Advances in Bioscience and Biotechnology
Vol.1 No.3(2010), Article ID:2360,10 pages DOI:10.4236/abb.2010.13021
In vitro demonstration of interactions among zinc-binding domains of cellulose synthases in Arabidopsis and aspen
1Biotechnology Research Center, School of Forest Resources and Environmental Science, Michigan Technological University, Houghton, USA;
2Department of Bioenergy Science and Technology, Chonnam National University, Gwangju, Korea.
Email: cpjoshi@mtu.edu
Received 16 April 2010; revised 25 May 2010; accepted 28 May 2010.
Keywords: Arabidopsis; Cellulose Synthase; Poplar; Protein-Protein Interaction; Yeast Two-Hybrid System; Zinc-Binding Domain
ABSTRACT
Plant cellulose synthases (CesAs) are the key enzymes necessary for cellulose biosynthesis. In Arabidopsis, two distinct groups of three CesAs each are necessary for cellulose synthesis during primary and secondary cell wall formation. It has also been suggested that such three CesAs interact with each other to form plasma-membrane bound rosette complexes that are functional during cellulose production. However, in vivo demonstration of such assemblies of three CesAs into rosettes has not been possible. We used yeast two-hybrid assays to demonstrate the possible interactions among several CesAs from Arabidopsis and aspen via their N-terminal zinc-binding domains (ZnBDs). While strong positive interactions were detected among ZnBDs from secondary wall associated CesAs of both Arabidopsis and aspen, the intergeneric interactions between Arabidopsis and aspen CesAs were weak. Moreover, in aspen, three primary wall associated CesA ZnBDs positively interacted with each other as well as with secondary CesAs. These results suggest that ZnBDs from either primary or secondary CesAs, and even from different plant species could interact but are perhaps insufficient for specificities of such interactions among CesAs. These observations suggest that some other more specific interacting regions might exist within CesAs. It is also possible that some hitherto unknown mechanism exists in plants for assembling the rosette complexes with different compositions of CesAs. Understanding how cellulose is synthesized will have a direct impact on utilization of lignocellulosic biomass for bioenergy production.
1. INTRODUCTION
Cellulose, the most abundant renewable biopolymer on earth, has a great potential to be used as a major raw material for bioenergy production. Cellulose synthases (CesA) play a pivotal role in the process of cellulose biosynthesis during the formation of primary and secondary cell walls of higher plants [1-2] . A better understanding of the process of cellulose biosynthesis will, therefore, have a far-reaching impact on agricultural and forest product industries that utilize cellulose as a main raw material. However, the precise molecular mechanism of cellulose biosynthesis has so far eluded scientists for a long time [3], and it is still unclear how plasma membrane bound CesA complexes regulate economically important characteristics of cellulose such as degree of polymerization, microfibril angle and crystallinity. It has been suggested that large enzyme complexes (> 500 kDa) consisting of 36 CesA subunits, arranged in the form of rosettes, synthesize about 36 glucan fibers-thick cellulose microfibrils at the plasma membrane [4]. Based on immunogold labeling experiments, CesAs appear to be a component of rosette complexes [5] although actual visualization of the 36 CesAs present in the rosettes has never been possible due to substantial technical limitations [6].
The Arabidopsis genome contains ten CesAs, at least six of which have been identified to be involved in cellulose biosynthesis based on mutant analysis: three (AtCesA1, 3 and 6) associated with primary cell wall formation and three (AtCesA4, 7, and 8) involved in secondary wall formation [1]. Such CesA mutant analysis in Arabidopsis has further delineated that the wild-type or unmutated status of these CesA proteins is essential for normal cellulose synthesis during primary or secondary wall formation, and that any such mutations in CesAs adversely affect the cellulose biosynthetic processes during the respective cell wall development [7-12] . Three secondary wall associated CesAs (designated as secondary CesAs here) have been shown to be coordinately expressed during the secondary wall-enriched xylem formation and appear to be interacting with each other based on pull-down assays [10]. Moreover, mutation in any one of these three secondary CesAs resulted in failure of interactions with the other two CesAs [13]. Using co-immunoprecipitation and bimolecular fluorescence complementation it has been proved that in primary wall, AtCesA1 and AtCesA3 were required for composing the complex and AtCesA6 was partially needed but redundant with AtCesA2 and AtCesA5 [14] . Similar suggestions of possible involvement of CesA trios in secondary wall formation have been reported in rice [15], barley [16], aspen [17-18] , and Eucalyptus [19] although such involvement has been suggested only on the basis of coordinate expression of three CesAs in respective cell wall forming plant cells.
The major question of how some of the three CesAs involved in secondary cell wall formation might be interacting with each other has been addressed in cotton using an in vitro approach with the help of yeast two-hybrid (Y2H) system [20]. Results of Y2H and co-immunoprecipitation assays showed that cotton GhCesA1 and GhCesA2 proteins, orthologous to Arabidopsis AtCesA8 and AtCesA4, respectively physically interact with each other to form homoor heterodimers via their N-terminal zinc-binding domains (ZnBDs). Furthermore, CesA dimerization is important for the stability of the CesA complexes so reduced monomers were degraded rapidly while oxidized CesA dimers were resistant to degradation during in vivo and in vitro conditions [21]. It has been recently reported that the secondary CesA trios from Arabidopsis can interact with each other, but it seems that only AtCesA4 can form homodimers [22]. However, Atanassov et al. [23] successfully purified secondary wall specific CesA complex without other contaminating proteins and also showed that all three subunits are able to form homodimers independently. No complete interactions among primary and/or secondary CesA trios through Y2H experiments have been reported in Arabidopsis or any other plants.
