American Journal of Plant Sciences
Vol.4 No.2(2013), Article ID:28350,7 pages DOI:10.4236/ajps.2013.42044
Wrinkled 1 (WRI1) Homologs, AP2-Type Transcription Factors Involving Master Regulation of Seed Storage Oil Synthesis in Castor Bean (Ricinus communis L.)
![]()
1Graduate School of Bioresource and Bioenvironmental Sciences, Kyushu University, Fukuoka, Japan; 2Fukuoka Agricultural Research Center, Chikushino, Japan; 3Department of Bioresource Sciences, Faculty of Agriculture, Kyushu University, Fukuoka, Japan.
Email: yuasa@agr.kyushu-u.ac.jp
Received December 29th, 2012; revised January 25th, 2013; accepted January 13th, 2013
Keywords: AP2; Castor Bean; Fatty Acid; Oil Seed; Ricinus communis L.; Transcription Factor; WRI1
ABSTRACT
Among APETALA2 (AP2)-type plant specific transcription factor family, WRINKLED1 (WRI1), has appeared to be a master gene transcriptionally regulating a set of carbon metabolismand fatty acid synthesis (FAS)-related genes responsible for seed specific triacylglycerols (TAGs) storage in oil plants. B3 type transcription factors, such as ABI3 and FUS3, are known to be involved in seed development, such as seed storage protein synthesis and maturation. Based on the recent whole genome sequence data of castor bean (Ricinus communis L.), putative WRI1 homologs (RcWRI1, RcWRI2) specifically expressed in castor bean seed have been identified by comparing organ specific expression profiles among seed development-related transcription factors, seed storage specific genes (Ricin, RcOleosin) and a set of FAS genes including genes for sucrose synthase (RcSUS2), biotin carboxyl carrier protein (a subunit of acetyl-CoA carboxylase, RcBCCP2) and ketoacyl-acyl carrier protein synthase (RcKAS1). Immunoreactive signals with WRI1, FUS3 and ABI5-related polypeptides were also detected in seed specifically, consistent with the expression profiles of seed development-related genes. The WRI1 binding consensus sites, [CnTnG](n)(7)[CG], designated as the AW-box, were found at the promoter region of RcBCCP2 and RcKAS1. Thus, RcWRI1 possibly play a pivotal role in seed specific TAGs storage during seed development by directly activating FAS -related genes.
1. Introduction
The castor bean plant is a tropical shrub that is one of the most important and valued oilseed crops. Castor bean was originated in Africa, but now is broadly cultivated in many tropical and subtropical regions [1]. Triacylglycerols (TAGs) are the major storage compound ranging from 40% to 60% (weight per dry weight) in the caster bean seed. About 90% of the seed TAGs is composed of ricinoleic acid (12-hydroxy-oleate), which is unusual hydroxylated fatty acid highly valued for chemical and pharmacological application [2]. It has been proposed that castor bean has a potential to produce biodiesel, because of its unique properties that the seed has high oil content, toxic storage protein (ricin) confers oil source without competition to food resource and the plant can tolerate to severe environments where food crops are unfavorable [3].
According to several lines of evidence of genetic regulation mechanisms involving seed development and seed storage accumulation biosynthesis [5-7]. Genetic and molecular biological studies using Arabidopsis thaliana have currently unveiled that a set of specific transcription factors are temporarily and spatially expressed during the seed development and its maturation and that those seed specific transcription factors collaborate in regulation of seed specific genes for storage proteins and TAGs synthesis [8,9]. Several genes responsible for A. thaliana mutants involved in the seed development and the early embryonic morphology have been identified, namely ABSCISIC ACID INSENSITIVE 3 (ABI3), FUS CA 3 (FUS3) and LEAFY COTYLDON 1 and 2 (LEC1, LEC2), etc. [10]. ABI3, FUS3 and LEC2 appeared to be key regulators belonging to B3-type transcription factor family, whereas LEC1 is a NFY-B, a HAP3 subunit of the CCAAT-binding transcription factors [7]. It is known that, at the downstream of LEC1, LEC2, ABI3 and FUS3 form a complicated network interacting to each other to promote embryogenesis, seed storage accumulation and establishment of dormancy [11]. Recently, WRINKLED 1 (WRI1) transcription factor has been identified to be an important regulator for a set of carbohydrate metabolism and FAS in maturing Arabidopsis seed [8,12]. TAGs contents in the WRI1 mutant seeds decreases by 80% compared with that in wild type Arabidopsis seeds, while WRI1 mutant seeds accumulate high sucrose [13]. It has been reported that WRI1 accelerates the expression of genes encoding enzymes of glycolysis, fatty acid elongation and the biotin and lipoic acid synthesis reactions [14]. Among LEC2, ABI3 and FUS3, WRI1 appeared to be regulated at the downstream of LEC2 [15]. It has been reported that WRI1 specifically binds to a conserved ciselement motif (AW-box) which are identified in upstream promoter regions of seed specific FAS related gene KAS1, BCCP2 and SUS2 in Arabidopsis [9]. Over expression of the WRI1 homologs in Arabidopsis, Brassica and maize resulted in significant increase of seed TAGs contents [16-18]. Recently, whole genome analysis projects with the next-generation DNA sequencer has unveiled a set of whole genome data in various oil crops (maize, soybean, castor bean, jatropha and palm tree), indicating that the genetic manipulation of oil crops has significant potential for improvement of vegetable oil production in an aspect of breeding [19-23].
