Advances in Bioscience and Biotechnology
Vol.3 No.8(2012), Article ID:25330,5 pages DOI:10.4236/abb.2012.38133
Vesicle-bound and free microRNAs in spent cell culture medium and biological fluids
![]()
Department of Medicine & Therapeutics, The Chinese University of Hong Kong, Hong Kong, China
Email: ccszeto@cuhk.edu.hk
Received 8 September 2012; revised 15 October 2012; accepted 20 November 2012
Keywords: Cell Signaling; Fibrosis; Cytokine
ABSTRACT
Background: In addition to the control of gene translation intra-cellularly, microRNAs (miRNAs) have been found to exist extracellularly. However, extracellular miRNA identified in previous studies were largely confined to microvesicles. It remains uncertain whether free extracellular miRNA exists. Methods: We quantify a panel of miRNAs (miRNA 200 family, miR-205, and miR-192) in spent culture medium of renal tubular epithelial cells, as well as serum and urine from patients with systemic lupus erythematosus and healthy controls. Microvesicle bound and free miRNA were separated by ultracentrifugation. Results: In spent cell culture medium, we found substantial amount of miR-200a, miR-200b, miR-200c, miR-141, miR-429, miR-205, and miR-192 in microvesicles, as well as the presence of miR-200a, miR-200b, miR-429, miR-205 and miR-192, but not miR-200c or miR-141 as nonvesicle bound free form. In healthy individuals, we found substantial amount of miR-200a, miR-200b, miR-200c, miR-141 and miR-192 in microvesicles from serum and urine, while only miR-141 exists as free form in both serum and urine. There was no significant difference in serum or urinary free miR-141 levels between SLE patients and healthy controls. In SLE patients, urinary free miR-141 level was significantly higher than serum. Neither serum nor urinary free miR-141 levels correlated with lupus disease activity. Conclusion: We found miRNA is present extracellularly, both within microvesicles and as free form, in spent cell culture medium, serum and urine. The biological role of extracellular miRNA requires further study.
1. INTRODUCTION
MicroRNAs (miRNAs) are a highly conserved family of short noncoding ribonucleic acid (RNA) molecules that regulate gene expression at the posttranscriptional level by degrading or repressing the translation of target messenger RNA (mRNA) [1,2]. The human genome encodes over 1000 miRNA species, which target over 60% of all protein-coding genes [3,4]. It is now clear that miRNAs are involved in almost every cellular process; dysregulation of miRNA has been associated with many human diseases [5].
Besides intracellular miRNAs with the traditional function of tranlation regulation, there is accumulating evidence that miRNAs exist extracellularly in body fluid [6,7]. Although the exact physiological function of extracellular miRNAs remains unclear, a number of studies suggested that circulating extracellular miRNAs may participate in intercellular communication and has the potential of being developed as biomarkers of various cancers and other diseases [8-12].
More recently, however, it is realized that there exists more than one form of extracellular miRNA in body fluids. Most of the circulating microRNAs described in previous studies actually locate in microvesicles or are combined with proteins attached to microvesicles [13]. On the other hand, other observations identified the existence of mature single-stranded miRNAs in serum that are not within microvesicles and do not bound to protein, suggesting a group of genuinely “free” miRNA [6,14]. However, the relationship between microvesicle-bound and free microRNAs, as well as their physiological significance, remains unknown.
We have previously identified miRNA 200 family, miR-205 and miR-192 in urine and serum of patients with systemic lupus erythematosus (SLE) and healthy controls, and found that these miRNA targets may participate in the pathogenesis of SLE [15]. However, the miRNAs we studied were probably microvesicle-related or protein-bound. In the present study, we quantify separately the vesicle-bound and free miRNA levels in spent culture medium and human.