In this study, we show that ZnBDs from AtCesA4, 7 and 8 from Arabidopsis positively interact with each other in the Y2H system and are capable of forming homodimers and heterodimers. We have previously cloned and characterized aspen orthologs of the same three secondary CesA genes (PtrCesA1, 2 and 3) that are coordinately expressed during xylem development [17-18] . We also demonstrate here that ZnBDs from PtrCesA1, 2 and 3 also strongly interact with each other. Furthermore, ZnBDs from 3 Arabidopsis secondary CesAs also interact with 3 aspen secondary CesAs, although much more weakly than the interactions among CesAs from the same species. Finally, we also demonstrate that ZnBDs from three aspen primary CesAs (PtrCesA4, 5 and 6), orthologous to three Arabidopsis primary CesAs (AtCesA1, 3 and 6), interact successfully with each other as well as with the three secondary CesAs from aspen, thus suggesting that ZnBDs are insufficient to confer specific interactions among CesAs .
2. MATERIALS AND METHODS
2.1. Plasmid Constructs
The cDNA regions encoding ZnBD regions of three secondary CesAs from Arabidopsis (GenBank accession nos. AtCesA4: AF458083, AtCesA7: AF088917, AtCesA8: AF267742) and six CesAs from aspen (Populus tremuloides) (GenBank accession nos. PtrCesA1: AF072131, PtrCesA2: AY095297, PtrCesA3: AF527387, PtrCesA4: AY162181, PtrCesA5: AY055724, PtrCesA6: AY196961) were PCR amplified and cloned into pGBKT7 DNA binding vector (BD) and pGADT7 activation vector (AD) from Matchmaker Two-Hybrid System 3 (Clontech, Palo Alto, CA). Similarly, the zinc binding domain-like region of one cellulose synthase-like D cDNA (PtrCSLD) from aspen (AY162184) was also cloned into these two AD and BD vectors. Since PtrCSLD does not contain a legitimate ZnBD, it was expected that inclusion of such PtrCSLD constructs will provide an additional negative control for Y2H experiments. All vector constructs prepared and primer pairs used in this study are summarized in Tables 1 and 2.
2.2. Yeast Two-Hybrid Assays
The GAL4 fusion protein-based Matchmaker Twohybrid System 3 from Clontech was used for yeast two-hybrid assays according to the manufacturer’s instructions. This system is a highly improved version of the original Y2H system that has been reengineered to significantly reduce the number of false positives. All BD-CesA plasmids were checked for the lack of autonomous activation of reporter genes in the yeast host strain AH109 provided by Clontech. Positive clones were selected by growth on a stringent medium (-AdeHis-Leu-Trp) and also by activation of α-galactosidase in the presence of X-α-Gal chromogenic substrate imparting blue color to yeast colonies.
Yeast transformations were performed by a smallscale lithium acetate method as recommended by the manufacturer. AH109 co-transformants containing empty GAL4AD and GAL4BD vectors as well as Lamin C and SV40 large T-antigen fusion constructs were used as negative controls, while vectors carrying p53 and the SV40 large T-antigen fusion constructs were used as positive controls.
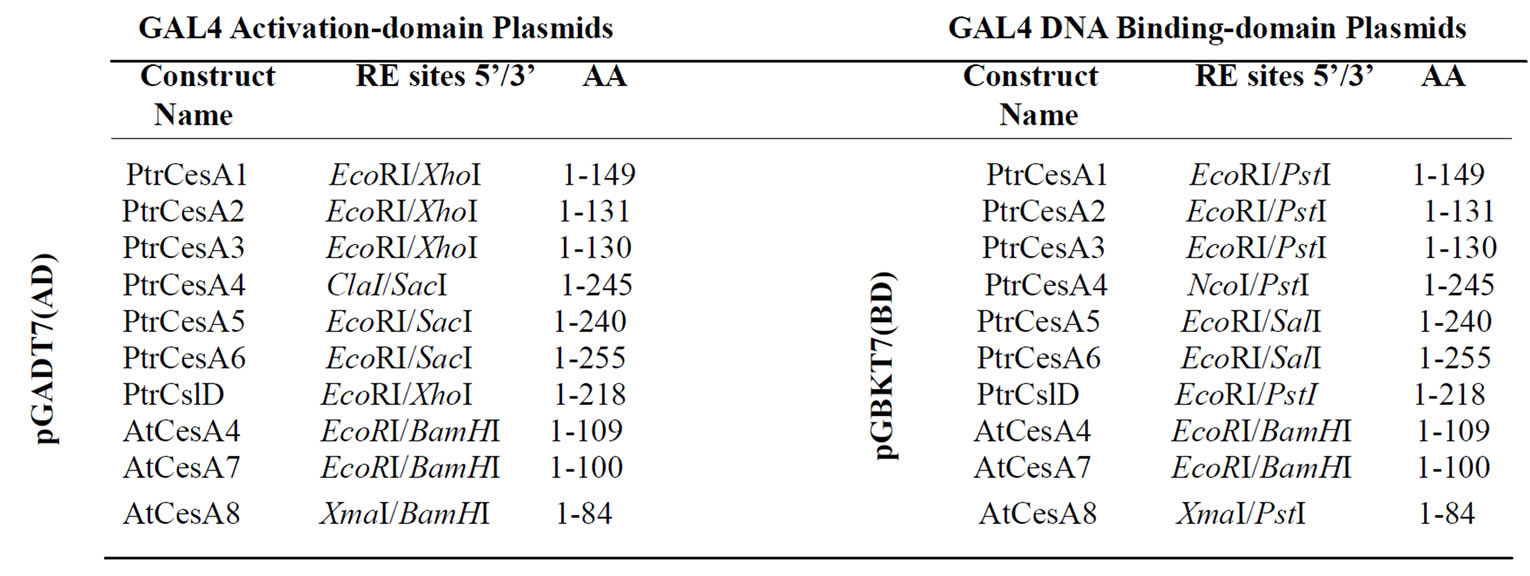
Table 1. Vector constructs for yeast two-hybrid assays of ZnBDs from CesAs and CSLD.

