American Journal of Plant Sciences
Vol.5 No.18(2014), Article
ID:49156,9
pages
DOI:10.4236/ajps.2014.518293
Transcript Level Analysis of Lignin and Flavonoid Biosynthesis Related Genes in Eucalyptus globulus
Tomotaka Shinya, Kazunori Hayashi, Shinichi Onogi, Akiyoshi Kawaoka*
Agri-Biotechnology Research Laboratory, Nippon Paper Industries Co. Ltd., Tokyo, Japan
Email: *akawaoka@nipponpapergroup.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 18 June 2014; revised 19 July 2014; accepted 12 August 2014
ABSTRACT
We have investigated a correlation of transcript abundances of key genes that influence the quality of wood and flavonoid biosynthesis, such as the two p-hydroxycinnamoyl-CoA:quinate shikimate p-hydroxycinnamoyl transferase (HCT) and the two chalcone synthases (CHS) from Eucalyptus globulus grown in a greenhouse. The EglHCT1 and EglHCT2 transcripts accumulated in stems of all ages, but to a lesser extent in leaves. On the other hand, EglCHS3 and EglCHS4 exhibited high transcript levels in leaves, roots and shoots, but low levels in the stem. A positive correlation (R2 > 0.70) was observed between the transcript levels of the EglHCT1, EglHCT2 genes and Klason lignin (KL) content. In addition, the sum of transcript levels of EglHCT1 and EglHCT2 genes were highly correlated to KL contents (R2 > 0.85). However, there is no relationship between transcript levels of two CHS genes and, KL or flavonoid contents. This may imply that lignin biosynthesis and flavonoid biosynthesis are independently regulated in E. globulus.
Keywords:HCT, CHS, 4CL, Transcript Level, Lignin Content, Eucalyptus globulus

1. Introduction
Eucalyptus species is the main hardwood used in intensively managed forest plantations in the temperate and subtropical regions of the world. Their wood is used as a raw material for the production of cellulose or energy source. Eucalyptus species have fast growth rates and ability to adapt to a broad range of geographic locations. E. globulus is one of the most important commercial temperate hardwoods for the pulp and paper industries because of its wood properties and pulp production characteristics.
Woody plant cell wall is mainly composed of cellulose, hemicellulose and lignin. For kraft pulp making, wood with higher contents of cellulose and hemicellulose is preferred. On the other hand, for the purpose of combustible energy production, higher lignin content is ideal, because lignin has relatively high calorie as compared with polysaccharides [1] . Therefore, understanding the mechanism of regulation of the individual components of wood is of great commercial importance. Many structural and regulatory genes involved in cell wall formation have been reported [2] . Xylem transcription profiles obtained using novel sequencing technologies in three Eucalyptus species revealed many genes for wood formation [3] . Most structural genes involved in lignin biosynthesis have been identified in many species, including in woody plants [4] . The aromatic lignin polymers commonly found in woody plants are primarily composed of two monolignols, namely coniferyl and sinapyl alcohols, which form guaiacyl (G) and syringyl (S) lignin when polymerized. These monolignols are synthesized via the phenylpropanoid pathway, which begins with the deamination of phenylalanine to form cinnamate, followed by a series of ring hydroxylations, O-methylations, and side-chain modifications. Lignin found in gymnosperms and ferns generally lack the S units [4] , suggesting that the branch leading to sinapyl alcohol biosynthesis may be a relatively recent addition to the angiosperms’ biochemical repertoire. Our understanding of the monolignol biosynthetic pathway has undergone multiple major revisions over the past decade due to the results generated from both in vitro kinetic studies on lignin biosynthetic enzymes and from genetic studies on mutants and transgenic plants with altered expression levels of phenylpropanoid pathway genes [4] [5] . Current thinking suggests that ten enzymes are required for monolignol biosynthesis. In the cell wall, peroxidases and laccases are believed to catalyze the dehydrogenative polymerization of the monolignols [4] ; however, the exact roles of these enzymes in the regulation of lignin biosynthesis have not been defined.
The enzyme p-hydroxycinnamoyl-CoA:quinate shikimate p-hydroxycinnamoyl transferase (HCT) catalyzes the transfer of p-coumaroyl CoA and caffeoyl CoA to shikimate and quinate for the biosynthesis of the corresponding shikimate and quinate esters. These enzyme reactions may be the entry points for the biosynthesis of methoxylated phenylpropanoids. Tobacco and alfalfa plants with downregulation of HCT show reduced lignin content and increased p-hydroxyphenyl units in lignin [6] [7] . Defects in phenylpropanoid biosynthesis arising from deficiency in HCT lead to reduced lignin and hyperaccumulation of flavonoids in Arabidopsis thaliana [8] . The biosynthetic pathways leading to lignin and flavonoids diverge at the common intermediate p-coumaroyl CoA (Figure 1). On the other end of the biosynthesis pathway, the catalysis of the enzyme chalcone synthase (CHS) serves as the initial step for flavonoid biosynthesis. Flavonoids are important plant secondary metabolites that serve various functions in higher plants and are considered natural regulators of cellular auxin efflux and consequent auxin polar transport [9] . Auxin is an important hormone for wood formation [10] .