2. MATERIALS AND METHODS
2.1. Cell Culture
Stabilized human proximal tubular epithelial cell (HK2) were purchased from the American Type Culture Collection (ATCC). They were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% heat inactivated fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 g/mL) antibiotic mixture at 37˚C under 5% CO2/95% air. All reagents were obtained from Invitrogen (Carlsbad, CA, USA). The culture medium was replaced every 2 days. Medium were collected in a 50 mL tube (BD Falcon) and stored at 4˚C for subsequent study.
2.2. Patients
We recruited 40 patients with systemic lupus erythematosus (SLE). All the patients were diagnosed according to the American College of Rheumatology diagnostic criteria and required maintenance immunosuppressive therapy. We also recruited 10 healthy subjects as controls. The study was approved by the Clinical Research Ethical Committee of the Chinese University of Hong Kong. All patients provided informed consent.
2.3. Sample Processing
A whole-stream early morning urine specimen and 5 mL of whole blood was collected from all recruited subjects. Cell medium, blood and urine samples were stored at 4˚C and processed within hours after collection. The specimens were centrifuged at 3000 g for 30 min at 4˚C. Then, 10 mL cell medium, 2 mL serum, or 2 mL urine supernatant was transferred in Eppendorf tubes and centrifuged at 12,000 g for 45 min at 4˚C. The resulting supernatant was further transferred to an ultrcentrifuge tube and centrifuged at 120,000 g for 2 hours at 4˚C. The final sediment (representing microvesicle pellets) and supernatant were separated with pipettor carefully and used for microRNA quantification. The microvesicle pellets were lysed by lysis buffer (Ambion, Inc., Austin, TX) in each ultracentrifuge tube and pooled together.
2.4. miRNA Preparation and Quantification
MirVana™ miRNA isolation kits (Ambion, Inc.) were used for the extraction of total RNA from microvesicle pellets according to the manufacturer’s protocol. The final ultrcentrifuge supernatant of HK2 cell medium, serum or urine was directly used as template for reverse transcription.
TaqMan® miRNA reverse transcription Kits (Applied Biosystems, Foster City, CA) were used for reverse transcription. Briefly, 1.67 μl total RNA from microvesicles or ultrcentrifuge supernatant was mixed with 1 μl specific primers, 0.05 μl 100 mM dNTPs (with dTTP), 0.5 μl 10× reverse transcription buffer, 0.33 μl (50 U) MultiScribe™ Reverse Transcriptase, 0.06 μl RNase inhibitor (20 U/μl), and made up to 5 μl with H2O. Reverse transcription was performed at 16˚C for 30 min, 42˚C for 30 min, and 85˚C for 5 min. The resulting cDNA was stored in −80˚C until use.
Levels of miR-200a, miR-200b, miR-200c, miR-141, miR-429, miR-205 and miR-192 were quantified by real time quantitative polymerase chain reaction (RT-QPCR) using the ABI Prism 7900 Sequence Detection System (Applied Biosystems). Commercially available Taqman primers and probes, including two unlabeled PCR primers and one FAM™ dye-labeled TaqMan® MGB probe were used for all the targets (all from Applied Biosystems). For RT-QPCR, 2.5 μl universal master mix, 0.25 μl primer and probe set, 0.33 μl cDNA, and 1.92 μl H2O were mixed to make a 5-μl reaction volume. Each sample was run in triplicate. RT-QPCR were performed at 50˚C for 2 min, 95˚C for 10 min, followed by 40 cycles at 95˚C for 15 s and 60˚C for 1 min. Because this study examined the miRNA level in cell-free and microvesicle-free biological fluids, which have no constant levels of any particular housekeeping RNA species, normalization of expression level by an endogenous control or housekeeping gene was not possible. As a result, identical volume of serum and urine supernatant was used for all samples, and the same baseline and threshold cycle were set for each target so that the level could be compared between samples. The levels of miRNA are therefore expressed as 50-CT as previously described [8].
