Applied Mathematics
Vol.06 No.09(2015), Article ID:58629,17 pages
10.4236/am.2015.69135
Mathematical Model of Seed Dispersal by Frugivorous Birds and Migration Potential of Pinyon and Juniper in Utah
Ram C. Neupane1, James A. Powell1,2
1Department of Mathematics and Statistics, Utah State University, Logan, USA
2Department of Biology, Utah State University, Logan, USA
Email: ram.neupane@aggiemail.usu.edu
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 16 June 2015; accepted 2 August 2015; published 6 August 2015
ABSTRACT
Seed dispersal of juniper and pinyon is a process in which frugivorous birds play an important role. Birds either consume and digest seeds or carry and cache them at some distance from the source tree. These transported and settled seeds can be described by a dispersal kernel, which captures the probability that the seed will move a certain distance by the end of the process. To model active seed dispersal of this nature, we introduce handling time probabilities into the dispersal model to generate a seed digestion kernel. In the limit of no variability in handling time the seed digestion kernel is Gaussian, whereas for uniform variability in handling time the kernel approaches a Laplace distribution. This allows us to standardize spatial movement (diffusion) and handling time (peak settling rate) parameters for all three distributions and compare. Analysis of the tails indicates that the seed digestion kernel decays at a rate intermediate between Gaussian and Laplace seed kernels. Using this seed digestion kernel, we create an invasion model to estimate the speed at which juniper and pinyon forest boundaries move. We find that the speed of seed invasion corresponding to the digestion kernel was faster than seeds resulting from Laplace and Gaussian kernels for more rapidly digested seeds. For longer handling times the speeds are bounded between the Laplace (faster) and Gaussian (slower) speeds. Using parameter values from the literature we evaluate the migration potential of pinyon and juniper, finding that pinyon may be able to migrate up to two orders of magnitude more rapidly, consistent with observations of pine migration during the Holocene.
Keywords:
Dispersal Kernel, Invasion Speed, Seed Handling Time, Pinus monophylla, Juniperus osteosperma