In this study, we focused on the two key genes HCT and CHS, and investigated the correlation between their expression and lignin and flavonoid biosynthesis. Our results show that transcript levels of two HCT genes were correlated to Klason lignin content in E. globulus grown in a greenhouse.
2. Materials and Methods
2.1. Plant Material
E. globulus plants were selected on the plantation of Bunbury Treefarm Project, Nippon Paper Resources,
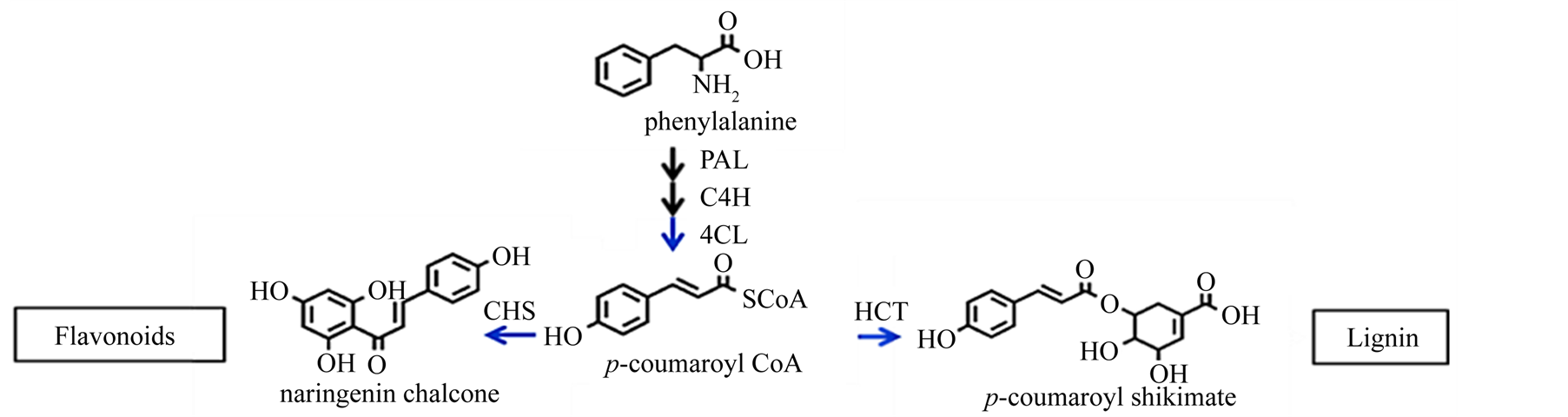
Figure 1. Junction part of monolignol biosynthesis and flavonoid biosynthesis pathway. p-coumaroyl CoA is a junction compound of metabolic routes leading either to monolignols or to flavonoids and sinapoylmalate. CHS and HCT activities control the metabolic flux entering the two routes.
Western Australia. The clonal E. globulus plants were produced by a photoautotropic culture method [11] and were grown in a greenhouse using culture pots (45 × 45 cm) under natural day length.
2.2. Isolation of HCT and CHS Genes
A cDNA library was constructed using mRNA purified from stems of four-month-old E. globulus grown in the greenhouse. From the Database of EucgenIE (http://eucgenie.org/) two orthologous HCTs gene (EglHCT1: scaffold_10:38338422-38343186, EglHCT2: scaffold_6:48055422-48058060) and two orthologous CHSs genes (EglCHS3: scaffold_8:40814969-40817241, EglCHS4: scaffold_8:56889409-56890937) were focused on because these gene transcripts existed in the xylem tissue. The cDNAs were isolated by PCR according to the nucleotide sequence information from E. grandis. The amplified DNA fragments were sequenced using the sequencer XL2000 (Beckmann, Fullerton, CA, USA).
2.3. RT-qPCR Analysis
RT-qPCR expression analysis was carried out on cDNA synthesized from total RNA from each organ. Total RNAs were extracted from shoots, young stems at 15 - 20 cm from top, middle stems at about 50 cm from top, base stems at 10 cm from bottom, leaves, and roots of E. globulus plants grown in a greenhouse [12] . Primers for RT-qPCR (Table 1) were designed with Primer 3 (http://frodo.wi.mit.edu/primer3/). Cycling conditions were 5 min of denaturation at 95˚C followed by 40 cycles of 95˚C, denaturation for 15 s, 60˚C annealing for 15 s and 72˚C elongation for 20 s. Following amplification, a dissociation stage was carried out to detect any complex products. Data analysis was performed with RQ manager software (Applied Biosystems, Foster city, CA, USA) and transcript abundance was determined relative to the E. globulus ubiquitin gene as an internal standard.
2.4. Cell Wall Composition Analysis
Lignin determination was performed on the dried insoluble cell wall residues (CWR) of samples Soxhlet extracted with toluene/ethanol, ethanol and water. Klason lignin was measured by the method of Effland [13] .
2.5. Total Phenolic Content and Total Flavonoid Content Determination
Basal parts of stem were air-dried, pulverized to a coarse powder in a mechanical grinder, passed through 40 mesh sieve and extracted with 80% (v/v) methanol for one month at room temperature. Total phenolic contents of the extracts were determined by Folin-Ciocalteu assay [14] . Three replicates were made for each test sample and gallic acid was used as a standard. Total flavonoid content was determined by the aluminum calorimetric method, using quercetin as a standard [15] . Three replicates were made for each test sample.