2.5. Statistical Analysis
Statistical analysis was performed by SPSS for Windows software version 15.0 (SPSS Inc., Chicago, IL, USA). All the results were presented in mean ± SD unless otherwise specified. Since the miRNA levels and SLEDAI data are highly skewed, the Mann-Whitney U test was used to compare miRNA levels between groups, and Spearman’s rank correlation coefficient was used to test the association between miRNA levels and clinical parameters. A p-value < 0.05 was considered statistically significant. All probabilities were 2-tailed.
3. RESULTS
3.1. Vesicle-Bound and Free miRNAs in Spent Cell Culture Medium
The vesicle-bound and free miRNA levels in spent cell culture medium are quantified and summarized in Figure 1. In essence, there was substantial amount of miR-200a, miR-200b, miR-200c, miR-141, miR-429, miR-205 and
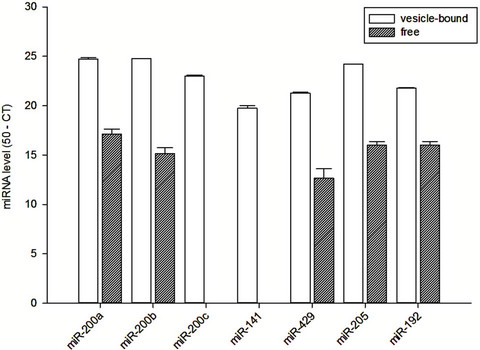
Figure 1. Vesicle-bound and free miRNA levels in the spent culture medium of HK-2 cells.
miR-192 in microvesicles from spent cell culture medium. In contrast, we found no detectable amount of free miR-200c and miR-141 from the spent cell culture medium, but substantial amount of free miR-200a, miR- 200b, miR-429, miR-205 and miR-192 could be identified. There was no significant correlation between the vesicle-bound and free miRNA levels in the spent culture medium (details not shown).
3.2. Vesicle-Bound and Free miRNAs in Healthy Subjects
The vesicle-bound and free miRNA levels in the serum and urine of healthy volunteers are quantified and summarized in Figure 2. In serum, there was substantial amount of miR-200a, miR-200b, miR-200c, miR-141 and miR-192 in microvesicles from serum. In contrast, only miR-141 could be found as free form in the serum (Figure 2(a)).
In urine, there was substantial amount of vesiclebound miR-200a, miR-200b, miR-200c, miR-141, miR- 429, miR-205 and miR-192 (Figure 2(b)). In contrast, only miR-141 could be found as free form in the urine. There was no significant correlation between the vesicle-bound and free miRNA levels in the serum or urine (details not shown).
3.3. Serum and Urinary Free miR-141 in SLE Patients
Since only free miR-141 could be detected in serum and urine, we went on and studied 40 SLE patients. Their baseline clinical data are compared to the healthy controls and summarized in Table 1. In essence, the SLE patients were older and had more proteinuria than the controls.
The serum and urinary free miR-141 levels of the two groups are summarized in Figure 3. We found no sig-
 (a)
(a) (b)
(b)
Figure 2. Vesicle-bound and free miRNA levels in (a) serum; and (b) urine of healthy individuals.

Table 1. Baseline demographic and clinical data.
nificant difference in the serum free miR-141 levels between SLE patients and controls (9.30 ± 0.81 vs 9.30 ±

Figure 3. Comparison of serum and urinary free miR-141 levels between patients with systemic lupus erythematosus (SLE) and healthy controls. Data are compared by Mann-Whitney U test.
2.05, p = 0.7). Similarly, there was no significant difference in urinary free miR-141 levels between SLE patients and controls (10.48 ± 0.78 vs 6.48 ± 2.46, p = 0.3). The level of miR-141 in urine supernatant without microvesicles was significantly higher than that in serum supernatant without microvesicles (9.82 ± 0.79 vs 9.30 ± 0.75, p = 0.03). When the SLE patients were analyzed alone, there was no significant correlation between serum or urinary free miR-141 levels and the degree of lupus activity, as represented by proteinuria, renal function, serum complement level, and anti-ds DNA antibody titre (details not shown).