1. Introduction
Forest boundaries change over time, and in favorable climates can expand as tree seeds spread beyond the range of the forest and germinate into new trees. Seeds may spread in a variety of ways. Common seed dispersal agents include wind, transportation in water, and transportation via birds and animals (either through being consumed and digested or being carried and cached). Because the diet of birds and some animals is often made up of fleshy-fruited plants, the pattern of seed dispersal and activities of vertebrate dispersers are closely related [1] [2] . Birds in particular contribute heavily to the spread of some plant populations [3] [4] .
A case in point is seed dispersal and forest migration in two southwestern tree species: pinyon (Pinus monophylla) and juniper (Juniperus osteosperma). In both species birds play a major, but different, role. Junipers produce seeds which are available most of the winter, and consequently many birds (particularly American robins, Turdus migratorius, and cedar waxwings, Bombycilla cedrorum) consume juniper berries [5] . The berries are then digested and seeds are deposited some time later by defecation. Digestion does not impede the seeds’ ability to germinate, particularly in the case of robins [6] , and juniper is thus dispersed while robins forage over scores of meters.
By contrast, pinyon seed dispersal by Clark’s nutcracker (Nucifraga columbiana) and pinyon jays (Gymnorhinus cyanocephalus) occurs primarily through seed caching in summer and fall, when the cones mature. Some seeds are consumed immediately, but the majority is placed in a sublingual pouch and carried several kilometers to remote cache sites, where they are buried in small groups [7] [8] . Most of the cache sites are found during the winter, but a substantial percentage of caches are never revisited and the cached seeds are in an ideal situation for germination, which determines pinyon distribution.
Variation in climate also influences the expansion of pinyon-juniper (P-J) woodland via impacts on germination and survival rates. The abundance of summer rainfall and the warming in the winter and spring have caused P-J boundaries to shift northwards [9] [10] . Juniper can sustain more severe drought than pinyon [11] [12] , making pinyon more sensitive to climate than juniper [13] . Although juniper is more drought tolerant than pinyon, both species have declined as their habitat shrinks in the southwestern United States because of accelerated global warming. Recent drought conditions in northern New Mexico, Arizona and southern Utah are more severe than any historic drought. Consequently, P-J habitat boundaries are shifting northwards rapidly [11] [14] [15] . On the other hand, climate change is creating new P-J habitat in central Nevada [12] , the central Great Basin [16] , northeastern Utah [17] and southeastern Oregon [18] . This begs the following research questions: 1) can either pinyon or juniper disperse far enough northward to colonize the new habitat? 2) How rapidly may we expect forest boundaries to move? We will contribute to answering these questions by developing a PDF (Probability Density Function) for seed distribution by active dispersers and using a population-level Integrodifference Equation (IDE) to evaluate the migration potential of these two species.
To model the spread of the seeds, we assume that seed cachers or frugivorous animals collect seeds (through consumption in the case of juniper berries or collection of pinyon seeds to cache at a distance) and then follow a random walk, using the modeling framework introduced by [19] . However, unlike previously considered “failure rates” or “hazard functions” (rates at which seeds are deposited on the ground), we note that the distribution of settling times for seed dispersal by birds should be modeled as distributions in time. Seeds defecated or cached at times sampled from a seed handling PDF will be distributed on the ground in a spatial PDF, or seed digestion kernel (SDK), which is different from any previously-considered dispersal kernel. We will derive the SDK from first principles and find that the mean handling time plays a crucial role in determining its form. The different handlings of juniper and pinyon seeds lead to very different dispersal behaviors. We will compare the SDK with two limiting kernels discussed by [19] , the Laplace and Gaussian kernels; the SDK behaves quite differently in the small handling time limit.
In this paper we numerically calculate the SDK based on PDFs of seed handling times. Once the kernel is determined, we will use it in a generic IDE population model to estimate invasion speeds and compare with speeds generated from Gaussian and Laplace kernels, which are limiting cases. Surprisingly, in some parameter regimes the SDK yields more rapid invasion speeds than either Laplace or Gaussian kernels. Predictions for juniper and pinyon migration rates will be generated using literature values for parameters. We find that pinyon has much higher potential to find and occupy new niches than juniper, consistent with observations of Holocene range expansion for the two species.
2. Methods
2.1. Model for Seed Dispersal
We begin with a common model of dispersal and settling of propagules, (any material that is used for propagating an organism) introduced by [19]
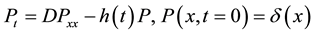 (1)
(1)
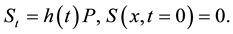 (2)
(2)
In this model  represents the density of seeds during dispersal by frugivorous birds and animals, which are assumed to follow a random walk with diffusion rate D. The function
represents the density of seeds during dispersal by frugivorous birds and animals, which are assumed to follow a random walk with diffusion rate D. The function 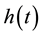 represents the hazard function or a failure rate of seeds (i.e. rate at which seeds are placed on the ground by either caching or defectation). The function
represents the hazard function or a failure rate of seeds (i.e. rate at which seeds are placed on the ground by either caching or defectation). The function 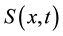 is the density of settled seeds (seeds on the ground) at time t. The Dirac delta function,
is the density of settled seeds (seeds on the ground) at time t. The Dirac delta function, 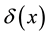 , places seeds initially at the origin, with no seeds yet on the ground. Because the system conserves the integral of all seeds at all locations, the sum,
, places seeds initially at the origin, with no seeds yet on the ground. Because the system conserves the integral of all seeds at all locations, the sum, 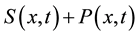 , is a PDF for seed location in space at time t. The SDK, K(x), is the long time limit of this process,
, is a PDF for seed location in space at time t. The SDK, K(x), is the long time limit of this process,
 .
.
An important modeling point is that, to be consistent with mechanisms of seed handling by vertebrates, the hazard function, 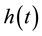 , must be a PDF in time. For example, when researchers measure times required for seed digestion and defecation, results are communicated as skewed frequency distributions with a strong mode and tails which decline to zero (e.g. [20] ). This is in direct contrast to failure rates considered in [19] , none of which are modal PDFs. Observed distributions of handling times are asymmetrical, with long tails, and consequently a minimal characterization of such PDFs requires three parameters (one controlling the shape to the left of the mode, one controlling the location of the mode, and one controlling the shape of the tail for large t). We therefore propose
, must be a PDF in time. For example, when researchers measure times required for seed digestion and defecation, results are communicated as skewed frequency distributions with a strong mode and tails which decline to zero (e.g. [20] ). This is in direct contrast to failure rates considered in [19] , none of which are modal PDFs. Observed distributions of handling times are asymmetrical, with long tails, and consequently a minimal characterization of such PDFs requires three parameters (one controlling the shape to the left of the mode, one controlling the location of the mode, and one controlling the shape of the tail for large t). We therefore propose
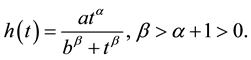 (3)
(3)
In this distribution the constant b scales the mean digestion time of seeds while a is a normalization constant (not free, since it depends directly on the other three parameters so that  integrates to one). The parameters
integrates to one). The parameters  and
and  determine the shape of the tails of
determine the shape of the tails of 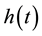 (shown in Figure 1 and Figure 2); if
(shown in Figure 1 and Figure 2); if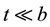 ,
, 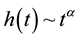 while
while 