Castor bean (Ricinus communis L.) is one of the most profitable oil crops and suitable for research of inductrial oil and drug such as castorol oil and ricinoleic acid, etc. In order to address the key regulating mechanism of TAG biosynthesis in castor bean, we searched for transcriptional factors involved in seed storage synthesis by inspecting the available Castor Bean Genome Database (http://castorbean.jcvi.org) [21,24]. We focused on the expression profiles of transcription factors (3 of WRI1s, 1/2 of B3-type and 1 of bZIP-type) and FAS-related genes in developing seeds. We profiled the expression levels of WRI1 homologs in castor bean organs and identified Rc WRI1 as a putaive master transcription factor regulating seed specific FAS. Immunoblot also indicated that seed specific transcription factors are coexpressed in developing seed with the WRI1-related polypeptides, suggesting that a seed specific transcription factor network composing B3-type transcription factors and WRI1 homolog is conserved in the seed storage oil synthesis mechanism between castor bean and Arabidopsis.
2. Materials and Methods
2.1. Plant Materials
Seeds of castor bean (Ricinus communis L.) were supplied from Matsuura Kampo Co. (Nagoya, Japan). Three to five seeds of castor bean were sown in the pots (16 cm × 20 cm) filled with soil in the Kaizuka field, Kyushu University on 10 April 2011, and the seedlings were thinned to one plant per pot upon germination in order to get uniform plant stand. Chemical compound fertilizer, mamekase (N:P2O5:K2O-3:10:10) (5 g pot-1) and lime (5 g pot-1) were well mixed with the soil before sowing.
2.2. RNA Extraction and Semi-Quantitative RT-PCR of Castor Bean WRI1 Homologs and Fatty Acid Synthesis(FAS)-Related Genes
BLAST search was conducted using WRI1 homologs, AINTEGUMENTA (AtANT) and other AP2-type transcription factors, DREB1A, ERF1, of Arabidopsis thaliana in the castor bean gene index at phytozome v.9.0 in JGI (http://www.phytozome.net/) identified castor bean orthologs. Gene index numbers and/or accession numbers and specific primer sequences of the castor bean genes were shown in Table l.
RNA preparation by the SDS/phenol/LiCl method, cDNA synthesis and PCR were carried out as described previously [25,26] using Rev Tra Ace reverse transcriptase (Invitrogen) and GoTaq kit (Promega) according to the manufacturer’s manuals and the thermal cycler PC-816 (ASTEC, Fukuoka, Japan). Thermal cycle conditions in PCR were as follows; initial denaturing at 94˚C for 2 min, followed by 30 cycles of denaturing at 94˚C for 30 s, annealing at 61˚C for 30 s and extending at 72˚C for 40 s, and then final extending at 72˚C for 5 min. The PCR products were separated in 1.5% agarose gel and visualized with FluorChem (AlphaInnotech, San Leandro, CA, USA) after ethidium bromide staining.
2.3. Preparation of Extracts from Castor Bean Tissues
Protein samples were prepared from various organs with minor modification as described previously [27]. Frozen tissues (2 g) were homogenized on an ice-cooled motor in 6 mL of lysis buffer containing 25 mM Tris-HCl [pH 7.4], 10 mM EDTA-Na, 10% glycerol, 0.1% β-mercaptoethanol, 1 mM PMSF, 5 mM n-aminocaproic acid, 1 mM benzamidine, 1 µg·mL−1 leupeptin and 1% Triton X-100. The resultant homogenates were centrifuged at 10,000 × g for 20 min at 4˚C. Protein concentrations of samples were measured by a Bio Rad protein assay kit (BioRad, Hercules, CA, USA) with bovine serum albumin (BSA) as a standard.