2.6. Statistics Analysis
Data analysis was performed using Microsoft Excel statistical tools.
3. Results
3.1. Organ-Specific Expression of HCTs and CHSs
Because the two genes, HCT and CHS, are involved in two different biosynthetic pathways, we first wanted to evaluate if their expressions were organ specific. We isolated cDNAs of the two HCT and two CHS genes from
E. globulus. The transcript levels of EglHCT1 and EglHCT2 in each organ were assayed by Real Time-quantitative PCR (RT-qPCR). The EglHCT1 mRNA levels were high in the middle and basal parts of stems and in roots (Figure 2(a)). High expression of EglHCT2 mRNA levels was also observed in the middle and basal parts of stems, but low in the roots (Figure 2(a)). The expression level of both genes was negligible in the leaves and young stem (Figure 2(a)). In contrast, EglCHS3 and EglCHS4 displayed high transcript levels in shooting buds, leaves, roots and young stem, but low levels in the middle and base of stems (Figure 2(b)).
3.2. Relative Transcript Levels of HCTs and CHSs in Ten Different E. globulus Lines
We then wanted to see the relative expression levels of the different genes involved in the lignin and flavonoid biosynthetic pathway. Transcript levels of HCTs and CHSs were measured in ten different E. globulus lines. Total RNAs were extracted from basal parts of the stems of 6-month-old plants (about 1 m high) and RT-qPCR was carried out. Relative expression levels of Egl4CL1 (the data were from previous work; [16] , EglHCT1, EglHCT2, EglCHS3 and EglCHS4 in ten different lines represented as A-J are shown in Figure 3. The transcript
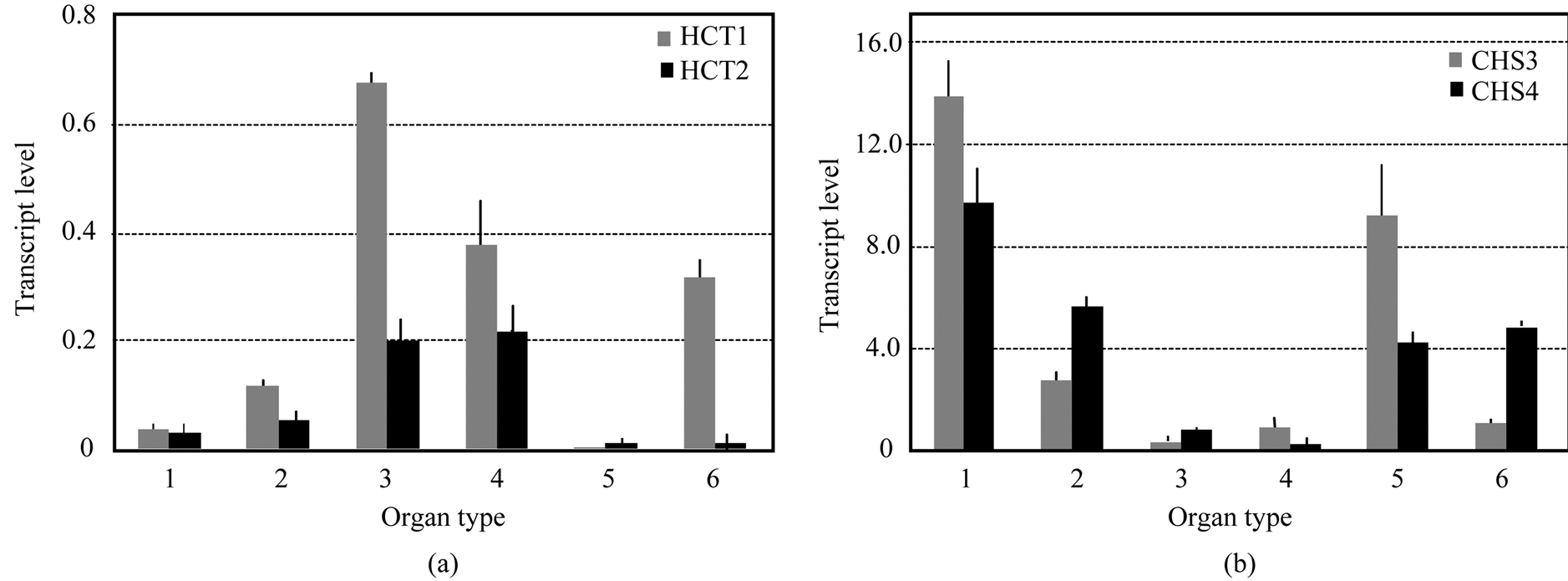
Figure 2. Characterization of the HCT and CHS genes in E. globulus. (a) Organ specific transcript abundances of EgHCT1 and 2; (b) Organ specific transcript abundances of EglCHS3 and 4. Each blot was replicated at least three times with fresh plant extracts. Total RNAs were extracted from different organs viz. shooting bud (1), young stem (2), middle stem (3), base stem (4), leaf (5) and root (6). Ubiquitin (EglUBI) gene was used as an internal control. Each blot was replicated at least three times with fresh plant extracts.