2.2. Solution Technique for Calculating SDK
We integrate the PDE directly and then approximate time integrals using the trapezoid rule. To begin, let

Equation (1) becomes

An integrating factor of 

Using Equation (6) in the model (2) then we get

Figure 1. This plot demonstrates the shape of the seed settling rate, h(t), over time with β = 55 and various values of α (α = 1(―), α = 7(− −), α = 25(− ∙ −) and α = 50(∙∙∙∙∙∙)). In this plot, we see that the left tail of the distribution is shifting to the right as α increases.
Figure 2. This plot demonstrates the shape of the seed settling rate, h(t), over time with α = 3 and various values of β (with β = 5(―), β = 10(− −), β = 25(− ∙ −) and β = 50(∙∙∙∙∙∙)). As in Figure 1, we also see the right tail shifts to the right as β increases.
Numerical approximations are then calculated using the trapezoid rule for numerical integration. Solutions generated this way were cross-checked against the (much) more time-consuming finite difference solution of (1) and (2) (see Appendix A) to ensure accuracy. For the same size of time steps we found that direct quadrature of integrals in (7) was substantially more accurate (and rapid) than solution of the PDEs using finite differences.
2.3. Standardizing the Three Kernels for Comparison
We wish to compare the SDK with the Gaussian and Laplace seed. The question is how to standardize the three kernels for comparison? Below we will show that the Laplace and Gaussian kernels arrive from different limiting choices for
For convenience, we assume 



We will call this time


Figure 3. This plot provides an initial comparison of the three types of seed digestion rate, the PDF of seed digestion times (―) and the PDFs leading to the Gaussian kernel (− − −) and Laplace kernel (∙∙∙∙∙∙). For this comparison we fixed β = 7.5, α = 5.5 and b = 5, it can be seen that the three kernels share the same maximum to standardize the three kernels.
Then

This gives

To generate a Laplace kernel we assume that the distribution of seed settling is a step function defined by

As is shown in Appendix B, the solution to (1) and (2) with the step function defined in Equation (11) is approximately the Laplace kernel

After this standardization, the Gaussian kernel (10) and the Laplace kernel (12) are ready for comparison with the seed digestion kernel. Figure 4 illustrates the shape of seed distribution on the ground for the standardized kernels. Seeds seem to disperse to the furthest for the Laplace (having fattest tail), 


Figure 4. Comparison of the seed digestion kernel (―), Gaussian kernel (− − −) and Laplace kernel (∙∙∙∙∙∙) with b = 10, a =3 and β = 7. The Gaussian tail decays more rapidly than tail of seed digestion and the seed digestion tail decays more slowly than Laplace (the fattest tail).
2.4. Analyzing the Tail of Seed Digestion Kernels
Here, we approximate the tail of the SDK using the steepest descent method [21] to compare tails with Gaussian and Laplace seed kernels. As in the previous section, we assume 

Define the exponent in (7) as

The critical point of the function 

with respect to t and evaluating at 

Let us suppose 

Equations (15) and (16) can be used with the generalized steepest descent theorem to approximate (7), giving

Analyzing K as 
2.5. Population Model for Evaluating Migration Potential
To determine how the shape of the SDK affects rates of invasion, the kernels must be imbedded in a population model. Below we present a simplification of the model used by [22] to describe the general behavior of an invasion by perennial plants. For xeric-adapted species like pinyon and juniper dispersing into the new regions, competition for water and space occurs primarily among seedlings. We take

where Nt represents the population density of adults in generation t. The function 