2.4. Raising Anti-WRI1, ABI3 and ABI5-Peptide Specific Antibodies and Immunoblot
To purpose to detect endogenous transcription factors in-
Table 1. Oligo DNA Primers used for RT-PCR analysis.


Numbers of Gene indexes were refrerred for Pytozome ver.9.
volved in seed development, specific peptide epitope sequences were designed as follows; H2N-CVSKYRGVARHHHNGRWEAR -COOH for WRI1, H2N-YHMLESAHSWPHSLFNVASC-COOH for ABI3 and H2N-ENGQLKLALADLERRRKQQC-COOH for ABI5, which appeared to be highly conserved amino acid sequences present in orthologs of WRI1, ABI3 and ABI5 among various higher plants, respectively. The anti peptide specific polyclonal antibodies were raised in rabbits with those synthetic oligopeptide antigens (SigmaAldrich Co., St. Louis, USA) [25]. After SDS-polyacrylamide electrophoresis (SDS-PAGE), separated polypeptides were electrotransferred onto a PVDF membrane at 10 V·cm−1 for 120 min in 25 mM Tris-base containing 0.05% SDS and 20% methanol) and then the PVDF membrane (Millipore) was blocked by incubating in 1 × TBS (25 mM Tris-HCl [pH 7.4], 150 mM NaCl) containing 3% skim milk (TBS-milk) for 60 min. The PVDF membrane was incubated with an appropriate antibody (anti-WRI1 antibody, anti-ABI3 antibody or ABI5 antibody) (one to 3000 dilution [v/v]) in TBS-milk containing 0.05% Tween 20 at room temperature for 2 hours. Subsequently, the PVDF membrane was incubated at room temperature for 60 min in TBS containing horseradish peroxidase (HRP)–conjugated protein G (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) (one to 10,000 dilution [v/v]), and 0.05% Tween 20. The immunodecorated protein bands were visualized by an ECL Plus kit (GE Healthcare Bio-Sciences) and FluorChem Imaging Analyzer.
3. Results
3.1. Phylogenic Analysis of WRI1-Related Transcription Factor Genes
A phylogenetic tree of WRI1 homologs in dicots and monocots, showed that WRI1-related genes can be classified into two subfamilies (Figure 1(a)). The phylogenic tree of WRI1 homologs and AP2 homologs indicates that the castor bean WRI1-related genes formed a distant group from other AP2-related genes including AtERF1, AtDREB1A and RAV1 (Figure 1(a)). The RcWRI1,2,3 and other WRI1 orthologs share a very similar domain structure, composing of two AP2-type domains, as expected. Closely associated to the WRI1 homolog clade, AtANT also contains two AP2 type domains, but appeaerd to be involved in floral development rather than seed storage regulation [28]. RcWRI1 encodes have a 49.3 kDa peptide relatively larger than RcWRI2 (37.3 kDa, 327 aa) and RcWRI3 (36.6 kDa, 330 aa). The predicted amino acid sequences of RcWRI1, RcWRI2 and RcWRI3 have similarities of 49%, 43% and 45%, respecttively, to that of AtWRI1. Comparison among AP2/ERF type transcription familes indicates that the WRI1 orthologs forms an unique subfamily in the amino acid sequence similaruty and the domain structural similarity.
3.2. The Expression Profiles of Seed Development-Related Transcription Factor Genes
A significant signal of RcWRI1 was detected specifically in filling seeds (2, 3.5 cm diameters) and weakly in matured seed, while the expression levels of RcWRI1 were not or marginal in leaf and pod tissues at 32 cycles (Figure 1(b)). RcWRI2 was detected at 35 cycles in seed specifically as like RcWRI1, but the expression levels were weaker than those of RcWRI1, accordingly to the cycle numbers. In contrast, RcWRI3 were detected at the highest level in leaf, and weakly in filling-seeds, but not in pod at 35 cycles (Figure 1(b)). The relative signals of RcABI3 was expressed in filling seed-specific manner as same as RcWRI1 (Figure 1(b)).
