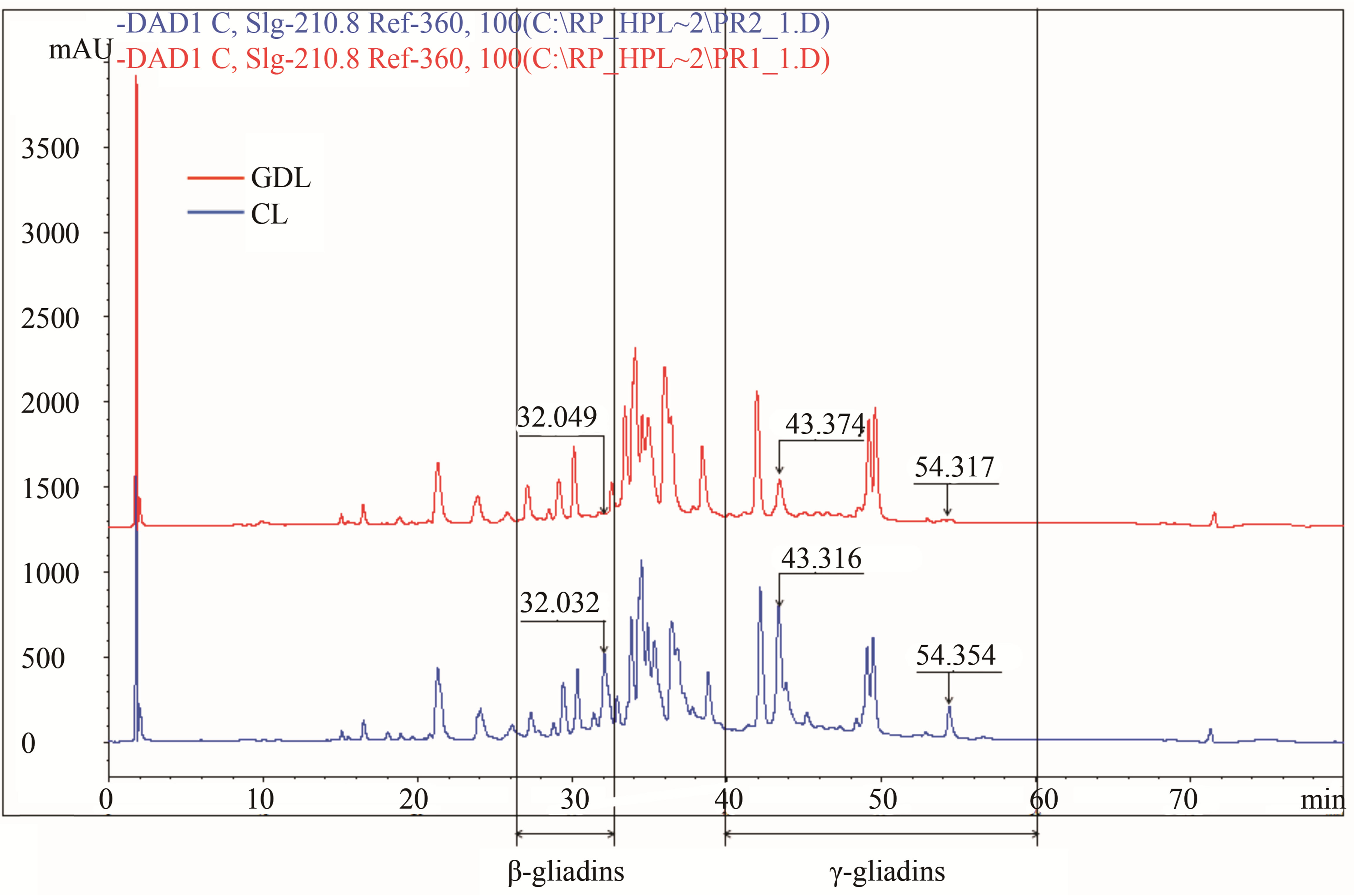 (b)
(b)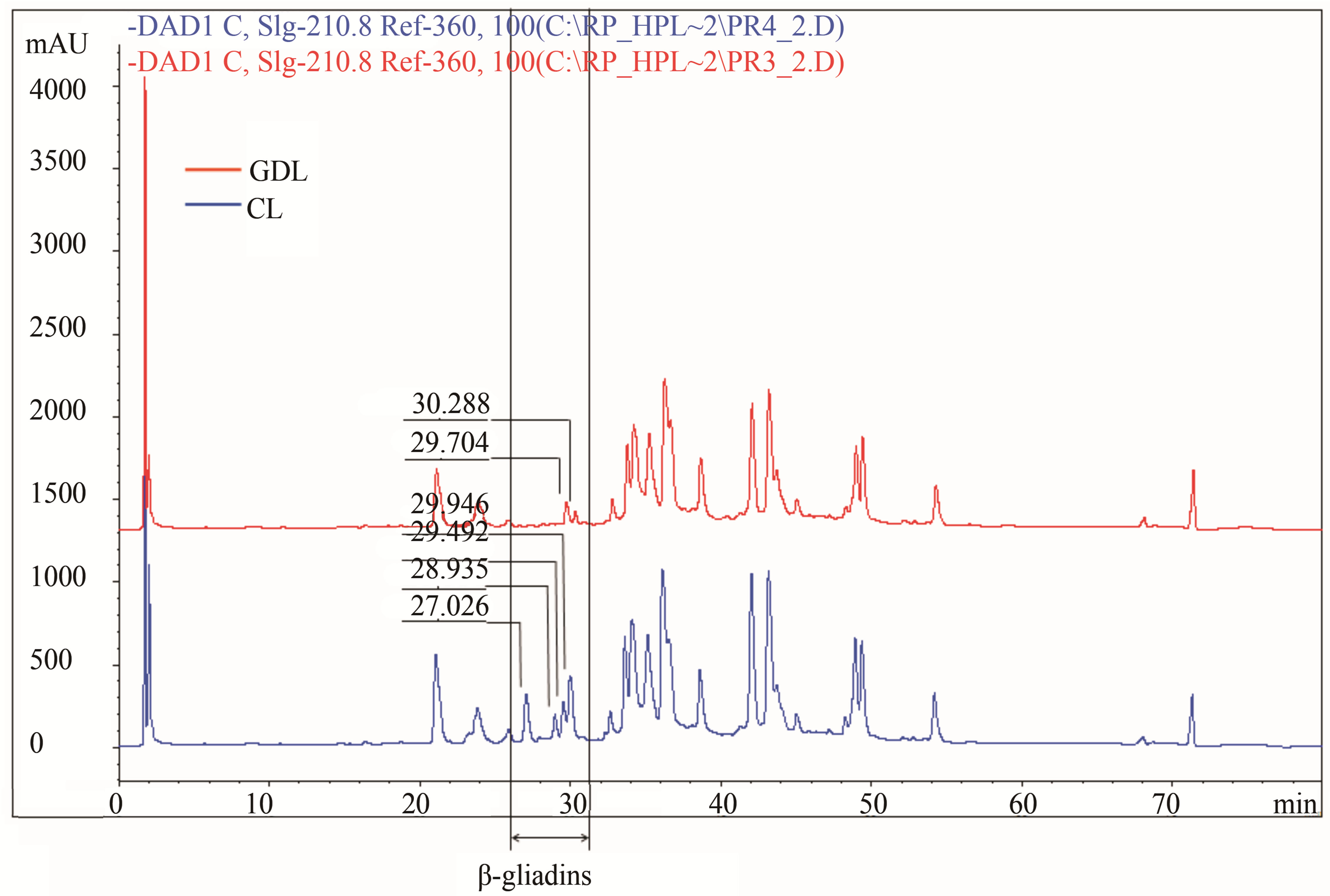 (c)
(c)
Figure 2. Gliadin protein of analyzed hybrid genotypes (CL in blue and GDL in red) separated by RP-HPLC. Gliadin peaks present in CL but lacking in GDL are marked by the arrows. Retention times are given above corresponding peaks. (a) Null allele observed in GDL is localized on the chromosome1D (locus Gli D1). Peaks for overexpressed proteins, observed in 1D GDL, and corresponding peaks in 1D CL are marked by the asterisks; (b) Null allele observed in GDL is localized on the chromosome 1B (locus Gli B1); (c) Null allele observed in GDL is localized on the chromosome 6B (locus Gli B2).