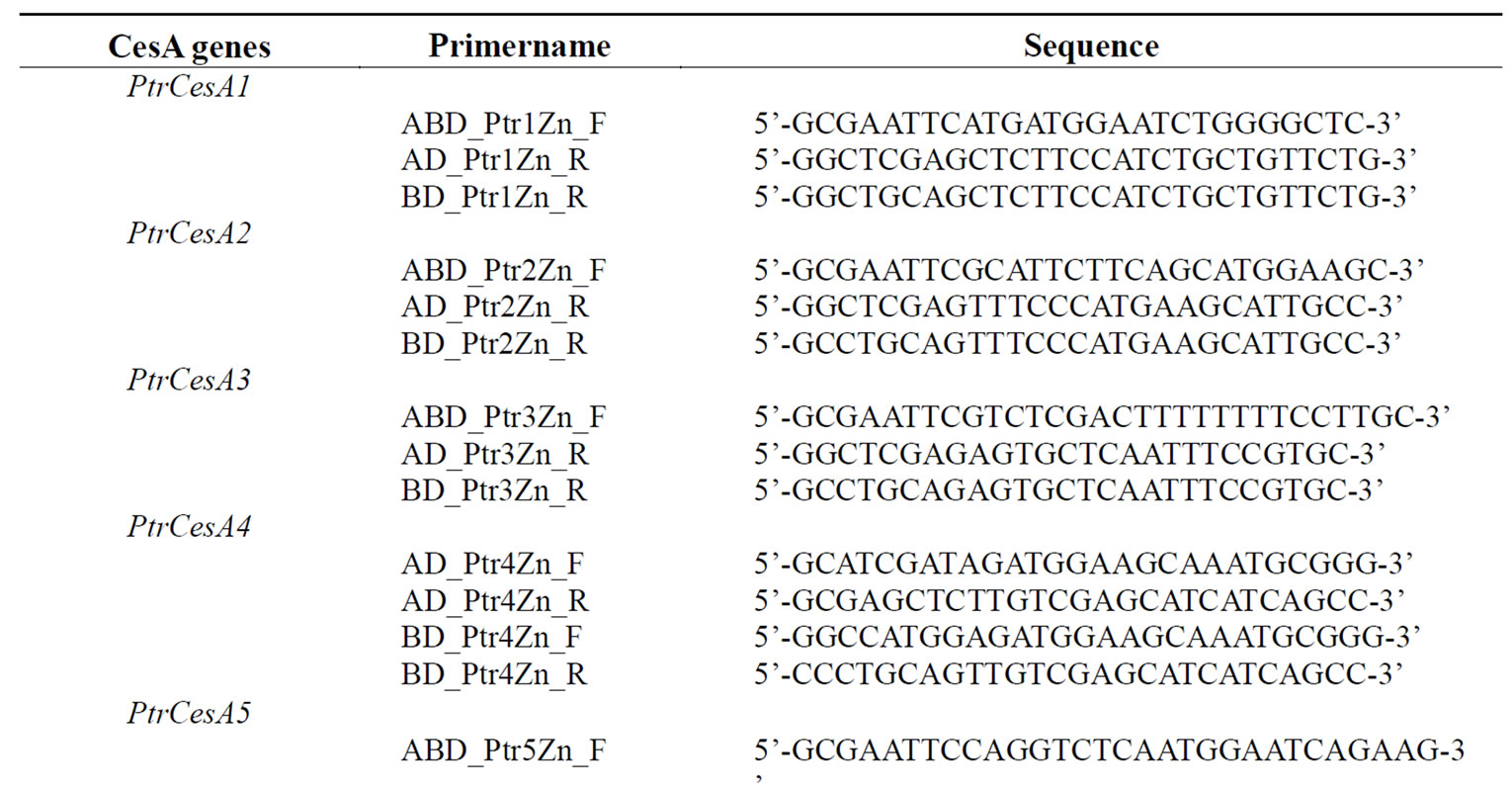
Table 2. Primers used for cloning ZnBDs of CesA cDNAs in AD and BD vectors in Y2H.
2.3. Analysis of interaction strengths between CesA pairs via α-Galactosidase activity assays
The relative strength of interactions among ZnBDs from CesAs was analyzed by a quantitative α-galactosidase (α-Gal) assay (Clontech). All combinations of CesAs used for measuring the strength of interactions were the same as described above. α-Gal activity [milliunits/(ml × cell)] was calculated according to Yeast Protocols Handbook from Clontech.
2.4. In Vitro Transcription/Translation
Two pairs of ZnBDs from primary and secondary CesA proteins (PtrCesA1 and PtrCesA4; PtrCesA3 and PtrCesA6) were synthesized by in vitro transcription/ translation using the TNT® T7 Quick Coupled Transcription/Translation System (Promega, Madison, WI) and the fusion constructs obtained from the Y2H assays as described above. Synthesized proteins from pGBKT7 and pGADT7 vectors carried c-Myc and HA tags, respectively.