3.3. The Expression Profiles of FAS-Related Genes
A metabolic cascade of FAS in oil storage tissue in Arabidopsis seed indicates that sucrose synthase (SUS) in cytosol plays an importa nt role in clevage of sucrose into UDP-glucose and fructose cytosol for subsequent FAS in plastids [14]. Among a set of plastid enzymes involved in FAS, a set of genes includeing biotin carboxyl carrier protein (BCCP) and ketoacyl-acyl carrier protein synthase (KAS) appeared to be regulated transcriptionaly by WRI1 [9]. We focused on castor bean orthologs of SUS2, BCCP2 and KAS1 to examine whether those FAS-related genes and RcWRI1 are coexpressed in developing seed specific manner. Among castor

Figure 1. Wrinkled 1 homologs and a B3-type transcription factor in various organs of castor bean plant. (a) A phylogenic tree of WRI1 homologs and AP2/ERF transcription factors from castor bean and other higher plantswas aligned by ClustalW (UPGMA). The genes used were follows: RcWRI1, 2, 3 (Table 1); AtWRI1, At3g54320; AtDREB1A, At4g25480; AtERF1; At3g23240; AtANT(AINTEGUMENTA), At4g37750; RAV1, At1g13260; Cotton-WRI1, Gorai. 011G225700; HvWRI1, TC250449; OsWRI1, Os12g0126300; SunflowerWRI1, TC52295; ZnWRI11, GRMZM2G124524. Gene index numbers and tentative consensus (TC) numbers were referred to Phytozome ver 9.0 (http://www.phytozome.net/) and TGI database (http://compbio.dfci.harvard.edu/tgi/plant.html), respectively. (b) The expression levels of mRNAs of castor bean WRI1 homologs and an ABI3 homolog in various organs are shown. Semi-quantitative RT-PCR was carried out at indicated numbers of thermal cycles as described in Materials and Methods. RcActin was used as a standard.
bean homlog genes, RcSUS2, RcBCCP2 and RcKAS1 were identified to have the highest similarites of 75%, 49% and 85% to AtSUS2 (At5g49190), AtBCCP2 (At 5g15530) and AtKAS1 (At5g46290), respectively.
Significant signal of RcSUS2 were detected broadly in leaf, filling seeds (2, 3.5 cm diameters), matured seed and pod tissues at 27 cycles (Figure 2). RcBCCP2 was expressed in developing seed at relatively high levels, while the expression was weak in leaf and pod but marginal in matured seed. RcKAS1 was also expressed in developing seeds (2, 3.5 cm diameters), while the expression was weak or marginal in leaf, matured seed and pod. In contrast to RcBCCP2 and RcKAS1, Oleosin involved in oil body formation were detected at high level both in developing seeds and matured seed at 24 cycles (Figure 2). Significant signals of Ricin was deteced at 27 cycles in developing seed-specific manner as same as RcBCCP2 and RcKAS1 (Figure 2).
3.4. Immunochemical Detection of Seed-Specific Transcription Factors in Vaious Organs of Castor Bean
Conserved amino acid sequence motives of WRI1 is shown with alignments of AP2/ERF-related proteins (Figure 3(a)). Alignments of the second AP2-type domains among WRI1 homologs revealed a highly con served motif of 19 amino acids
(VSKYRGVARHHHNGRWEAR) that is specific to WRI1 orthologs but not to other AP2/ERF with single AP2-domain (Figure 3(a)). AtANT has different 6 amino acids in the region of the 19 amino acid motif for WRI1 specific epitope. Specificities of the antibodies were con-
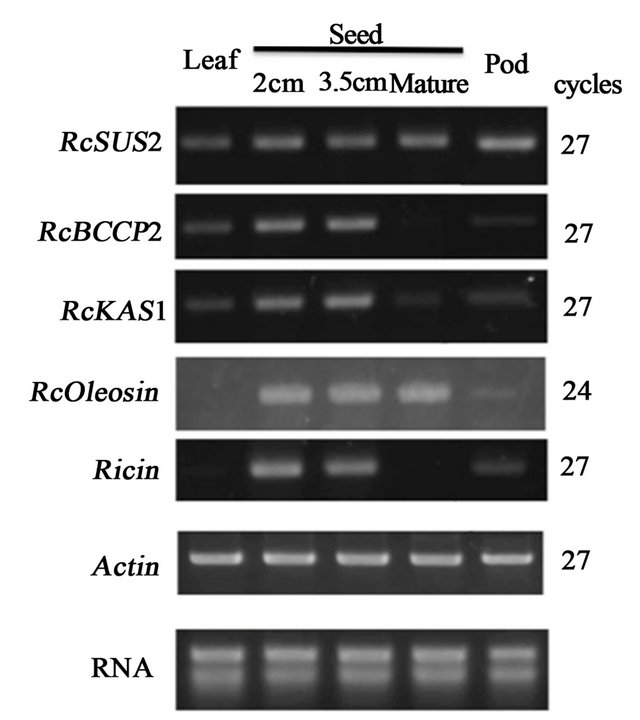
Figure 2. The expression profiles of FAS-related and seed storage protein genes. The expression levels of SUS2, BCCP, KAS Oleosin and Ricin in various organs were analyzed by semi-quantitative RT-PCR at indicated numbers of thermal with the specific primer sets (Table 1). RcActin was used as a standard.