Table 3. Decreasing of peaks high and area under the peaks for gliadin groups observed in GDL as compared with CL based on RP-HPLC separations.
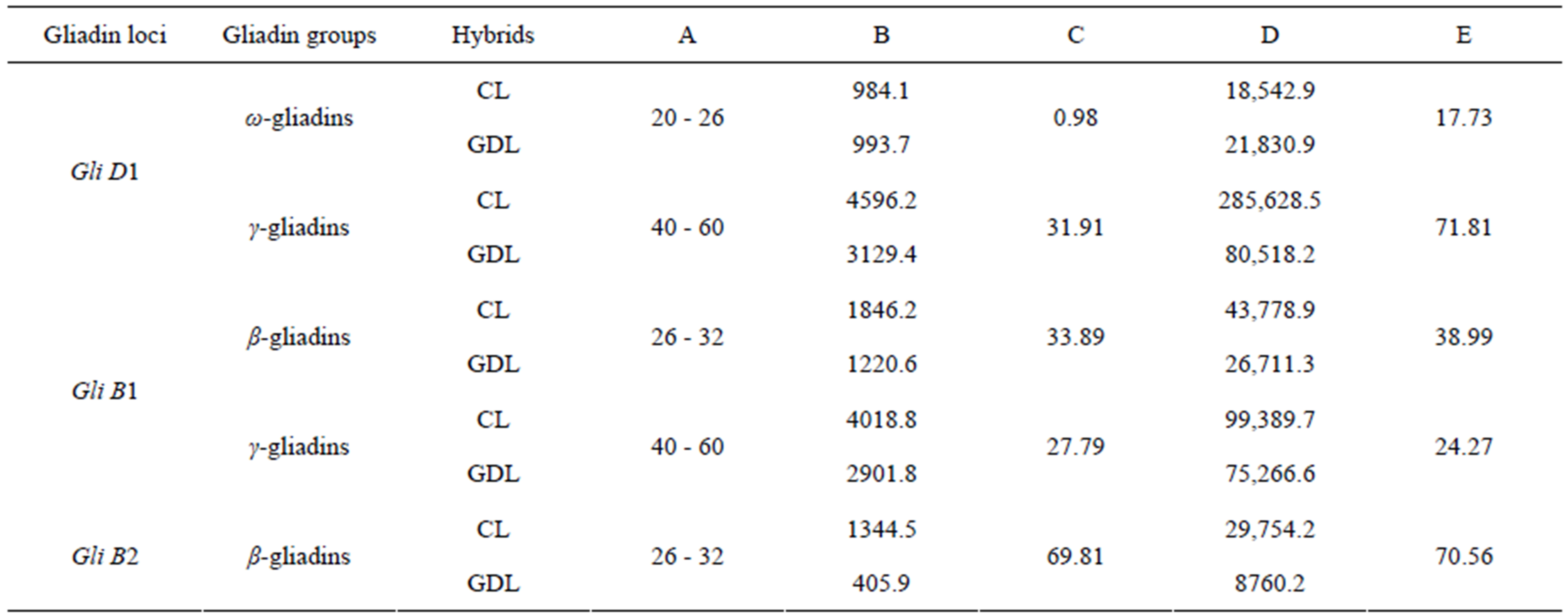
CL: Control lines; GDL: Gliadin deletion lines; A: Retention times (min); B: Peaks high (mAU); C: Proportional decreasing of peak high observed for GDL in relation to CL; D: Area under the peaks (mAUxs) ; E: Proportional decreasing of area under the peaks observed for GDL in relation to CL (%).