4. DISCUSSION
In this study, we investigated the expression of miR-200 family, miR-205, and miR-192 in microvesicles from spent cell culture medium, as well as serum and urine of human subjects. In line with previous studies [16-18], we found a substantial amount of miRNAs in microvesicles. We also found that miRNA species in microvesicles originate from different liquid are not the same. For instance, while all seven species of studied miRNAs are detected in microvesicles from spent cell culture medium as well as urine, two of them (miR-429 and miR-205) were not present in microvesicles from serum. The result suggests microvesicles derived from serum and urine have different functions. Our result also implies that microvesicle-related miRNA in urine does not come from glomerular filtration of the systemic circulation, but, rather, are locally produced from renal parenchymal cells (for example, renal tubular epithelial cells). Our observation also provides indirect evidence to suppport the hypothesis that microvesicle-related miRNA are possible mediators of intercellular communication.
In this study, we identified five miRNAs (i.e. miR200a, miR200b, miR429, miR205, miR-192) in the microvesicle-free supernatant of spent cell culture medium. In contrast, only miR-141 was detected in the microvesicle-free supernatant of serum and urine. Although the CT values of these targets were high (around 31 to 38), they had typical amplification plots and, therefore, false positive is unlikely. These results suggest the microvesiclefree miR-141 we detected in serum and urine is not from renal tubular cells. The origin of the microvesicle-free miR-141 in serum and urine needs further investigation.
The nature and origin of extracellular microvesiclefree miRNA remain unknown. Since not every miRNA target in microvesicle presents as free form, it is unlikely that free miRNA in serum and urine supernatant originates from the passive of damaged microvesicles. A recent study showed that most of the extracellular miRNA outside microvesicles are combined with Ago2 protein and are most likely by-products of dead cells [13]. However, the possible existence of free single-stranded miRNA outside cells and microvesicles could not be excluded [13]. There is evidence suggesting that some cells could actively secret free miRNAs [7]. This hypothesis is further supported by a previous study that used serum as direct template for polymerase chain reaction [6]. More recently, Zhang et al. [14] showed that exogenous plant miRNAs are present in the sera and tissues of various animals. More importantly, these exogenous plant miRNAs are primarily acquired orally through food intake, and are therefore unlikely to be microvesicle-bound miRNA. Further studies are needed to test the existence of free single-stranded miRNAs in other body fluids and determine how single-stranded miRNA can survive abundant RNase degradation without the protection by microvesicles or proteins.
In this series of work, we further explored the potential biological relevance of free miR-141, which is the only miRNA that can be detected in urine and serum from the first part of our study. In short, we found there is no significant difference between serum free miR-141 levels between SLE patients and healthy controls, and there was no significant correlation between serum or urinary free miR-141 levels and any clinical parameter. However, SLE patients actually had a marginally higher urinary free miR141 levels than the control group (see Figure 3), and a genuine difference may be missed because of the small sample size in our study. In addition, for each individual patient, urinary free miR-141 level was almost always slightly higher than the corresponding serum level, again suggesting that cells in the kidney and/or urinary tract are involved in production of extracellular free miR-141 in the urine.
In summary, we found miRNA is present extracellularly, both within microvesicles and as free form, in spent cell culture medium, serum and urine. The biological role of extracellular miRNA requires further study.
5. ACKNOWLEDGEMENTS
GW, BCHK and CCS designed the experiments; GW and KBL performed the laboratory experiments; GW, KMC and CCS performed data analysis; GW and CCS wrote the manuscript; PKTL took care of the administrative and regulatory aspect of the study. This work was supported in part by the CUHK research accounts 6901031 and 7101215. All authors declare we have no conflict of interest.