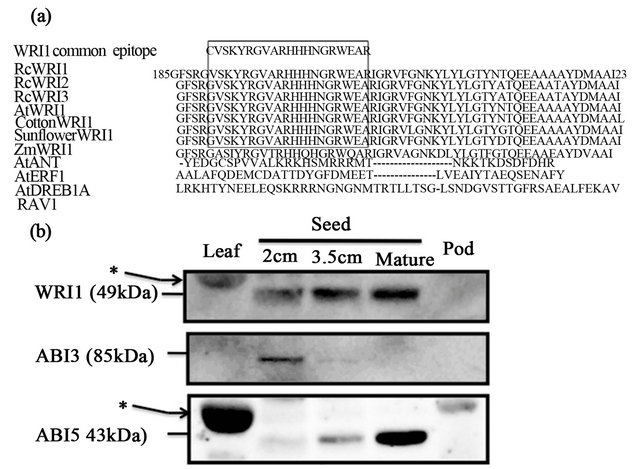
Figure 3. Immunoblot of WRI1-related polypeptides and seed specific transcription factor-related polypeptides in various organs of castor bean. (a) Alignments of deduced amino acid sequences of WRI1 homologs and other AP2/ ERF homologs. A highly conserved amino acid motif (VSKYRGVARHHHNGRWEAR) present in the second AP2-type domain was used as an epitope for raising antiWRI1 specific antibody. (b) Immunoblot of tissu extracts prepared from leaf, developing seed, mature seed and pod of castor bean plant was carried out by anti-WRI1 antibody, anti-ABI3 antibody and anti-ABI5 antibody. Each 50 μg protein per lane was subjected to SDS-PAGE (12.5% acrylamide gels) and immunoblot with specific antibodies as indicated. (*) indicates nonspecific signal of RuBisCO protein.
firmed by immunoblot using recombinant proteins of castor bean WRI1, ABI3 and ABI5 homologs (data not shown).
Immunoblot with the anti-WRI1 antibody showed a significant signal in filling seeds and the matured seed with a relative molecular mass of about 50 kDa (Figure 3(b)). Weak signals with the antibody were also detected in leaf at about 55 kDa, indicating a nonspecific signal possibly derived from RuBisCO large subunit. Immunoreactive signals with anti-ABI3 antibody were detected significantly in 2 cm seed, but not or marginally in leaf, 3.5 cm seed, matured seed and pod. In contrast to the anti-ABI3 antibody, immunoreactive signals with antiABI5 antibody were detected significantly in matured seed, weakly in 3.5 cm seed but not or marginally in leaf, 2 cm seed and pod.
4. Discussion
The caster bean WRI1 homologs formed a monophyletic cluster separated from other clusters of AP2/ERF groupe, AtERF1, RAV1 and AtDREB1A. The phylogenic tree or WRI1 homologs indicates that WRI1 cluster have three clade. RcWRI1 and AtWRI1 belong to one clade of the WRI1 cluster and has the highest similarity (62%) in amino acid sequence to that of cotton WRI1 homolg, and relatively high similarity (66%) in amino acid sequences to those of AtWRI1. Other two castor bean WRI1 homologs, RcWRI2 and RcWRI3, belongs to another clade of WRI1 cluster, and have similarity (%) in amino acid sequences to those of HvWRI1. It is possible that RcWRI1 belonging to the same clade of AtWRI1 and ZmWRI1 plays an important role in seed specific expression of FAS genes, because AtWRI1 and ZmWRI1 appeared to promote the master regulation of a set of FAS-related genes [8,18]. It was reported that AtWRI1 binds to a specific sequence motif, AW-box, in the promoter regions of BCCP2 and KAS1 and other FAS genes [9]. According to castor bean genome sequence data base, the 5’-promoter regions within 300 bp from starting Met of RcBCCP2 and RcKAS1 genes possesses 2 and 3 boxes similar to the AW-box consensus motif, respectively. It is reasonable that RcWRI1 is expressed specifically in seed and that RcWRI1 plays pivotal roles in master regulation of seed specific FAS genes via binding to AW-box as same as AtWRI1 does. In contrast to RcWRI1 and RcWRI2, RcWRI3 is expressed mainly in leaf but not reproductiove organs, seed and pods, suggesting that RcWRI3 regulates FAS genes involved in membrane lipid synthesis in vegetative organs such as, glycerolipids (DGDG, MGDG and SQDG) in thylakoid apparatus indispensable for photosynthesis.