3.3. ELISA
Gliadin immunoreactive properties of analyzed wheat genotypes were evaluated by ELISA using pooled sera of five patients allergic to gluten proteins. The obtained results showed minor effects of immunoreactivity decrease for all of three examined GDL as compared with corresponding CL (Figure 3). Proportional decrease of OD values (immunoreactivity indicators) observed in GDL ranged from 6.14% for genotype with deletion of gliadins coded by Gli D1 locus to 18.02% in the case of genotype with decreased number of gliadins coded by Gli B1 locus.
4. Discussion
In the present work the effects of spontaneous mutations at three gliadin coding loci (Gli D1, Gli B1 and Gli B2) on wheat allergenic properties were analyzed. The evidenced mutations caused two types of changes in gliadin protein composition: complete suppression of all components belonging to one gliadin block in the Gli D1 locus (gliadin block Gli D1-1.2) and partial suppression of protein block in Gli B1 and Gli B2 loci. Plant materials were grouped in three sets of genotypes, each set comprised a doublet of hybrid lines: the wild type parental line containing a whole set of protein fractions (CL) and mutated, gliadin deletion line (GDL) showing lack of proteins controlled by particular loci. Gliadin composition of the three analyzed doublets were characterized by electrophoresis A-PAGE and chromatography RP-HPLC. Both methods are complementary to one another in the research on gliadins polymorphism [19]. The observed differences between GDL and CL showed complete or partial inhibition of genes among particular Gli loci. Complete inhibition of all of the genes was observed for Gli D1 complex locus coded for proteins belonging to ω-1.2 and γ gliadins. These proteins make the Gli D1-1.2 gliadin block composed of five bands on the A-PAGE electrophoregram. A-PAGE of GDL showed lack of all the five bands as compared with CL while RP-HPLC showed a loss of only two peaks. The first of two peaks with RT of 21 and the latter 42 min. The former peak belongs to ω-gliadins that are proteins of the lowest hydrophobicity and elute in the beginning of the RP-HPLC separation. The latter peak of RT = 42 min belongs to γ-gliadins according to the classification of gliadin fractions with regard to their hydrophobicity. Thus three electrophoretic bands of ω-1.2 gliadins correspond to only one peak (RT = 21 min) while the γ-gliadin bands correspond to the second RP-HPLC peak (RT = 42 min). This suggests that three ω-1.2 gliadin proteins exhibiting the same hydrophobicity differ in electrophoretic mobility (result of molecular mass and electric charge). Another phenomenon was observed for lines differing with regard to Gli B1 locus. A-PAGE showed a loss of only one, weakly stained protein band localized in ω-5 gliadin zone. However, RP-HPLC analysis showed a drastic decrease of protein amount for three main peaks localized in γ and β-gliadin zone. This may suggest that the three peaks mentioned above constitute a new, not detected by A-PAGE, components of the Gli B1-1 block. Another possible explanation is that the observed mutation in Gli B1 locus caused also changes in other Gli loci.