2.5. Co-Immunoprecipitation (Co-IP) and Immunoblotting
Matchmaker CO-IP kit (Clontech, Palo Alto, CA) was used for the co-immunoprecipitation following the manufacture’s protocol. Protein-protein interactions were confirmed based on the presence of BD constructs with c-Myc tags in the CO-IP reactions that were incubated with HA antibodies. Therefore, c-Myc antibody was used as the primary antibody for immunoblotting analysis. C-Myc-tagged proteins were visualized using secondary alkaline phosphatase (AP)-conjugated anti-mouse IgG (Sigma) antibody and BCIP/NBT as substrate.
3. RESULTS AND DISCUSSION
3.1. Interactions among Secondary Cell Wall-Specific CesAs
Previous genetic experiments with three cellulose synthesis-defective mutants from Arabidopsis, namely, irx1, irx3, and irx5 exhibiting irregular xylem phenotypes have suggested that the presence of at least three secondary CesAs is essential for cellulose biosynthesis in secondary cell wall forming cells [9-11] . Two independent studies based on publicly available microarray data have further established that AtCesA4, 7 and 8 coordinately express during secondary cell wall development [24-25] . However, it is currently unknown whether these three Arabidopsis CesAs can actually form homoas well as heterodimers in vitro via their ZnBDs as was suggested by Y2H results from using two cotton CesAs, GhCesA1 and GhCesA2 [20]. Such understanding has a direct impact on our concepts of rosette formation including the number and types of CesAs interacting during cellulose biosynthesis in higher plants. Moreover, no cotton ortholog of AtCesA7 has yet been reported and such interactions among three CesAs using the Y2H system have never been studied either in cotton or in Arabidopsis or any other plant. Therefore, we first set out to determine whether ZnBDs from AtCesA4, 7 and 8 can physically interact with each other and whether they are capable of forming homoas well as heterodimers in the Y2H system. In order to examine the existence of such interactions, ZnBD regions from AtCesA4, AtCesA7 and AtCesA8 were cloned in both activation domain (AD) and binding domain (BD) vectors from Clontech’s Matchmaker system 3 (Tables 1 and 2). In all co-transformants Gal4-fusion proteins containing AtCesA ZnBD regions formed protein-protein interactions resulting in the activation of transcription of the a-galactosidase reporter gene and expression of histidine and adenine (Figure 1). These results show that all three secondary AtCesAs were able to interact with themselves to form homodimers and interacted with the remaining two CesAs to form heterodimers. Kurek et al. have previously confirmed the validity of such interactions through pull-down assays with recombinant GhCesA1 and GhCesA2 [20]. Therefore, rosettes in secondary wall forming Arabidopsis cells most likely contain at least up to 3 CesAs.