Here M is the maximum number of surviving seeds, g is the germination rate, and 

and the integral represents the total number seeds arriving at location x from all possible locations, y. Therefore, the first term on the right hand of the invasion model (18) predicts the distribution of new trees depending on the available sources and the second term provides the surviving number of old trees so that the total is the population of trees in the next generation.
2.6. Analysis of Invasion Speeds
To analyze invasion speeds for models like (18), we follow the analysis of [23] . Because the population density of a tree population approaches zero in advance of the invasion front, we can assume that as

with

Combining Equations (19) and (20)

Plugging this into Equation (18),

Taking only leading order terms,

where 
Writing the convolution of Equation (23) in terms of an integral, we have

where the moment generating function, M(u), is defined by

Differentiating Equation (24) with respect to u and setting to zero to find the extremal invasion speed gives

Using Equation (26) to eliminate c in (24) gives

To find the invasion speed, 

3. Results
3.1. Shape of Kernels Based on Mean Digestion Time Scaling Parameter
The mean digestion time scaling parameter b plays a major role in determining seed dispersal. Changing the value of b generates different shapes of solutions (see Figure 5) with larger values of b corresponding to broader dispersal, as we expected. If digestion or caching takes longer, birds have more time to travel before depositing seeds, resulting in seeds traveling further from the source.
3.2. Comparison of Tails
We would like to characterize the shape of the tail of the SDK and place it in the context of the well-known Gaussian (10) and Laplace kernels (12). The exponents of the exponential functions for both kernels determine
Figure 5. Comparison of seed digestion kernels for various mean seed handling times. In this figure, b = 1(―), b = 3(− −), b = 5(− ∙ −) and b = 9(∙∙∙∙∙∙), illustrating broader dispersal for larger mean digestion times.
the shapes of the corresponding tails. To analyze the tails of these kernels, we consider large x and assume other parameter values are bounded. The dominant terms in the exponents of the Gaussian and Laplace kernels are


than the tail of the Laplace kernel when
In the case of the most slowly-decaying PDF of seed handling times, 

where 


and therefore

The exponents of Gaussian kernel and Laplace kernels are

and

Observing
we conclude that the tail of Gaussian kernel decays to zero most rapidly while the tail of Laplace kernel is the slowest. The tail of the SDK is intermediate between the other two.
3.3. Relationship between Invasion Rate and Mean Digestion Time
We have not chosen scales for generation time, population density and space yet. To compare the speeds of invasion from the population IDE (18), we may therefore, without loss of generality, choose mortality







Figure 6. Speeds of invasion calculated for the seed digestion kernel, the Gaussian kernel and Laplace kernel. The solid line indicates the speed with seed digestion kernel, the dashed line indicates the speed with Gaussian kernel and the dotted line is the speed with Laplace kernel. The figure shows that the invasion speed produced from all three kernels always increasing in different rates as the increase of mean digestion time scaling parameter.
There is a strong relationship between the characteristic handling time, b, and invasion speed, c. The longer it takes to digest a seed, the faster forest migration. For small b, the SDK invasion speed is higher than the speeds corresponding to the Gaussian and Laplace kernels. On the other hand, for bigger b values, the speed of invasion with the Laplace kernel is fastest, the speed with a Gaussian kernel is slowest and the speed corresponding to the SDK stays between the other two, as might be expected from comparing tails.
As





4. Migration Potential of Pinyon and Juniper
To quantify invasion speeds in terms of yearly distance covered for both pinyon and juniper, we need specific data such as the mean generation time (G), mean dispersal space step




where G is the generation time (duration from seedling to getting matured tree for producing seeds), n is the initial number of trees and N is the total number of trees at the end of the study. To estimate the diffusion rate D for birds, we use

where 
In order to calculate yearly invasion speed, c, we need to rescale both space and time, since we fixed D = 1 (equivalent to nondimensionalized space) in the numerical calculations. Additionally, each step in our IDE is a generation, which must be scaled back to years for comparison purposes. Assuming the dimensional diffusion rate, D, is in m2 per minute and the mean handling time 



Table 1. Parameters used to estimate seed dispersal kernels and migration rates of juniper and pinyon in Utah. References for parameter values are provided.