Gliadin extracts from flour of the analyzed CLs and GDLs were immunoprobed by ELISA. Human IgE class antibodies present in pooled sera of five patients allergic
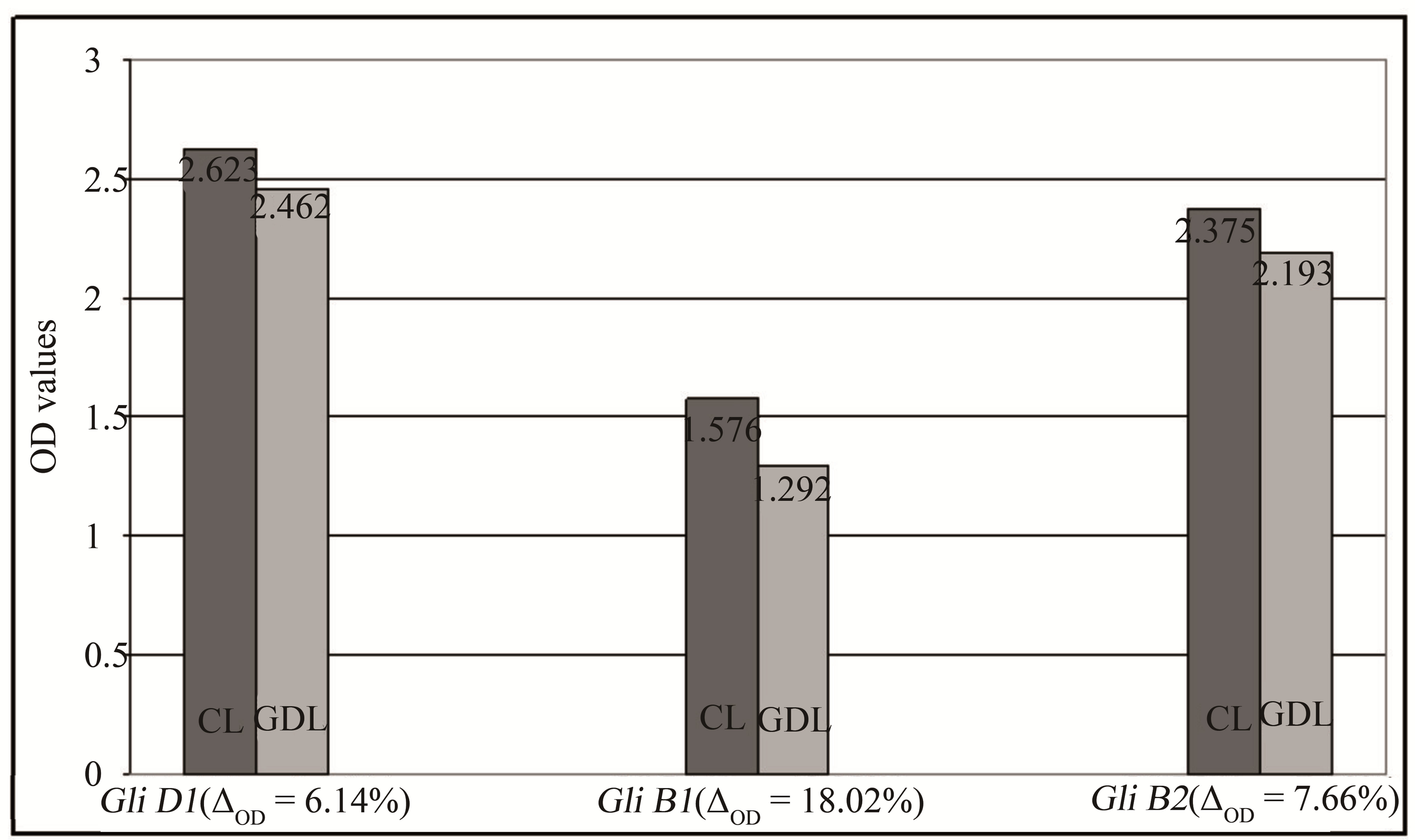
Figure 3. Differentiation of gliadins immunoreactivity observed for analyzed CLs and GDLs estimated by ELISA. Average OD values for individual hybrids are given at the top of each pile. Proportional decreasing of the OD values observed in GDLs are given at the bottom of the piles.