REFERENCES
- Bartel, D.P. (2009) MicroRNAs: Target recognition and regulatory functions. Cell, 136, 215-233. doi:10.1016/j.cell.2009.01.002
- Bartel, D.P. (2004) MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116, 281-297. doi:10.1016/S0092-8674(04)00045-5
- Bentwich, I., Avniel, A., Karov, Y., Aharonov, R., Gilad, S., Barad, O., et al. (2005) Identification of hundreds of conserved and nonconserved human microRNAs. Nature Genetics, 37, 766-770. doi:10.1038/ng1590
- Lewis, B.P., Burge, C.B. and Bartel, D.P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120, 15-20. doi:10.1016/j.cell.2004.12.035
- Jiang, Q., Wang, Y., Hao, Y., Juan, L., Teng, M., Zhang, X., et al. (2009) miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Research, 37, D98-D104. doi:10.1093/nar/gkn714
- Chen, X., Ba, Y., Ma, L., Cai, X., Yin, Y., Wang, K., et al. (2008) Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research, 18, 997-1006. doi:10.1038/cr.2008.282
- Zen, K. and Zhang, C.Y. (2012) Circulating MicroRNAs: A novel class of biomarkers to diagnose and monitor human cancers. Medicinal Research Reviews, 32, 326-348. doi:10.1002/med.20215
- Gilad, S., Meiri, E., Yogev, Y., Benjamin, S., Lebanony, D., Yerushalmi, N., et al. (2008) Serum microRNAs are promising novel biomarkers. PLoS One, 3, e3148. doi:10.1371/journal.pone.0003148
- Mitchell, P.S., Parkin, R.K., Kroh, E.M., Fritz, B.R., Wyman, S.K., Pogosova-Agadjanyan, E.L., et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences USA, 105, 10513-10518.
- Hunter, M.P., Ismail, N., Zhang, X., Aguda, B.D., Lee, E.J., Yu, L., et al. (2008) Detection of microRNA expression in human peripheral blood microvesicles. PLoS One, 3, e3694. doi:10.1371/journal.pone.0003694
- Yuan, A., Farber, E.L., Rapoport, A.L., Tejada, D., Deniskin, R., Akhmedov, N.B., et al. (2009) Transfer of microRNAs by embryonic stem cell microvesicles. PLoS One, 4, e4722. doi:10.1371/journal.pone.0004722
- Valadi, H., Ekstrom, K., Bossios, A., Sjostrand, M., Lee, J.J. and Lotvall, J.O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, 654-659. doi:10.1038/ncb1596
- Turchinovich, A., Weiz, L., Langheinz, A. and Burwinkel, B. (2011) Characterization of extracellular circulating microRNA. Nucleic Acids Research, 39, 7223-7233. doi:10.1093/nar/gkr254
- Zhang, L., Hou, D., Chen, X., Li, D., Zhu, L., Zhang, Y., et al. (2012) Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Research, 22, 107-126. doi:10.1038/cr.2011.158
- Wang, G., Tam, L.S., Li, E.K., Kwan, B.C., Chow, K.M., Luk, C.C., et al. (2011) Serum and urinary free microRNA level in patients with systemic lupus erythematosus. Lupus, 20, 493-500. doi:10.1177/0961203310389841
- Camussi, G., Deregibus, M.C. and Tetta, C. (2010) Paracrine/endocrine mechanism of stem cells on kidney repair: Role of microvesicle-mediated transfer of genetic information. Current Opinion in Nephrology and Hypertension, 19, 7-12. doi:10.1097/MNH.0b013e328332fb6f
- Hata, T., Murakami, K., Nakatani, H., Yamamoto, Y., Matsuda, T. and Aoki, N. (2010) Isolation of bovine milkderived microvesicles carrying mRNAs and microRNAs. Biochemical and Biophysical Research Communications, 396, 528-533. doi:10.1016/j.bbrc.2010.04.135
- Muralidharan-Chari, V., Clancy, J.W., Sedgwick, A. and D’Souza-Schorey, C. (2010) Microvesicles: Mediators of extracellular communication during cancer progression. Journal of Cell Science, 123, 1603-1611. doi:10.1242/jcs.064386