Now we turn to specific parameter values for pinyon and juniper.
For pinyon we use a generation time G ~ 20 years from [26] . To estimate R0 we refer to [24] , who determined that only 6 pinyon survived the nineteenth-century fire on their site. The number of pinyon pines increased to 1051 over the next 145 years giving R0 = 2.04/generation for pinyon from equation (33). [7] observed that Clark’s Nutcracker fly from 4000 to 5000 meters while caching seeds, taking 15 - 30 minutes. We therefore take
λ = 4500 meters and use τ = 22.5 minutes, giving
three phases. Nutcrackers spend 45 minutes collecting seeds to fill their pouch, 15 - 30 minutes to travel to the caching area and 5 - 10 minutes to cache all seeds carried in their pouch. Averaging and summing, we estimate
seed mean handling time to be 

timate annual mortality at 0.08% - 0.23% for common pinyon. Taking the mean and converting to a rate per generation gives ω = 0.00155. Taken together, these parameters give a minimum speed of 518.97 m/year and a maximum of 946.1 m/year for pinyon, with an average of 773.31 m/year.
On the other hand, for juniper we use a generation time G ~ 50 years from [28] . To estimate R0 we follow [24] , who observed that only 109 junipers survived the nineteenth-century fire on their site. The number increased to 172 over the next 145 years giving R0 = 1.17/generation for juniper from Equation (33). [6] observed that American robins forage in the range of 10 - 100 meters with mean 55 meters and average 4 minutes between trees. We therefore take λ = 55 meters and time step τ = 4 minutes, giving D = 189.1 m2/min. [20] report that the cedar waxwing takes between 7.35 and 22.45 minutes to digest red cedar (Juniperus virginiana) seeds; we
therefore use a handling time 