to wheat proteins were used for immunodetection. Such a technique is often applied in the research on wheat allergenicity [20]. Immunological datas evidenced a decrease in OD values for all of the analyzed GDLs as compared with CLs. Ability of IgE to bind gliadins extracted from GDLs genotypes lacking proteins controlled by Gli D1, Gli B1 and Gli B2 loci was lower by about 6%, 18% and 7% respectively. The highest decrease of immunoreactivity was observed in GDL lacking the ω-5 gliadin band confirming the highest allergenicity and immunodominant character of ω gliadins in allergies to wheat, especially ω-5 fractions that may trigger WDEIA [21,22]. However, decrease of IgE binding with GDL of reduced number of β fractions confirms our previous results on immunoreactivity of individual, purified gliadins indicating that other gliadin groups (besides the ω gliadins) may also share allergenicity of the protein complex [23].
The frequency of allergies in human societies increases along with technological and industrial progress that increases susceptibility of people to allergic diseases. This unnatural process is of special importance in highly developed European and American countries. Hence, a number of research projects on plant proteins were directed toward decreasing their allergenic potential. In the research on suppression of plant allergens, conducted more often by scientist in different countries, molecular biology methods are usually used. The gene silencing technology seems to be especially effective. Tada et al. succeeded in suppression of gene expression of the low molecular weight protein of molecular mass 14 - 16 kDa from rice using antisense gene strategy [24]. This small, allergenic protein provokes symptoms of asthma and eczema. Another example of a successful allergen suppression is the silencing of gene coding for Gly m Bd 30 K-the immunodominant soyabean allergen, a member of papain superfamily of cysteine proteases [25]. This protein causes gastric distress, atopic dermatitis and anaphylaxis. Authors evidenced complete suppression of Gly m Bd 30 K accumulation using immunoblotting technique with a mixture of IgE antibodies from six soyabean sensitive individuals. More recently Altenbach et al. reported effective silencing of genes coding for ω-5 gliadins in US bread wheat cultivar Butte 86 aiming to decrease a risk of WDEIA [26]. Although the expected effect was not supported by immunological research it is highly probable in the light of relationships between gliadin ω-5 fractions and WDEIA development. In the present work on wheat genotypes lacking some allergenic fraction of gliadins, were developed using non-molecular, conventional plant breeding methods.
Individual protein fractions, where suppression was observed in the present work, were only a small portion of allergenic gliadin protein complex. The question is, to what extent inhibition of individual allergens may decrease the allergenicity of the whole protein complex. In our research the inhibition of individual proteins decreased allergenicity of gliadin complex from 6% to 18%. This is probably not enough to accept the modified wheat genotypes as hypoallergenic plants. However, inhibited loci suppressed accumulation of only a few protein fractions (composed probably of dozen of monomeric polypeptides) while gliadin complex is comprised of, on average (depending on wheat genotype), while gliadin complex comprised, on average, hundreds of polypeptides. So, in spite of the fact, that individual GDLs are still allergenic, they are important as a natural source for development of hipoallergenic wheat genotypes. The obtained results allow us to expect that a combination of a number of suppressed gliadin genes in one plant genotype may amplify the effects of allergenicity decrease caused by individual null alleles. Cumulate of several null alleles is theoretically possible as they are localized on different chromosomes (inherited independently) and may appear in all possible combinations in hybrid populations as a result of genetic recombinations, according to Mendelian laws. Development of such genotypes will be the object of our further research.
5. Acknowledgements
This work was carried out with financial support from The Polish National Center of Science by research grant No. N N310/1622/38.
REFERENCES
- P. R. Shewry, N. G. Halfornd, A. S. Tatham, Y. Popineau, D. Lafiandra and P. S. Belton, “The High Molecular Weight Subunits of Wheat Glutenin and Their Role in Determining Wheat Processing Properties,” Advances in Food & Nutrition Research, Vol. 45, 2003, pp. 219-302. doi:10.1016/S1043-4526(03)45006-7
- H. Wieser, “Chemistry of Gliadins,” European Journal of Gastroenterology & Hepatology, Vol. 3, No. 2, 1991, pp. 102-107.