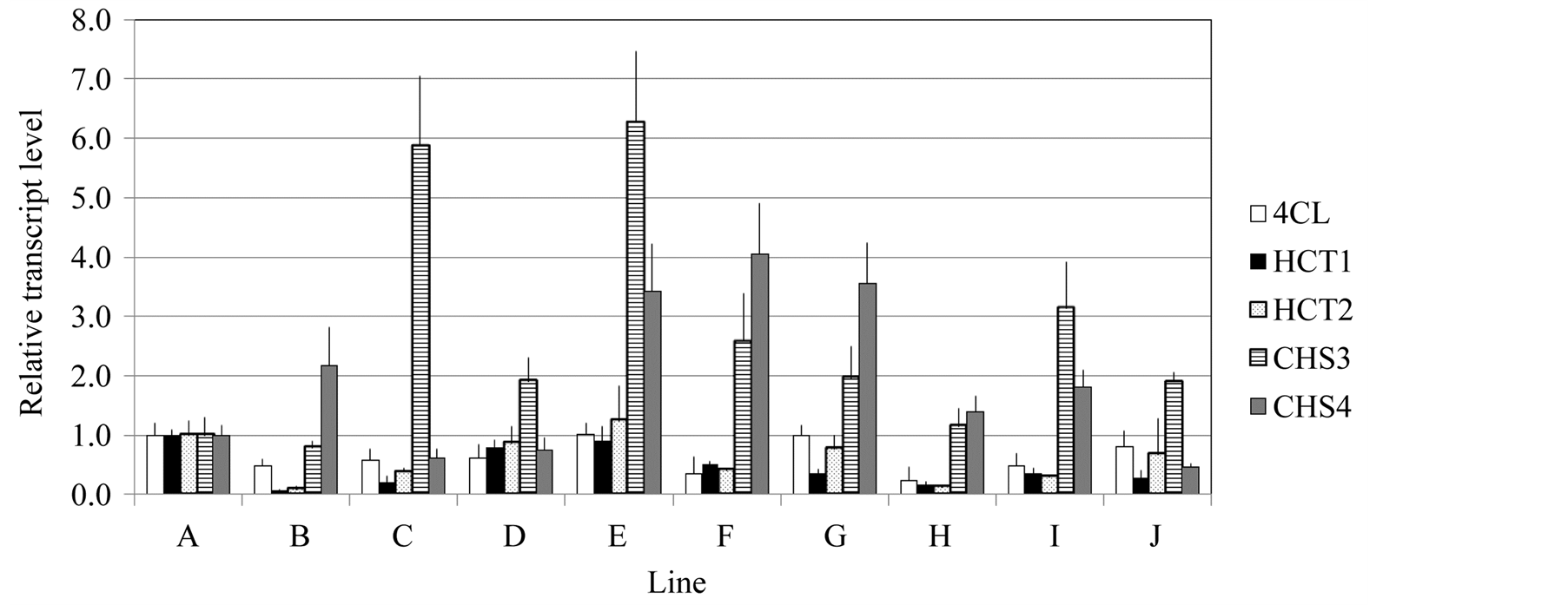
Figure 3. Relative expression levels of the 4CL1, HCT1, HCT2, CHS3 and CHS4 genes. Total RNA was extracted from stems of ten independent lines and RT-qPCR was carried out using gene-specific primers. All transcript levels were indicated as 1.0 of transcript level each gene in line A. The Eucalyptus ubiquitin gene was used as an internal control. Each data point represents the average of three independent replicates and the error bars represent standard error values.
levels of EglHCT1and EglHCT2 were similar to the levels of Egl4CL1, respectively across all lines tested (Figure 3). The transcript levels of EglCHS3 and EglCHS4 were highly variable across the lines (Figure 3).
3.3. Correlation between Lignin Content and Transcript Levels of HCTs and CHSs
Next, we investigated if there was a correlation between the expression levels of these enzymes and the wood quality of the plant, as measured by the Klason lignin (KL) content. We measured KL content in 6-month-old plants of ten different E. globulus lines and investigated if there is a relationship between the expression levels of the two EglHCT genes and KL content. On an average, the KL content in the plants ranged from 21.7% - 25.6%. A highly positive correlation was observed between KL content and the expression levels of the EglHCT1 (R2 = 0.70) and EglHCT2 (R2 = 0.76). The sum of transcript levels of EglHCT1 and EglHCT2 was also highly correlated to KL content (R2 = 0.86) (Figure 4). However, no correlation was observed between transcript levels of two CHS genes and KL content (Figure 5(a) and Figure 5(b)). This may imply that lignin biosynthesis and flavonoid biosynthesis are independently regulated in E. globulus of younger age.