estimate annual mortality at 0.01% - 0.07% for Utah juniper (Juniperus osteosperma). Taking the mean and converting to a rate per generation gives ω = 0.0004. Using these parameter ranges we find that juniper spreads with minimum speed 0.42 m/year and maximum speed 7.3 m/year with an average of 3.3 m/year, two orders of magnitude more slowly than pinyon.
These results match up well with what is known about these two species and their relative movements during the Holocene. Juniper seems to have been present in the Great Basin area for at least 30,000 years, based on evidence from fossilized packrat middens [29] . While its range contracted due to climatic shifts there were no significant expansions. By contrast, pinyon pine was limited to Arizona and New Mexico up to 9000 years ago, but migrated up the Wasatch front to the northeastern corner of Utah in the next 1000 - 1500 years [30] , which would have required speeds in excess of 500 m/year.
5. Conclusions
Mechanisms of plant migration vary based on the source plant and the dispersal process delivering seeds to new locations for germination. Juniper berries mainly disperse after being eaten by vertebrates who deposit seeds after digestion. Birds, particularly robins, may be the biggest dispersers. Seeds of pinyon trees, on the other hand, are commonly spread while animals cache, and corvids (jays and nutrcrackers), which cache at large distances, are the largest contributors. In this paper, we introduced a PDF of seed-handling to reflect the effects of digestion/caching on dispersal of pinyon and juniper seeds. We connected this distribution to hazard functions or failure rates in an existing random-walk dispersal model to determine a seed digestion kernel modeling the probable location of seeds after active dispersal. As expected, if birds or animals take more time to handle seeds, those seeds are dispersed further away from the source tree. While no closed-form solution for the SDK is available, it is easy to calculate numerically (and would only have to be calculated once, in advance, for implementation in an IDE model for population invasion).
To evaluate migration potential for pinyon and juniper we introduced an IDE model with competition among seedlings, which is appropriate for desert-adapted trees in the xeric environment of the American Southwest. The SDK was compared with well-known Laplace and Gaussian kernels (L(x) and G(x)). After standardizing the associated PDFs for handling time, the speed of invasion for the SDK was the fastest for shorter handling times (rapidly digesting seeds). As handling times increased, however, the speeds for the SDK fell between the Laplace kernel’s (faster; based on an assumption of constant seed deposition) and the Gaussian kernel’s (slower; based on the assumption of instantaneous seed deposition), as would be expected from the relative behavior of the tails.
Using the SDK and median parameter values estimated from the literature it turned out that pinyon has migration potential at least two orders of magnitude larger than juniper due to avian dispersal. Along with changing temperatures and diminishing moisture levels the favorable environment for P-J is moving northwards through Utah. Over time, these trees will not be able to survive in the southern limits of their current habitat. The large migration potential of pinyon means that it is most likely to occupy new habitats opening to the north.
Of course, juniper already occupies much of the available northern habitat, and with longer generation times and much stronger adaptation to variable moisture regimes juniper can be expected to flourish in northern Utah for the foreseeable future. Moreover, juniper may have much higher migration potential than our analysis indicates. For the slower juniper we can probably not ignore mammalian dispersers [31] and passive dispersal agents (such as runoff and streams for dispersing juniper berries, see [5] ). The two main avian juniper dispersers, American robins and cedar waxwings, both forage and defecate locally and therefore do not seem to make a large contribution to juniper spread. However, mammals such as foxes, bears and coyotes may disperse juniper seeds long distances since they have much longer gut-retention times and can travel more than 10 kilometer per day [32] . Since juniper seeds persist through winter, dispersal by spring runoff can also contribute substantially. Nevertheless, dispersers like pinyon jay and Clark’s nutcracker likely give pinyon the dispersal advantage over juniper.
The largest factor ignored in our study is spatial variability. As [22] pointed out, vertebrate dispersers move rapidly through some habitats and linger in others, and western landscapes are comprised of highly variable habitat, particularly at the leading edge of invasions. One would expect step sizes in the random walks that dispersers follow, and therefore their diffusions rates, to vary strongly with habitat type. Recent advances in the use of homogenization [33] make integration of reaction-diffusion models with highly variable constants surprisingly easy, so building SDKs with variable diffusion and applying asymptotic techniques like homogenization will be our future concentration of research.