RT-PCR and immunoblot indicates that mRNA and protein of RcABI3 are expressed specifically in developing seed, suggesting that a network of B3 type transcripttion factors play pivotal roles in regulation of seed development and seed storage proteins in castor bean as same as in Arabidopsis. Furthermore, the observation that a significant signal of ABI5-related polypeptiude is detected specifically in in matured seed is consistent with previous studies that ABI5 is required for establishing and maintaining seed dormancy [11].
The present study revealed a line of evidence about seed specific transcription factors and FAS-related genes in castor bean: 1) Among WRI1 homologs in castor bean, RcWRI1 mRNA is expressed specifically in developing seed.; 2) The RcABI3 and ABI5-related proteins function in different stages of seed development; 3) The expression of seed specific FAS-related genes, RcBCCP2 and RcKAS1 possessing AW-box motives in the promoter regions, are tightly associates with those of Rc WRI1 and RcKAS1. These observations suggest that RcWRI1 is one of master transcription factors possibly regulating seed specific FAS-related genes. It is necessary for clarifying direct interaction of RcWRI1 with AW-box and transcriptional activation of RcBCCP2 and RcKAS1 by RcWRI1 recombinant protein and/or chromatin-immunoprecipitation assay with anti-WRI1 antibody. While both WRI1 and ABI3-related polypeptides were detected in developing seeds by immunoblot, it is possible that other B3-type transcription factors (including LEC2 orthologs) are required for direct regulation of RcWRI1 expression rather than ABI3. It remains to be clarified whether other novel master regulation factors are involved in regulation of key steps of seed oil synthesis in addition of WRI1-FAS genes cascade.
Increasing evidence suggests that the network of various transcription factors, such as AP2-, B3- and bZIPtype transcription factors regulate seed development and storage compound synthesis. Therefore, identifications and molecular application technique of the signaling moleculaes regulating seed development and oil synthesis are important for and improvement of vegetative oil crops.
5. Acknowledgements
This work was supported in part by Grant-in-Aids from the Ministry of Education, Sports, Culture, Science and Technology of Japan, No. 19380022 (to T. Yuasa), Astep feasibility study program in Japan Science and Technology Agency. (JST), (#AS2111128D) (to T. Yuasa) and 2011th Itoh Himashiyu Research Foundation (to T. Yuasa).
REFERENCES
- T. A. McKeon, J. T. Lin and A. E. Stafford, “Biosynthesis of Ricinoleate in Castor Oil,” Advances in Experimental Medicine and Biology, Vol. 464, 1999, pp. 37-47. doi:10.1007/978-1-4615-4729-7_4
- A. Cagliari, M. Margis-Pinheiro, G. Loss, A. A. Mastroberti, J. E. de Araujo Mariath and R. Margis, “Identification and Expression Analysis of Castor Bean (Ricinus communis) Genes Encoding Enzymes from the Triacylglycerol Biosynthesis Pathway,” Plant Science, Vol. 179, No. 5, 2010, pp. 499-509. doi:10.1016/j.plantsci.2010.07.015
- G. Q. Chen, C. Turner, X. He, T. Nguyen, T. A. McKeon and D. Laudencia-Chingcuanco, “Expression Profiles of Genes Involved in Fatty Acid and Triacylglycerol Synthesis in Castor Bean (Ricinus communis L.),” Lipids, Vol. 42, No. 3, 2007, pp. 263-274. doi:10.1007/s11745-007-3022-z
- N. A. Jolliffe, C. P. Craddock and L. Frigerio, “Pathways for Protein Transport to Seed Storage Vacuoles,” Biochemical Society Transactions, Vol. 33, No. 5, 2005, pp. 1016-1018.