3.4. Correlation between Flavonoid, Total Phenolic Contents and Transcript Levels of CHSs
Next, we examined a possible correlation between the expression of CHS and the flavonoid and total phenolic content. p-coumaroyl CoA is the common substrate for the two enzymes, CHS and HCT in the monolignol and flavonoid biosynthesis pathway (Figure 1). CHS catalyzes the formation of the flavonoid skeleton by condensation of p-coumaroyl CoA with three malonyl CoA molecules. We investigated if there was a correlation between transcript levels of EglCHS3 and EglCHS4 and flavonoid and total phenolic contents. As mentioned above, total RNAs of basal part of stem in ten Eucalyptus lines were extracted and RT-qPCR was carried out. Simultaneously, wood samples from the same areas were taken and analyzed for flavonoid and total phenolic contents. A moderate correlation was observed between CHS transcription and flavonoid content (Figure 5(g) and Figure 5(h)) and weak to no correlation was obtained between CHS transcription and phenolic content (Figure 5(d) and Figure 5(e)). All comparisons of transcript levels between individual genes and the KL content as well as that of flavonoid and phenolic content yielded very low coefficients of determination (R2 < 0.2). We compared the sum of transcript levels of EglCHS3 and EglCHS4, and flavonoid content (Figure 5(f)) or total phenolic content (Figure 5(i)), but did not observe any correlation (R2 < 0.2).
4. Discussion
In this study, we investigated the possible correlation between expression patterns of certain key genes and wood properties, such as lignin content, flavonoid and polyphenol contents in E. globulus. We focused on five genes, Egl4CL1, EglHCT1, Egl4HCT2, EglCHS3 and EglCHS4, which are known to be involved in lignin and flavonoid pathways. First, we compared the transcript levels of EglHCT1, Egl4HCT2, EglCHS3 and EglCHS4 across various parts of the plants, such as shoot apex, top, middle and basal parts of stems, leaves and roots. EglHCT1 and Egl4HCT2 showed robust expression in the basal and mid parts of the stem (Figure 2). This strong expression in the middle and lower parts of the stem is in accordance with the notion that HCT is the specific gene for lignin biosynthesis [2] . The reason for transcript abundance of EglHCT1 in roots and the differential expression between the two HCT genes is unclear at this time. Further research on the implications of this differential expression could reveal more insight into this.
In contrast, both CHS genes were expressed mainly in the shooting bud, upper stem and leaves. This could be explained by the fact that the CHS genes are involved in flavonoid biosynthesis. Flavonoids are important plant secondary metabolites that serve various functions in higher plants. These include pigmentation, UV protection, fertility, antimicrobial defense and the recruitment of nitrogen-fixing bacteria [17] . We were unable to find any correlation between the transcript abundances of HCTs and CHSs between different organs (Figure 2) and in lower stems of the ten different lines (Figure 3). These results suggest that HCT and CHS gene expressions are independently regulated. Such a dichotomy in regulation of lignin and flavonoid biosynthesis has been reported in Arabidopsis, where HCT-RNA interference (RNAi) was found to repress lignin biosynthesis, but did not affect flavonoid production [9] . The expression patterns of 4CL and HCTs were similar in the ten E. globulus lines (Figure 3).
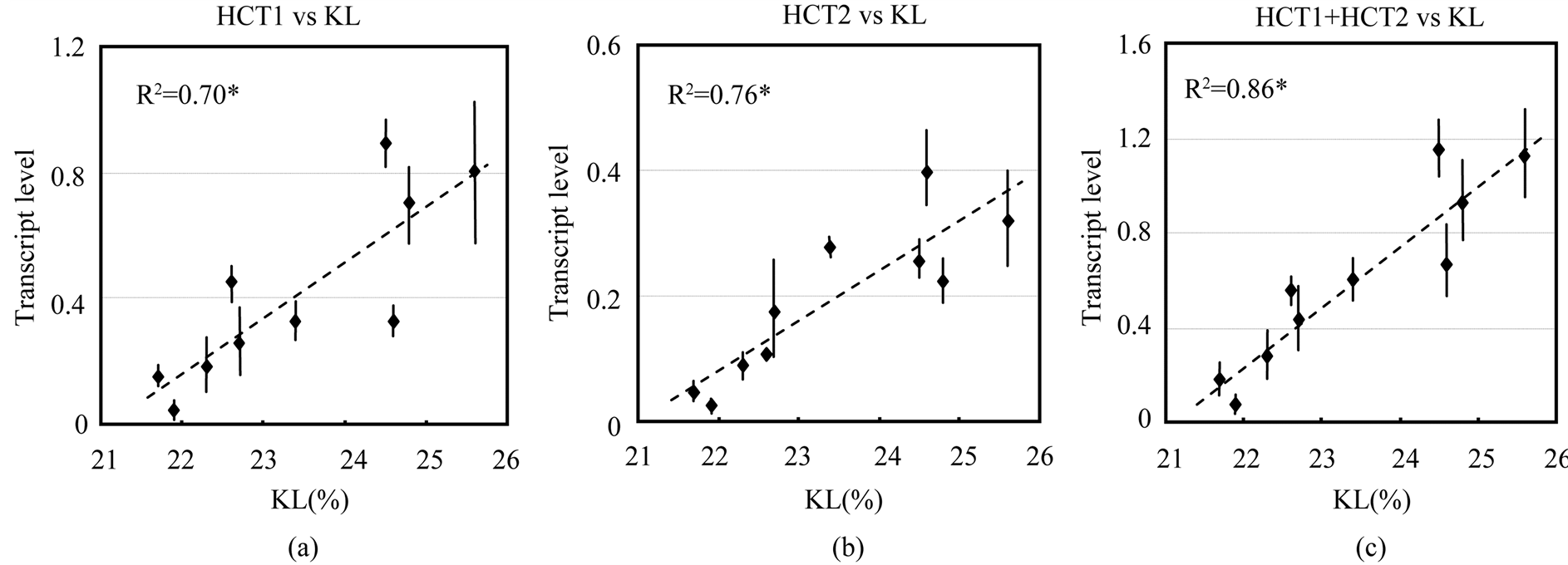
Figure 4. Relationship between transcript levels and wood quality parameters (KL content). Scatter plots showing transcript levels relative to ubiquitin as determined by RT-qPCR. (a) HCT1 transcript level vs. KL; (b) HCT2 transcript level vs. KL; (c) sum of transcript levels of HCT1 and HCT2 vs. KL. Error bars represent standard error values. *P < 0.01.