Acknowledgements
The authors would like to thank Tom Edwards and Jacob Gibson for discussion of life-history traits of pinyon and juniper and assistance with background literature. Luis Gordillo, Martha Garlick and USU’s MathBio group gave RCN much helpful feedback. RCN was partially supported by a grant from USU’s School of Graduate Studies. JAP would also like to thank the NSF for support under DEB grant 0918756 as well as the Western Wildland Environmental Threat Assessment Center (WWETAC).
Cite this paper
Ram C.Neupane,James A.Powell, (2015) Mathematical Model of Seed Dispersal by Frugivorous Birds and Migration Potential of Pinyon and Juniper in Utah. Applied Mathematics,06,1506-1523. doi: 10.4236/am.2015.69135
References
- 1. Corlett, R.T. (1998) Furgivory and Seed Dispersal by Vertebrates in the Oriental (Indomalayan) Region. Cambridge Philosophical Society, 73, 413-448.
http://dx.doi.org/10.1017/S0006323198005234 - 2. Wenny, D.G. (2001) Advantage of Seed Dispersal: A Re-Evaluation of Directed Dispersal. Evolutionary Ecology Research, 3, 51-74.
- 3. Clark, C.J., Poulsen, J.R. and Parker, V.T. (2001) The Role of Arboreal Seed Dispersal Groups on the Seed Rain of a Lowland Tropical Forest. Biotropica, 33, 606-620.
http://dx.doi.org/10.1111/j.1744-7429.2001.tb00219.x - 4. Herrera, C.M. (1995) Plant-Vertebrate Seed Dispersal Systems in the Mediterranean: Ecological Evolutionary and Historical Determinants. Annual Review of Ecology and Systematics, 26, 705-727.
http://dx.doi.org/10.1146/annurev.es.26.110195.003421 - 5. Chambers, J.C., Vander Wall, S.B. and Schupp, E.W. (1999) Seed and Seedling Ecology of Pinyon and Juniper Species in the Pygmy Woodland of Western North America. Botanical Review, 65, 1-38.
http://dx.doi.org/10.1007/BF02856556 - 6. Chavez-Ramirez, F. and Slack, R.D. (1994) Effects of Avian Foraging and Post-Foraging Behavior on Seed Dispersal Patterns of Ashe Juniper. Nordic Society Oikos, 71, 40-46.
http://dx.doi.org/10.2307/3546170 - 7. Vander Wall, S.B. and Balda, R.P. (1977) Coadaptations of the Clark’s Nutcracker and the Pinon Pine for Efficient Seed Harvest and Dispersal. Ecology, 47, 89-111.
http://dx.doi.org/10.2307/1942225 - 8. Balda, R.P. and Bateman, G.C. (1971) Flocking and Annual Cycle of Pinon Jay, Gymnorhinus cyanocephalus. Cooper Ornithological Society, 73, 287-302.
- 9. Neilson, K.P. (1987) On the Interface between Current Ecological Studies and the Paleobotany of Pinyon-Juniper Woodlands. In: Everett, R.L., Ed., Proceeding of the Pinyon-Juniper Conference, General Technical Report INT-215, US Department of Agriculture, Forest Service, Intermountain Research Station, Reno, 93-98.
- 10. Miller, R.F. and Wigand, P.E. (1994) Holocene Changes in Semiarid Pinyon-Juniper Woodlands. BioScience, 44, 465-474.
http://dx.doi.org/10.2307/1312298 - 11. Breshears, D.D., Cobb, N.S., Rich, P.M., Price, K.P., Allen, C.D., Balice, R.G., Romme, W.H., Kastens, J.H., Floyd, L.M., Belnap, J., Anderson, J.J., Myers, O.B. and Meyer, C.W. (2005) Regional Vegetation Die-Off in Response to Global-Change-Type Drought. Proceedings of the National Academy of Sciences of the United States of America, 102, 15144-15148.
http://dx.doi.org/10.1073/pnas.0505734102 - 12. Weisberg, P.J., Lingua, E. and Pillai, R.B. (2007) Spatial Patterns of Pinyon-Juniper Woodland Expansion in Central Nevada. Rangeland Ecology & Management, 60, 115-124.
http://dx.doi.org/10.2111/05-224R2.1 - 13. Mueller, R.C., Scudder, C.M., Porter, M.E., Trotter III, T.R., Gehring, C.A. and Whitham, T.G. (2005) Differential Tree Mortality in Response to Severe Drought: Evidence for Long-Term Vegetation Shifts. Journal of Ecology, 93, 1085-1093.
http://dx.doi.org/10.1111/j.1365-2745.2005.01042.x - 14. Breshears, D.D., Myers, O.B., Johnson, S.R., Meyer, C.W. and Martens, S.N. (1997) Differential Use of Spatially Heterogeneous Soil Moisture by Two Semiarid Woody Species: Pinus edulis and Juniperus monosperma. Journal of Ecology, 85, 289-299.
http://dx.doi.org/10.2307/2960502 - 15. Allen, C.D. and Breshears, D.D. (1998) Drought-Induced Shift of a Forest-Woodland Ecotone: Rapid Landscape Response to Climate Variation. Proceedings of the National Academy of Sciences of the United States of America, 95, 14839-14842.
http://dx.doi.org/10.1073/pnas.95.25.14839 - 16. Bradley, B.A. and Fleishman, E. (2008) Relationships between Expanding Pinyon-Juniper Cover and Topography in the Central Great Basin, Nevada. Journal of Biogeography, 35, 951-964.
http://dx.doi.org/10.1111/j.1365-2699.2007.01847.x - 17. Gray, S.T., Betancourt, J.L., Jackson, S.T. and Eddy, R.G. (2006) Role of Multidecadal Climate Variability in a Range Extension of Pinyon Pine. Ecology, 87, 1124-1130.