- S. Kurup, H. D. Jones and M. J. Holdsworth, “Interactions of the Developmental Regulator ABI3 with Proteins Identified from Developing Arabidopsis Seeds,” Plant Journal, Vol. 21, No. 2, 2000, pp. 143-155. doi:10.1046/j.1365-313x.2000.00663.x
- E. M. Söderman, I. M. Brocard, T. J. Lynch and R. R. Finkelstein, “Regulation and Function of the Arabidopsis ABA-Insensitive4 Gene in Seed and Abscisic Acid Response Signaling Networks,” Plant Physiology, Vol. 124, No. 4, 2000, pp. 1752-1765. doi:10.1104/pp.124.4.1752
- M. Suzuki and D. R. McCarty, “Functional Symmetry of the B3 Network Controlling Seed Development,” Current Opinion in Plant Biology, Vol. 11, No. 5, 2008, pp. 548- 553. doi:10.1016/j.pbi.2008.06.015
- S. Baud, S. Wuillème, A. To, C. Rochat and L. Lepiniec, “Role of WRINKLED1 in the Transcriptional Regulation of Glycolytic and Fatty Acid Biosynthetic Genes in Arabidopsis,” Plant Journal, Vol. 60, No. 6, 2009, pp. 933- 947. doi:10.1111/j.1365-313X.2009.04011.x
- K. Maeo, T. Tokuda, A. Ayame, N. Mitsui, T. Kawai, H. Tsukagoshi, S. Ishiguro and K. Nakamura, “An AP2-Type Transcription Factor, WRINKLED1, of Arabidopsis Thaliana Binds to the AW-Box Sequence Conserved among Proximal Upstream Regions of Genes Involved in Fatty Acid Synthesis,” Plant Journal, Vol. 60, No. 3, 2009, pp 476-487. doi:10.1111/j.1365-313X.2009.03967.x
- S. A. Braybrook and J. J. Harada, “LECs Go Crazy in Embryo Development,” Trends in Plant Science, Vol. 13, No. 12, 2008, pp. 624-630. doi:10.1016/j.tplants.2008.09.008
- M. Santos-Mendoza, B. Dubreucq, S. Baud, F. Parcy, M. Caboche and L. Lepiniec, “Deciphering Gene Regulatory Networks That Control Seed Development and Maturation in Arabidopsis,” Plant Journal, Vol. 54, No. 4, 2008, pp. 608-620. doi:10.1111/j.1365-313X.2008.03461.x
- A. Cernac and C. Benning, “WRINKLED1 Encodes an AP2/EREB Domain Protein Involved in the Control of Storage Compound Biosynthesis in Arabidopsis,” Plant Journal, Vol. 40, No. 4, 2004, pp. 575-585. doi:10.1111/j.1365-313X.2004.02235.x
- N. Focks and C. Benning, “Wrinkled1: A Novel, LowSeed-Oil Mutant of Arabidopsis with a Deficiency in the Seed-Specific Regulation of Carbohydrate Metabolism,” Plant Physiology, Vol. 118, No. 1, 1998, pp. 91-101. doi:10.1104/pp.118.1.91
- A. To, J. Joubès, G. Barthole, A. Lécureuil, A. Scagnelli, S. Jasinski, L. Lepiniec and S. Baud, “WRINKLED Transcription Factors Orchestrate Tissue-Specific Regulation of Fatty Acid Biosynthesis in Arabidopsis,” Plant Cell, 2012, in Press. doi:10.1105/tpc.112.106120
- S. Baud, M. S. Mendoza, A. To, E. Harscoët, L. Lepiniec and B. Dubreucq, “WRINKLED1 Specifies the Regulatory Action of LEAFY COTYLEDON2 towards Fatty Acid Metabolism during Seed Maturation in Arabidopsis,” Plant Journal, Vol. 50, No. 5, 2007, pp. 825-838. doi:10.1111/j.1365-313X.2007.03092.x
- Sanjaya, T. P. Durrett, S. E. Weise and C. Benning, “Increasing the Energy Density of Vegetative Tissues by Diverting Carbon from Starch to Oil Biosynthesis in Transgenic Arabidopsis,” Plant Biotechnology Journal, Vol. 9, No. 8, 2011, pp. 874-883. doi:10.1111/j.1467-7652.2011.00599.x
- J. Liu, W. Hua, G. Zhan, F. Wei, X. Wang, G. Liu and H. Wang, “Increasing Seed Mass and Oil Content in Transgenic Arabidopsis by the Overexpression of WRI1-Like Gene from Brassica napus,” Plant Physiology and Biochemistry, Vol. 48, No. 1, 2010, pp. 9-15. doi:10.1016/j.plaphy.2009.09.007