Similar to Arabidopsis secondary CesAs, ZnBDs of all three aspen homologous secondary CesAs were also able to interact with each other, and thus were able to form homoand heterodimers (Table 3). This agrees with the coordinate expression of three secondary CesAs that we observed in aspen ADDIN EN.CITE ADDIN EN.CITE.DATA [17-18] . There is also a general opinion that Y2H leads to too many false positives. However, the Y2H system that we have currently used is a highly improved version of traditional Y2H system and this advanced system drastically reduces false positives. The current version 3 utilizes four strict nutritional selection markers and successful interactions can be monitored by expression of a visible marker gene. The general impression of high rate of false positives stems from the library screening of Y2H assays used to identify unknown members interacting with the bait proteins. In our case, however, both the bait and prey proteins are already well-characterized and we are confirming whether such CesA interactions occur via ZnBDs or not. We successfully used all the positive and negative controls that were provided with the Matchmaker Two-hybrid System 3 (Figure 1). In addition, the negative controls using empty AD/BD vectors showed that none of the interactions were caused by self-activation. Furthermore, failure of interactions between the PtrCslD and PtrCesAs demonstrated that the zinc binding-like domain in PtrCslD was unable to form dimers with the CesA ZnBDs (Table 3), thus serving as another biological negative control. Collectively, use of all these control experiments strongly support the notion that ZnBDs of
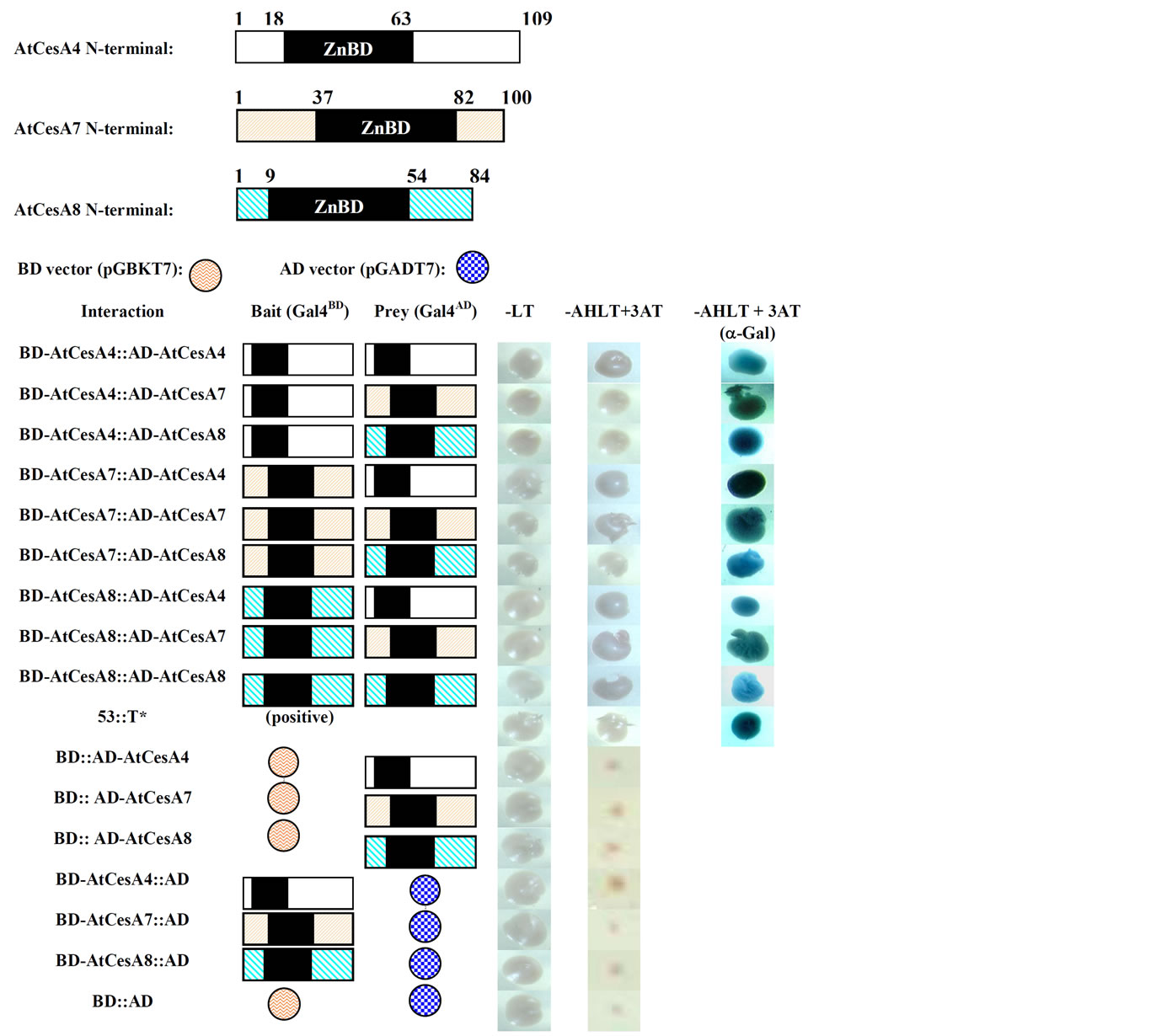
Figure 1. Interactions among the ZnBDs of AtCesA4, AtCesA7 and AtCesA8. Schematic representation of the AtCesA4, AtCesA7 and AtCesA8 N-terminal regions containing ZnBDs is shown at the top. Yeast strains transformed with the plasmids containing the GAL-4 binding domain (GAL4BD: bait) and the GAL-4 activating domain (GALAD: prey), as indicated, were grown on synthetic dropout media lacking Leu and Trp (-LT), and lacking Ade, His, Leu, Trp and 5mM 3-amino1, 2, 4-triazole (-AHLT+3AT), respectively. a-Galactosidase activity (blue colonies) was assayed on –AHLT medium.
these three aspen secondary CesAs are able to physically interact with each other in vitro and these domains might be involved informing a functional terminal complex in vivo. Unlike in Arabidopsis, isolation of CesA mutations has not been reported in aspen and doing in vivo experiments of mutant complementation or protein-protein interactions are very difficult to perform using aspen system because of its long life cycle.
High similarity of ZnBDs of Arabidopsis and aspen secondary CesAs also prompted us to examine whether ZnBDs from Arabidopsis and aspen secondary CesAs could interact with each other in the Y2H system. As shown in Table 3, CesAs from aspen and Arabidopsis were also able to positively interact with each other. However, the time required to develop the blue color indicative of a-galactosidase activity was much longer in the case of intergeneric CesA interactions than the CesA interactions within the same species. Thus these qualitative estimates suggested that intergeneric CesA interactions may be weaker than the intraspecies CesA interactions.