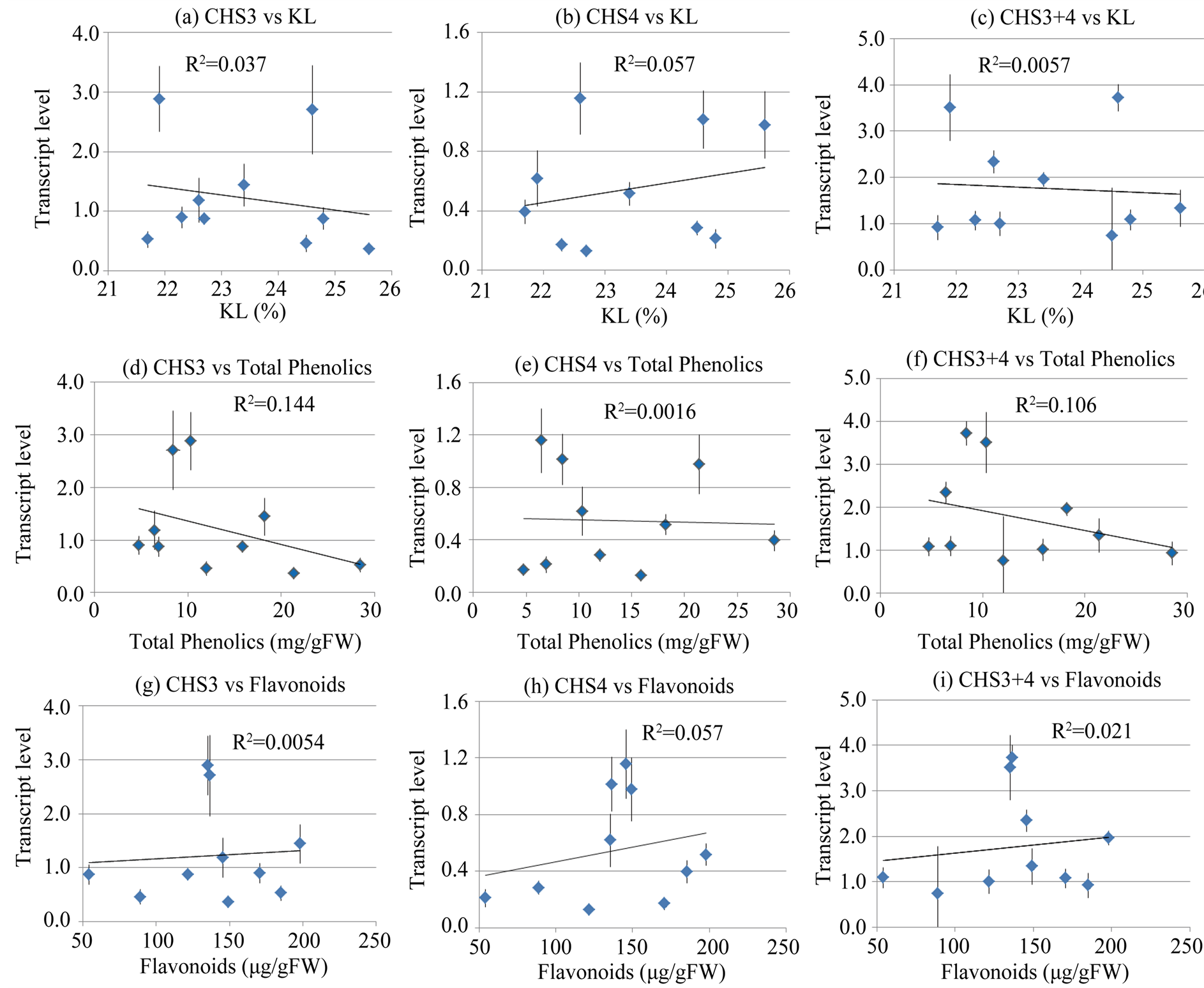
Figure 5. Correlation between CHS3, CHS4 transcript levels and KL, total phenolics or flavonoids. (a)-(c) Relationship between CHS3 (a), CHS4 (b) and CHS3 + 4 (c) transcript levels and KL content; (d)-(e) Relationship between CHS3 (d) CHS4 (e) and CHS3 + 4 (f) transcript levels and total phenolics; (g)-(i) Relationship between CHS3 (g), CHS4 (h) and CHS3 + 4 (i) transcript levels and flavonoids. Each data point represents the average of three replicates. P > 0.05 in every case (a)-(i).