- H. Gall, M. Steinert and R. U. Peter, “Exercise-Induced Anaphylaxis to Wheat Flour,” Allergy, Vol. 55, No. 11, 2000, pp. 1096-1097. doi:10.1034/j.1398-9995.2000.00812.x
- A. S. Tatham and P. R. Shewry, “Allergens in Wheat Related Cereals,” Clinical & Experimental Allergy, Vol. 38, No. 11, 2008, pp. 1712-1726.
- E. V. Metakovsky, “Gliadin Allele Identification in Common Wheat. II. Catalogue of Gliadin Alleles in Common Wheat,” Journal of Genetics and Breeding, Vol. 45, 1991, pp. 325-344.
- J. A. Bietz, “HPLC of Cereal Endosperm Storage Proteins,” In: K. M. Gooding and F. E. Regnier, Eds., HPLC of Biological Macromolecules, 2nd Edition, Marcel Dekker, Inc., New York, 2002, pp. 547-587.
- P. I. Payne, “Genetics of Wheat Storage Proteins and the Effect of Allelic Variation on Bread-Making Quality,” Annual Review of Plant Physiology, Vol. 38, 1987, pp. 141-152. doi:10.1146/annurev.pp.38.060187.001041
- S. M. Gilbert, N. Wallner, P. S. Belton, J. A. Greenfield, G. Siligardi, P. R. Shewry and A. S. Tatham, “Expression and Characterisation of a Highly Repetitive Peptide Derived from Wheat Seed Storage Proteins,” Biochimica et Biophysica Acta, Vol. 1479, No. 1-2, 2000, pp. 135-146. doi:10.1016/S0167-4838(00)00059-5
- A. A. Sozinov and F. A. Poperella, “Genetic Classification of Prolamins and Its Use for Plant Breeding,” Annales de Technologie Agricole, Vol. 28, 1980, pp. 229- 245.
- A. De Bustos, P. Rubio and N. Jouve. “Molecular Characterization of the Inactive Allele of the Gene Glu A1 and the Development of a Set of AS-PCR Markers for HMW Glutenins of Wheat,” Theoretical and Applied Genetics, Vol. 100, No. 7, 2000, pp. 1085-1094. doi:10.1007/s001220051390
- O. D. Anderson and F. C. Greene, “The α-Gliadin Gene Family. II. DNA and Protein Sequence Variation, Subfamily Structure and Origins of Pseudogenes,” Theoretical and Applied Genetics, Vol. 95, No. 1-2, 1997, pp. 59- 65. doi:10.1007/s001220050532
- G. Branlard and M. Dardevet, “A null Gli-D1 Allele with a Positive Effect on Bread Wheat Quality,” Journal of Cereal Science, Vol. 20, No. 3, 1994, pp. 235-244. doi:10.1006/jcrs.1994.1063
- W. J. Rogers, E. J. Sayers and K. L. Ru, “Deficiency of Individual High Molecular Glutenin Subunits Affords Flexibility in Breeding Strategies for Bread-Making Quality in Wheat Triticum aestivum L,” Euphitica, Vol. 117, No. 2, 2001, pp. 99-109. doi:10.1023/A:1004152027155
- S. Węgrzyn and J. Waga, “Relationships between the High Molecular Weight Glutenin Subunits and Variation of the Important Useful Traits of Winter Wheat Cultivars and Strains,” Biuletyn—Instytutu Hodowli i Aklimatyzacji Roslin, Vol. 211, 1999, pp. 55-69.