3.2. Relative Strength of Interactions between Secondary CesAs
In order to quantitatively determine the relative strength of interactions between various secondary CesAs from Arabidopsis and aspen, the expression level of the MEL1 reporter gene was analyzed through measuring a-galactosidase (a-Gal) activity secreted in yeast cell supernatants (Figure 2). The α-Gal activity ranged from 229 to 354
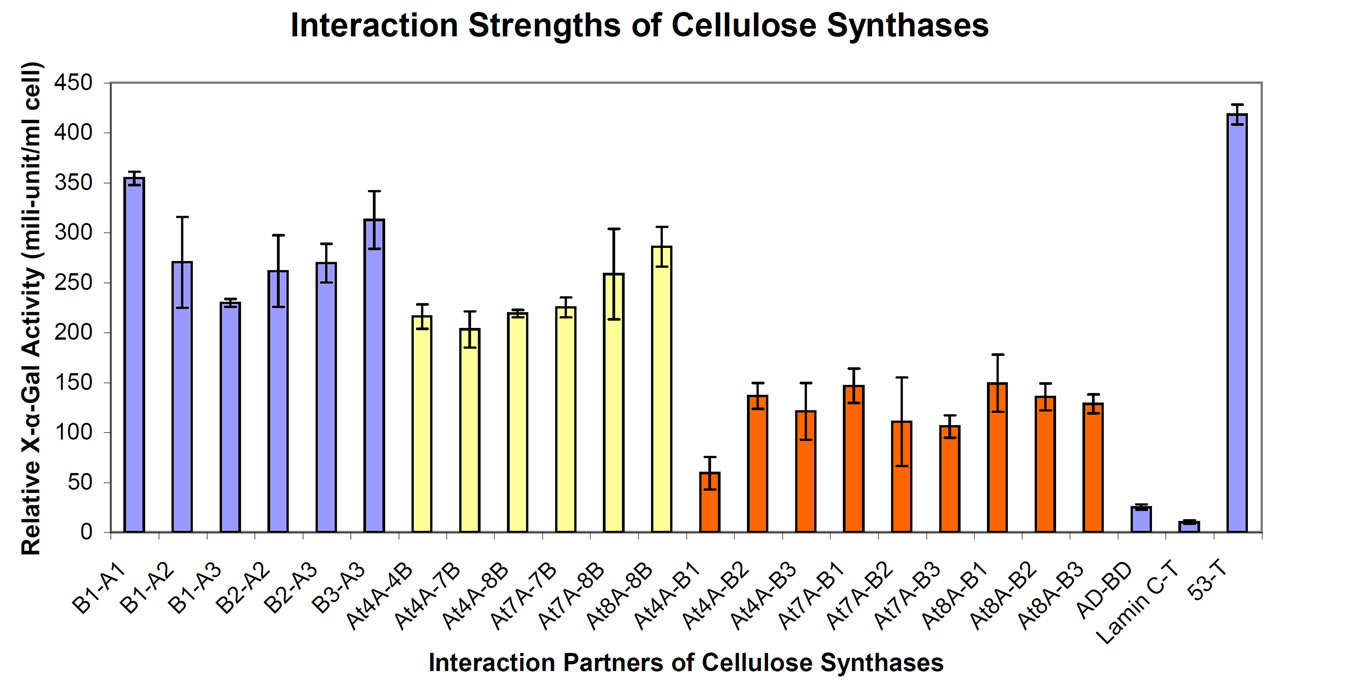
Figure 2. Quantitative estimation of interaction strength between secondary CesA ZnBDs. Results shown are the average α-Gal activity of three cultures for each paired construct, assayed in triplicate. Error bars are standard errors (S.E.). B1-A1: BD-PtrCesA1 and AD-PtrCesA1; B1-A2: BD-PtrCesA1 and AD-PtrCesA2; B1-A3: BD-PtrCesA1 and AD-PtrCesA3; B2-A2: BD-PtrCesA2 and AD-PtrCesA2; B2-A3: BD-PtrCesA2 and AD-PtrCesA3; B3-A3: BD-PtrCesA3 and AD-PtrCesA3; At4A-4B: AD-AtCesA4 and BD-AtCesA4; At4A-7B: AD-AtCesA4 and BD-AtCesA7; At4A-8B: AD-AtCesA4 and BD-AtCesA8; At7A-7B: AD-AtCesA7 and BD-AtCesA7; At7A-8B: AD-AtCesA7 and BD-AtCesA8; At8A-8B: AD-AtCesA8 and BD-AtCesA8; At4A-B1: AD-AtCesA4 and BD-PtrCesA1; At4A-B2: AD-AtCesA4 and BD-PtrCesA2; At4A-B3: AD-AtCesA4 and BD-PtrCesA3; At7A-B1: AD-AtCesA4 and BD-PtrCesA1; At7A-B2: AD-AtCesA7 and BD-PtrCesA2; At7A-B3: AD-AtCesA7 and BD-PtrCesA3; At8A-B1: AD-AtCesA8 and BD-PtrCesA1; At8A-B2: AD-AtCesA8 and BD-PtrCesA2; At8A-B3: AD-AtCesA8 and BD-PtrCesA3; AD-BD (negative control): pGADT7 and pGBKT7; Lamin C-T (negative control): AD-Lamin C and BD-SV40 large T-antigen; p53-T (positive control): AD-p53 and BD-SV40 large T-antigen.
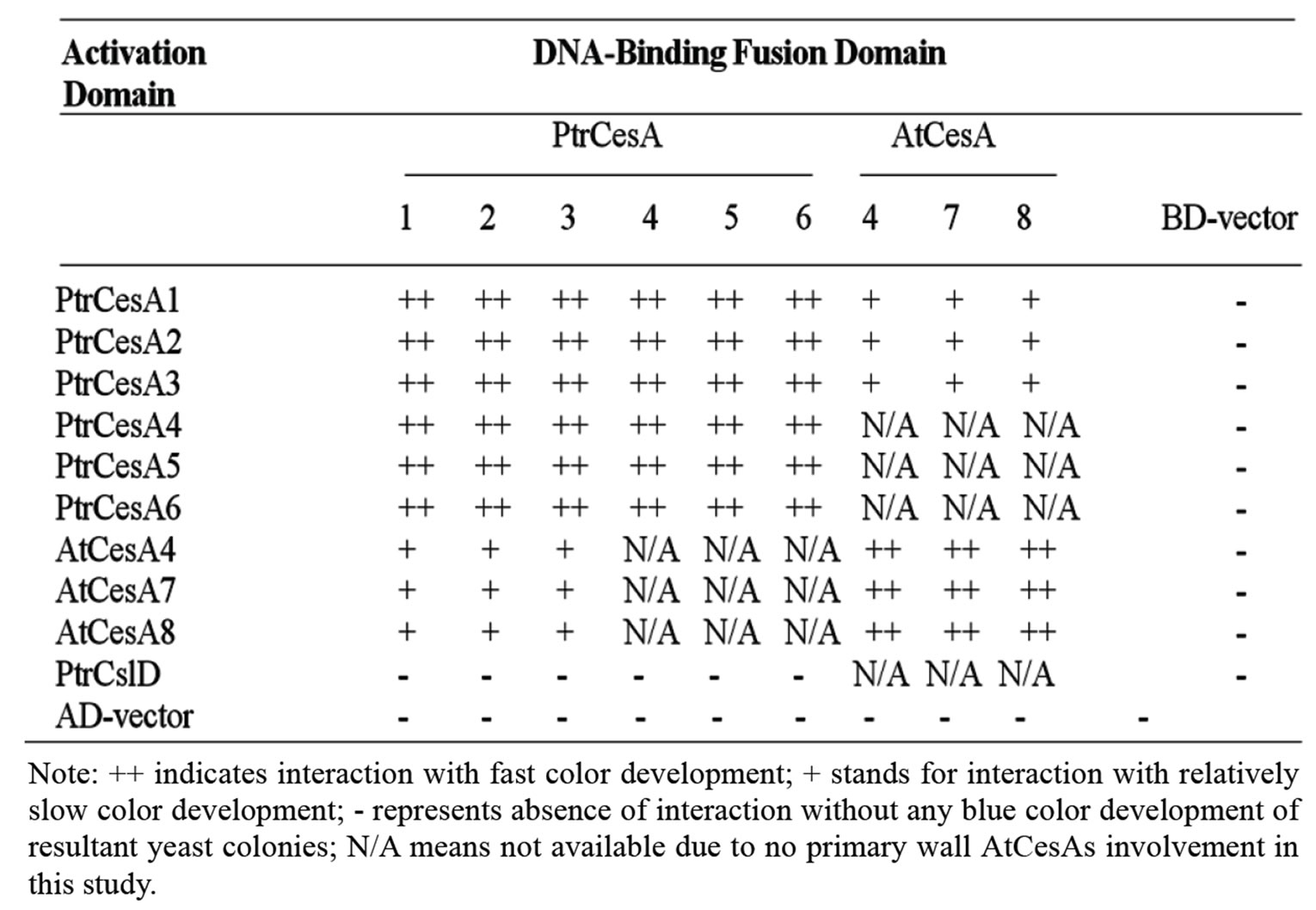
Table 3. Interactions among ZnBDs of PtrCesA1-6 and AtCesA4, 7, 8 with the Y2H system.
milliunits/(ml × cell) for the yeast cultures containing CesA interaction partners from aspen, from 216 to 285 milliunits /(ml × cell) for interacting Arabidopsis CesAs, and only from 59 to 148 milliunits /(ml × cell) for the yeast cultures containing CesAs from both Arabidopsis and aspen. These results suggested that the α-Gal activity was much stronger in the yeast cultures containing the interaction pairs from the same species as compared to the interaction pairs from two different plant species. Moreover, interaction strengths among Arabidopsis secondary CesAs were less than those for aspen CesAs. The negative controls, AD-BD and Lamin C-T co-transformants, showed very weak α-Gal background activity. The known positive control, p53-T combination, showed about 16-fold induction in this study as compared to empty negative controls. The CesA interaction partners for aspen members showed 9 to 14-fold inductions, 8 to 11-fold inductions were seen for Arabidopsis members, and only 2.5 to 5-fold inductions were seen in combinations of CesAs from both the species. These results suggested