We also observed a strong positive correlation between the expression of the individual HCT genes and the KL content (Figure 4). This result concurs with our previous report that HCT plays a role in the determination of lignin content as well as 4CL [16] . The combined coefficient of determination value (R2) of HCT1 and HCT2 in that study was 0.84 and this value was higher than the values determined for the association between the individual HCT gene and the percentage of KL values or 4CL. These two HCT genes are generally coexpressed in lignified tissue of stems. HCT is the most important enzyme in lignin biosynthesis pathway as it channels the phenylpropanoids towards the biosynthesis of the methoxylated monolignols, coniferyl alcohol and sinapyl alcohol, as shown in Arabidopsis [6] . The strong positive correlation we observed between the expression of these HCT genes and the lignin content in the plant emphasizes the regulatory roles of these genes in the monolignol pathway.
The transcript levels of the CHS gene, on the other hand, did not exhibit a strong correlation with flavonoid content, even though we used woody plants grown in a greenhouse (Figure 5). These results suggest that EglCHS3 and EglCHS4 are not the main regulatory enzymes of flavonoid biosynthesis. CHS gene is shown to be constitutively expressed in parsley and its expression is known to be facilitated by light, pathogens, elicitors and wounding [18] . CHS is known as the key entry enzyme committed to the production of the polyketide phenylpropanoids in plants. It seems that all plants contain at least one CHS gene and often CHS gene families exhibit different expression patterns in plants. E. gransdis has 13 CHS genes in the EucagenIE data base. Among them, two ortholog genes were expressed in xylem tissue. CHS catalyzes the first step in flavonoid biosynthesis from p-coumaroyl CoA. These flavonoids function as sunscreen, protecting against UV radiation and they also play a role in auxin transport regulation [19] .
Industrial interests provide a strong incentive for research on wood formation. Here, we described how two structural genes involved in lignin biosynthesis (HCTs) affect lignin content in E. globulus. These results suggest a strategy to increase the final fiber yield. Cellulose fiber production by kraft pulping process requires the removal of lignin through sodium hydroxide and sodium sulfide treatment. Thus, woody plants with low lignin contents and high cellulose content are desirable for efficient cellulose fiber production. Eucalyptus plantation and breeding companies are working on effective selection of superior trees that have higher growth rates, basic density and cellulose fiber yield. Our results indicate that lower transcript abundances of the HCT genes, could provide useful selection markers for lower lignin contents. In this study, we used greenhouse-grown, younger aged Eucalyptus plants that might not be affected by environmental factors. However, it has been reported that there are phenotypic differences between woody plants that are field-grown and greenhouse-grown. Voelker et al. [20] found physiological abnormalities in field-grown, low-lignin poplars. In the present study, we investigated correlations between the gene transcript abundances and wood qualities using woody plants grown in a greenhouse, where plants are mostly shielded from environmental stress. It would be interesting to see if similar results are obtained in the field. Finally, our results provide a strong dataset for selecting elite candidate trees by monitoring transcript levels of several useful genes such as 4CL [16] and HCTs.
Acknowledgements
We thank Toshiaki Takano of Kyoto University for technical support, and Kenji Yoshinaga, Phillip Uren of Nippon Paper Resources for collection of samples. This research was supported by Nippon Paper Industries Co. LTD.
References
- White, R.H. (1986) Effect of Lignin Content and Extractives on the Higher Heating Value of Wood. Wood and Fiber Science, 19, 446-452.