- E. Witkowski, J. Waga, K. Witkowska, M. Rapacz, M. Gut, A. Bielawska, H. Luber and A. J. Lukaszewski, “Association between Frost Tolerance and the Allele of High Molecular Weight Glutenin Subunits Present in Polish Winter Wheats,” Euphytica, Vol. 159, No. 3, 2008, pp. 377-384. doi:10.1007/s10681-007-9537-9
- J. Waga, “Catalogue of Gliadin Protein Blocks Occurring in Polish Common Wheat Cultivars and Strains (Triticum aestivum L.),” Biuletyn—Instytutu Hodowli i Aklimatyzacji Roslin, Vol. 243, 2007, pp. 3-23.
- J. Waga, J. Zientarski, M. Szaleniec and A. Skoczowski, “Efficiency of Poroshell Type Chromatographic Columns for Wheat Gliadin Separation Using RP-HPLC,” Biuletyn—Instytutu Hodowli i Aklimatyzacji Roslin, Vol. 260- 261, 2011, pp. 5-20.
- J. Waga, J. Zientarski, K. Obtułowicz and B. Bilo, “Flour Quality and Binding of Immunoglobulin E by Gliadin Proteins of Two Winter Wheat Genotypes,” Polish Journal of Food And Nutrition Sciences, Vol. 15, No. 56, 2006, pp. 305-310.
- G. L. Lookhart, S. R. Bean and J. A. Bietz, “HPLC of Gluten Monomeric Proteins,” In: P. R. Shewry and G. L. Lookhart, Eds., Wheat Gluten Protein Analysis, American Association of Cereal Chemists, St. Paul, 2003, pp. 91- 111.
- H. Wieser and P. Koehler, “The Biochemical Basis of Celiac Disease,” Cereal Chemistry, Vol. 85, No. 1, 2008, pp. 1-13. doi:10.1094/CCHEM-85-1-0001
- I. Bouchez-Mahiout, J. Snegaroff, M. Tylichova, C. Pecquet, G. Branlard and M. Lauriere, “Low Molecular Weight Glutennins in Wheat Dependant, Exercise-Induced Anaphylaxis: Allegenicity and Antigenic Relationships with Omega 5-Gliadins,” International Archives of Allergy and Immunology, Vol. 153, No. 1, 2010, pp. 35-45. doi:10.1159/000301577
- K. Palosuo, E. Varionen, O. M. Kekki, T. Klemola, N. Kalkkinen, H. Alenius and T. Reunala, “Wheat ω-5 Gliadin Is a Major Allergen in Children with Immediate Allergy to Ingested Wheat,” Journal of Allergy and Clinical Immunology, Vol. 108, No. 4, 2001, pp. 634-638. doi:10.1067/mai.2001.118602
- J. Waga, K. Obtułowicz, J. Zientarski, E. Czarnobilska and A. Skoczowski, “Purified Wheat Gliadin Proteins as Immunoglobulin E Binding Factors in Wheat Mediated Allergies,” American Journal of Plant Sciences, Vol. 2, No. 3, 2011, pp. 476-483. doi:10.4236/ajps.2011.23056
- Y. Tada, M. Nakase, T. Adachi, R. Nakamura, H. Shimada, M. Takahashi, T. Fujimura and T. Matsuda, “Reduction of 14-16 kDa Allergenic Proteins in Transgenic rice Plants by Antisense Gene,” FEBS Letters, Vol. 391, No. 3, 1996, pp. 341-345. doi:10.1016/0014-5793(96)00773-9
- E. M. Herman, R. M. Helm, R. Jung and A. J. Kinney, “Genetic Modification Removes an Immunodominant Allergen from Soybean,” Plant Physiology, Vol. 132, No. 1, 2003, pp. 36-43. doi:10.1104/pp.103.021865
- S. B. Altenbach and P. V. Allen, “Transformation of the US Bread Wheat ‘Butte 86’ and Silencing of Omega-5 Gliadin Genes,” GM Crops, Vol. 2, No. 1, 2011, pp. 66- 73. doi:10.4161/gmcr.2.1.15884
NOTES
*Corresponding author.