that stronger interactions may occur between the CesAs from the same plant species and weaker interactions may occur between CesAs from different genera. Additionally, the interaction strengths of ZnBDs from different genera could be different as observed here for the first time in Arabidopsis and aspen. It is well known that physical characteristics of natural celluloses vary depending on the botanical origin of their source plants. Transfer of CesA genes from one species to another will possibly be attempted in the future to introduce novel cellulose characteristics in economically important plants [2]. An Y2H screen of compatibility of CesAs might be useful before beginning such cross-species CesA gene transfers.
It is currently unknown whether CesAs from one species can be functional in another species. We have attempted to complement three irx mutants (irx3, 1 and 5) with corresponding secondary CesAs from aspen to study functional conservation of these genes/proteins across the eudicots but such complementation experiments were not successful (data not shown). The weak interactions among Arabidopsis and aspen CesAs as reported here corroborate such observations. It is possible that such heterologous CesAs may not form the functional rosettes during genetic engineering experiments of expressing heterologous CesAs in transgenic plants resulting in either rapid turnover of the CesA transgenes [26] or ineffective impact of ectopic expression of CesAs in transgenic plants. Experimental proof for such hypothesis is currently missing but could be obtained in the future.
3.3. Interactions among Primary Cell Wall-Specific Cellulose Synthases
A phylogenetic tree developed on the basis of protein similarities among 52 CesAs indicated that higher plant CesAs could be divided into six groups, three primary and three secondary CesAs [27]. In Arabidopsis mutant studies, AtCesA1, AtCesA3, and AtCesA6 have been shown to be required for the cellulose synthesis in primary cell wall [1,7,12,28-31] , indicating the potential of their interactions with each other. Earlier, we also isolated three cDNAs encoding aspen CesAs, PtrCesA4, 5 and 6, orthologs of primary CesAs from Arabidopsis, AtCesA1, 3 and 6 [27,32-33] . Y2H results using ZnBDs from these three aspen primary CesAs also showed strong positive interactions (Table 3). This demonstrates the in vitro interactions among primary poplar CesAs, suggesting that these three primary PtrCesAs may also be required during cellulose synthesis in the primary cell wall for aspen trees. Our results were consistent with the report by Desprez et al. [14] that AtCesA1, AtCesA3 and AtCesA6 or AtCesA6 partially redundant AtCesA2 and AtCesA5 are able to biophysically interact with each other based on the results of co-immunoprecipitation and bimolecular fluorescence complementation.