- B. Pouvreau, S. Baud, V. Vernoud, V. Morin, C. Py, G. Gendrot, J. P. Pichon, J. Rouster, W. Paul and P. M. Rogowsky, “Duplicate Maize Wrinkled1 Transcription Factors Activate Target Genes Involved in Seed Oil Biosynthesis,” Plant Physiology, Vol. 156, No. 2, 2011, pp. 674- 686. doi:10.1104/pp.111.173641
- C. Soderlund, A. Descour, D. Kudrna, M. Bomhoff, L. Boyd, J. Currie, A. Angelova, K. Collura, M. Wissotski, E. Ashley, et al., “Sequencing, Mapping, and Analysis of 27,455 Maize Full-Length cDNAs,” PLoS Genetics, Vol. 5, 2009, e1000740. doi:10.1371/journal.pgen.1000740
- J. Schmutz, S. B. Cannon, J. Schlueter, J. Ma, T. Mitros, W. Nelson, D. L. Hyten, Q. Song, J. J. Thelen, J. Cheng, D. Xu, U. Hellsten, G. D. May, Y. Yu, T. Sakurai, T. Umezawa, M. K. Bhattacharyya, D. Sandhu, et al., “Genome Sequence of the Palaeopolyploid Soybean,” Nature, Vol. 463, No. 7278, 2010, pp. 178-183. doi:10.1038/nature08670
- A. P. Chan, J. Crabtree, Q. Zhao, H. Lorenzi, J. Orvis, D. Puiu, A. Melake-Berhan, K. M. Jones, J. Redman, G. Chen, E. B. Cahoon, M. Gedil, M. Stanke, B. J. Haas, J. R. Wortman, C. M. Fraser-Liggett, J. Ravel and P. D. Rabinowicz, “Draft Genome Sequence of the Oilseed Species Ricinus communis,” Nature Biotechnology, Vol. 28, No. 9, 2010, pp. 951-956. doi:10.1371/journal.pone.0021743
- S. Sato, H. Hirakawa, S. Isobe, E. Fukai, A. Watanabe, M. Kato, K. Kawashima, C. Minami, A. Muraki, N. Nakazaki, et al., “Sequence Analysis of the Genome of an Oil-Bearing Tree, Jatropha curcas L.,” DNA Research, Vol. 18, No. 1, 2011, pp. 65-76. doi:10.1093/dnares/dsq030
- E. K. Al-Dous, B. George, M. E. Al-Mahmoud, M. Y. Al-Jaber, H. Wang, Y. M. Salameh, E. K. Al-Azwani, S. Chaluvadi, A. C. Pontaroli, J. DeBarry, V. Arondel, J. Ohlrogge, I. J. Saie, K. M. Suliman-Elmeer, J. L. Bennetzen, R. R. Kruegger and J. A. Malek, “De Novo Genome Sequencing and Comparative Genomics of Date Palm (Phoenix dactylifera),” Nature Biotechnology, Vol. 29, No. 6, 2011, pp. 521-527. doi:10.1038/nbt.1674
- M. Rivarola, J. T. Foster, A. P. Chan, A. L. Williams, D. W. Rice, X. Liu, A. Melake-Berhan, H. H. Creasy, D. Puiu, M. J. Rosovitz, H. M. Khouri, S. M. BeckstromSternberg, G. J. Allan, P. Keim, J. Ravel and P. D. Rabinowicz, “Castor Bean Organelle Genome Sequencing and Worldwide Genetic Diversity Analysis,” PLoS One, Vol. 6, No. 7, 2011, e21743. doi:10.1371/journal.pone.0021743
- J. Nakamura, T. Yuasa, H. T. Tran, K. Harano, S. Tanaka, T. Iwata, T. Phan and M. Iwaya-Inoue, “Rice Homologs of Inducer of CBF Expression (OsICE) Are Involved in Cold Acclimation,” Plant Biotechnology, Vol. 28, No. 3, 2011, pp. 303-309. doi:10.5511/plantbiotechnology.11.0421a
- M. Okuda, M. P. Nang, K. Oshima, Y. Ishibashi, S. H. Zheng, T. Yuasa and M. Iwaya-Inoue, “The Ethylene Signal Mediates Induction of GmATG8i in Soybean Plants under Starvation Stress,” Bioscience Biotechnology Biochemistry, Vol. 75, No. 7, 2011, pp. 1408-1412. doi:10.1271/bbb.110086
- T. Yuasa, Y. Ishibashi and M. Iwaya-Inoue, “A Flower Specific Calcineurin B-Like Molecule (CBL)-Interacting Protein Kinase (CIPK) Homolog in Yomato Cultivar Micro-Tom (Solanum lycopersicum L.),” American Journal of Plant Sciences, Vol. 3 No. 6, 2012, pp. 753-763. doi:10.4236/ajps.2012.36091
- B. Krizek, “AINTEGUMENTA and AINTEGUMENTALIKE6 Act Redundantly to Regulate Arabidopsis Floral Growth and Patterning,” Plant Physiology, Vol. 150, No. 4, 2009, pp. 1916-1929. doi:10.1104/pp.109.141119