http://dx.doi.org/10.1890/0012-9658(2006)87[1124:ROMCVI]2.0.CO;2 - 18. Miller, R.E. and Rose, J.A. (1995) Historic Expansion of Juniperus occidentalis (Western Juniper) in Southeastern Oregon. Great Basin Naturalist, 55, 37-45.
- 19. Neubert, M.G., Kot, M. and Lewis, M.A. (1995) Dispersal and Pattern Formation in a Discrete-Time Predator-Prey Model. Theoretical Population Biology, 48, 7-43.
http://dx.doi.org/10.1006/tpbi.1995.1020 - 20. Holthuijzen, A.M.A. and Adkisson, C.S. (1984) Passage Rate, Energetics, and Utilization Efficiency of the Cedar Waxwing. The Wilson Bulletin, 96, 680-684.
- 21. Marsden, J.E. and Hoffman, M.J. (1987) Basic Complex Analysis. W.H. Freeman and Company, New York.
- 22. Powell, J.A. and Zimmermann, N.E. (2004) Multiscale Analysis of Active Seed Dispersal Contributes to Resolving Reid’s Paradox. Ecology, 85, 490-506.
http://dx.doi.org/10.1890/02-0535 - 23. Kot, M., Lewis, M.A. and van den Driessche, P. (1996) Dispersal Data and Spread of Invading Organisms. Ecology, 77, 2027-2042.
http://dx.doi.org/10.2307/2265698 - 24. Tausch, R.J. and West, N.E. (1988) Differential Establishment of Pinyon and Juniper Following Fire. American Midland Naturalist, 119, 174-184.
http://dx.doi.org/10.2307/2426066 - 25. Turchin, P. (1998) Quantitative Analysis of Movement: Measuring and Modeling Population Redistribution of Plants and Animals. Sinauer Associates, Sunderland, MA.
- 26. Suzan-Azpiri, H., Sanchez-Ramos, G., Martinez-Avalos, J.G., Villa-Melgarejo, S. and Franco, M. (2002) Population Structure of Pinus nelsoni Shaw, an Endemic Pinyon Pine in Tamaulipas, Mexico. Forest Ecology of Management, 165, 193-203.
http://dx.doi.org/10.1016/S0378-1127(01)00617-X - 27. Shaw, J.D., Steed, B.E. and DeBlander, L.T. (2005) Forest Inventory and Analysis (FIA) Annual Inventory Answers the Question: What Is Happening to Pinyon-Juniper Woodlands? Journal of Forestry, 103, 280-285.
- 28. Li, Z., Zou, J., Mao, K., Lin, K., Li, H., Liu, J., Kallman, T. and Lascoux, M. (2011) Population Genetic Evidence for Complex Evolutionary Histories of Four High Altitude Juniper Species in the Qinghai-Tibetan Plateau. Evolution, 66, 831-845.
- 29. Nowak, C.L., Nowak, R.S., Tausch, R.J. and Wigand, P.E. (1994) Tree and Shrub Dynamics in Northwestern Great Basin Woodland and Shrub Steppe during the Late-Pleistocene and Holocene. American Journal of Botany, 81, 265-277.
http://dx.doi.org/10.2307/2445452 - 30. Lanner, R.M. and Van Devender, T.R. (1998) The Recent History of Pinyon Pines in the American Southwest. In: Richardson, D.M., Ed., Ecology and Biogeography of Pinus, The Press Syndicate of the University of Cambridge, Cambridge, 171-182.
- 31. Vander Wall, S.B. and Balda, R.P. (1997) Dispersal of Singleleaf Pinon Pine (Pinus monophylla) by Seed-Caching Rodents. Journal of Mammalogy, 78, 181-191.
http://dx.doi.org/10.2307/1382651 - 32. Willson, M.F. (1993) Mammals as Seed-Dispersal Mutualists in North America. Oikos, 67, 159-176.
- 33. Garlick, M.J., Powell, J.A., Hooten, M.B. and McFarlane, L.R. (2010) Homogenization of Large-Scale Movement Models in Ecology. Society of Mathematical Biology, 73, 2088-2108.
http://dx.doi.org/10.1007/s11538-010-9612-6 - 34. Ascher, U.M. and Greif, C. (2011) A First Course in Numerical Methods. SIAM, Philadelphia.
Appendix A: Finite Difference Approximation
We will use two techniques to solve the dispersal model given by Equations (1) and (2) with corresponding seed digestion rate given by the Equation (3). First, we will solve this PDE numerically using finite difference approximations

where


Using these discretizations and approximating 

with variance σ chosen to be very small, the system (1) and (2) can be solved numerically provided 
Appendix B: Error Calculation for Seed Digestion Kernel with Step Function h(t)
The Laplace kernel used for comparison in the main text arises from solving Equations (1) and (2) with a con-
stant hazard function
lace kernel with the seed digestion kernel with step function 
approximation of the systems (1) and (2) with a step function failure rate
To calculate the difference between the Laplace and actual kernels, we first find the error in P and use it to calculate the error in S from the models (1) and (2). We denote the actual solution for P (with stepped hazard function) as 



equal when 



The solution for 



when 


when 


The error in P as




Thus, the error in S is

Finally we must calculate the error in the kernel (


We use the trapezoid rule to approximate the integral and note that we have not chosen scalings of time and space, so without loss of generality D = 1 and b = 20. In a spatial domain −30 < x < 30 the estimated 



Figure 7. Comparison of the Laplace kernel (solid line) and seed digestion kernel with step function h(t) (dash-dot line). The error is the difference between two graphs.
Figure 8. Calculation of the pointwise error generated using Laplace kernel approximation. The error is high near the center and it is decreasing towards both tails, but is always <10% of the calculated dispersal kernel.