- Vonholme, R., Morreel, K., Ralph, J. and Boerjan, W. (2008) Lignin Engineering. Current Opinion in Plant Biology, 11, 278-285. http://dx.doi.org/10.1016/j.pbi.2008.03.005
- Salazar, M.M., Nascimento, L.C., Camargo, E.L.O., Danieli Cristina Gonçalves, D.C., Neto, J.L., Marques, W.L. et al. (2013) Xylem Transcription Profiles Indicate Potential Metabolic Responses for Economically Relevant Characteristics of Eucalyptus Species. BMC Genomics, 14, 201-214. http://dx.doi.org/10.1186/1471-2164-14-201
- Boerjan, W., Ralph, J. and Baucher, M. (2003) Lignin Biosynthesis. Annual Review of Plant Biology, 54, 519-546. http://dx.doi.org/10.1146/annurev.arplant.54.031902.134938
- Humphreys, J.M. and Chapple, C. (2002) Rewriting the Lignin Roadmap. Current Opinion in Plant Biology, 5, 224-229. http://dx.doi.org/10.1016/S1369-5266(02)00257-1
- Hoffmann, L., Besseau, S., Geoffroy, P., Ritzenthaler, C., Meyer, D., Lapierre, C., Pollet, B. and Legrand, M. (2004) Silencing of Hydroxycinnamoyl-Coenzyme A Shikimate/Quinate Hydroxycinnamoyltransferase Affects Phenylpropanoid Biosynthesis. Plant Cell, 16, 1446-1465. http://dx.doi.org/10.1105/tpc.020297
- Chen, F., Srinivasa Reddy, M.S., Temple, S., Jackson, L., Shadle, G. and Dixon, R.A. (2006) Multi-Site Genetic Modulation of Monolignol Biosynthesis Suggests New Routes for Formation of Syringyl Lignin and Wall-Bound Ferulic Acid in Alfalfa (Medicago sativa L.). The Plant Journal, 48, 113-124. http://dx.doi.org/10.1111/j.1365-313X.2006.02857.x
- Xu, L., Bonawitz, N.D., Weng, J.-K. and Chappel, C. (2010) The Growth Reduction Associated with Repressed Lignin Biosynthesis in Arabidopsis thaliana Is Independent of Flavonoids. Plant Cell, 22, 1620-1632. http://dx.doi.org/10.1105/tpc.110.074161
- Besseau, S., Hoffmann, L., Geoffroy, P., Lapierre, C., Pollet, B. and Legrand, M. (2007) Flavonoid Accumulation in Arabidopsis Repressed in Lignin Synthesis Affects Auxin Transport and Plant Growth. Plant Cell, 19, 148-162. http://dx.doi.org/10.1105/tpc.106.044495
- Nilsson, J., Karlberg, A., Antti, H., Lopez-Vernaza, M., Mellerowicz, E., Perrot-Rechenmann, C., Sandberg, G. and Bhalerao, R.P. (2008) Dissecting the Molecular Basis of the Regulation of Wood Formation by Auxin in Hybrid Aspen. Plant Cell, 20, 843-855. http://dx.doi.org/10.1105/tpc.107.055798
- Nagae, S., Takamura, T., Tanabe, T., Murakami, A., Murakami, K. and Tanaka, M. (1996) In Vitro Shoot Development of Eucalyptus citriodora on Rockwool in the Film Culture Vessel under CO2 Enrichment. Journal of Forest Research, 1, 227-230. http://dx.doi.org/10.1007/BF02348330
- Chomczynski, P. and Sacchi, N. (1987) Single-Step Method of RNA Isolation by Acid Guanidiniumthiocyanate-phenol-chloroform Extraction. Analytical Biochemistry, 162, 156-159. http://dx.doi.org/10.1016/0003-2697(87)90021-2
- Effland, M.J. (1977) Modified Procedure to Determine Acid Insoluble Lignin in Wood and Pulp. Journal of Korea Technical Association of the Pulp and Paper Industry, 60, 143-144.
- Meda, A., Lamien, C.E., Romio, M., Millogo, J. and Nacoulma, O.G. (2005) Determination of the Total Phenolic, Flavonoid and Praline Contents in Burkina Fusan Honey, as Well as Their Radical Scavenging Activity. Food Chemistry, 91, 571-577. http://dx.doi.org/10.1016/j.foodchem.2004.10.006
- Quettier-Deleu, C., Gressier, B., Vasseur, J., Dine, T., Brunet, C., Luyckx, M., et al. (2000) Phenolic Compounds and Antioxidant Activities of Buckwheat (Fagopyrum esculentum Moench) Hulls and Flour. Journal of Ethnopharmacology, 72, 35-42. http://dx.doi.org/10.1016/S0378-8741(00)00196-3
- Negishi, N., Nanto, K., Hayashi, K., Onogi, S. and Kawaoka, A. (2011) Transcript Abundances of LIM Transcription Factor, 4CL, Cald5H and CesAs Affect Wood Properties in Eucalyptus globulus. Silvae Genetica, 60, 288-296.
- Cain, C.C., Saslowsky, D.E., Walker, R.A. and Shirley, B.W. (1997) Expression of Chalcone Synthase and Chalconeisomerase Proteins in Arabidopsis Seedlings. Plant Molecular Biology, 35, 377-381. http://dx.doi.org/10.1023/A:1005846620791
- Schulze, L.P., Becker, A.M., Schulr, W., Hahlbrock, K. and Dangl, J.L. (1989) Functional Architecture of the Light-Responsive Chalcone Synthase Promoter from Parsley. The Plant Cell, 1, 707-714.http://dx.doi.org/10.1105/tpc.1.7.707
- Brown, D.E., Rashotte, A.M., Murphy, A.S., Normanley, J., Tague, B.W., Peer, W., Taiz, L. and Muday, G.K. (2001) Flavonoids Act as Negative Regulators of Auxin Transport in Vivo in Arabidopsis. Plant Physiology, 126, 524-535. http://dx.doi.org/10.1104/pp.126.2.524
- Voelker, S.L., Lachenbruch, B., Meinzer, F.C., Jourdes, M., Ki, C., Patteen, A.M., et al. (2010) Antisense Down-Regulation of 4CL Expression Alters Lignification, Tree Growth, and Saccharification Potential of Field-Grown Poplar. Plant Physiology, 154, 874-886. http://dx.doi.org/10.1104/pp.110.159269
Abbreviations
CHS: chalcone synthases;
HCT: p-hydroxycinnamoyl-CoA:quinateshikimate p-hydroxycinnamoyltransferase;
KL: Klason lignin;
R2: coefficient of determination;
RNAi: RNA interference;
NOTES

*Corresponding author.