3.4. Interactions between Secondary and Primary Cell Wall-Specific Cellulose Synthases
It has been widely acknowledged that the secondary and primary cell wall-specific CesAs are differentially expressed in plants [24]. However, we do not know whether they are able to interact physically with each other during the transition from cessation of primary wall deposition and initiation of secondary wall formation. In order to explore such possible interactions between aspen secondary and primary cell wall-specific CesAs, yeast two-hybrid experiments were carried out to explore their interaction potentials via their Zn-binding domains. Our result shows that any two aspen CesA ZnBDs from these two categories were able to interact while the negative controls showed no interactions (Table 3). In order to validate these Y2H-mediated interactions among primary and secondary CesAs, we also utilized co-immunoprecipitation assays to examine interactions of some selected interacting partner pairs. By coimmunoprecipitating ZnBDs from secondary PtrCesA1 and primary PtrCesA4 with antibodies against HA, we observed that the c-Myc protein complex is present in an immunoblot hybridized with c-Myc antibodies (Figure 3). The positive and negative controls confirmed that the reaction conditions that we used were able to discriminate between a positive reaction and a negative reaction. The similar result was also obtained in a pair of secondary wall associated PtrCesA3 and primary wall associated PtrCesA6 ZnBDs, confirming that primary and secondary CesAs can interact with each other in vitro via ZnBDs (Figure 3). These results suggest that primary and secondary CesAs might also be able to physically interact with each other in vivo but experimental proof of such a claim is currently unavailable.
It is generally believed that secondary CesA genes involved in secondary wall formation begin their transcription only after cessation of primary cell wall deposition [34]. It is, however, not known whether primary CesAs continue to transcribe and translate during secondary wall formation or whether primary and secondary CesAs productions are temporally separated in a plant cell since primary CesAs do not appear to be coordinately expressed with secondary CesAs [24]. Although we have previously reported the presence of primary and secondary CesA transcripts in the developing xylem tissue [35], use of a mixture of primary and secondary wall forming tissues in the developing xylem (such as rays and developing vessels/fibers) precludes specific conclusions about temporal and spatial separation of primary and secondary CesA formation during aspen xylem development. Based on the data presented here it is possible that during the transition from primary to secondary cell wall formation, both primary and secondary CesAs might be co-expressed and might interact with each other in intact plants. This is further supported by two studies on the involvement of a secondary wall specific AtCesA7 in the cellulose synthesis during primary cell wall formation. Over-expression of one mutant form of AtCesA7, fra5, under the CaMV 35S promoter reduced cell wall thickness and cell elongation via affecting cellulose synthesis during primary wall formation as well as causes a dominant negative effect on cellulose synthesis during secondary wall formation [36] . This suggests that the fra5 mutant protein may also be incor-
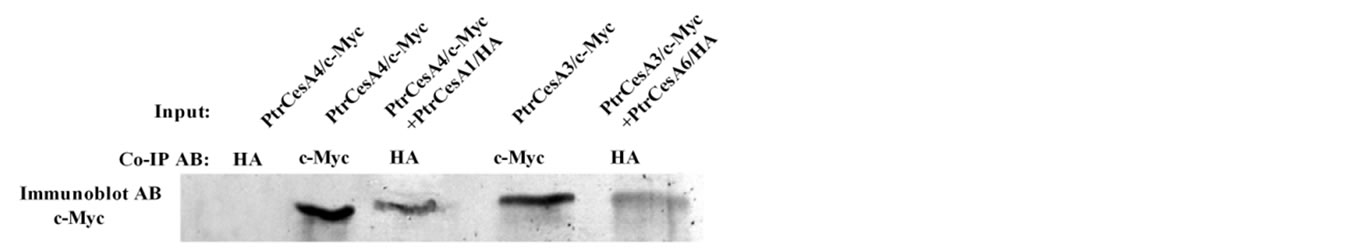
Figure 3. Co-immunoprecipitation of PtrCesA4/PtrCesA1 and PtrCesA3/PtrCesA6 ZnBDs representing a pair of primary and secondary CesAs, and secondary and primary CesAs, respectively. The first lane shows lack of immunoprecipitation of PtrCesA4 ZnBD with HA antibody; lane 2 shows immunoprecipitation of PtrCesA4 ZnBD with c-Myc antibody; lane 3 shows interaction among ZnBDs from PtrCesA4 and PtrCesA1 that were co-immunoprecipitated with HA antibodies; lane 4 shows immunoprecipitation of PtrCesA3 ZnBD with c-Myc antibody; and lane 5 shows co-immunoprecipitation of PtrCesA3 and PtrCesA6 ZnBDs with HA antibody.
porated into cellulose synthase terminal complex in the fra5-overexpressing plants, in elongating cells, so that affecting the normal function of CesA proteins involved in primary wall synthesis. Moreover, it is now still unclear whether and how two processes of primary and secondary wall formation are interrelated, and Bosca et al. [37] also reported that a widely recognized secondary wall-specific AtCesA7 (MUR10) is required for normal primary cell wall deposition and remodeling as well as plant growth, hypocotyl strength, and fertility. They further pointed out that there may exist a mechanism sensing secondary cell wall integrity and regulating primary cell wall structure and cellular differentiation [37]. It is, therefore, inferred that there may exist a stage in which primary wall-specific CesAs co-exist with some of secondary wall-specific CesAs if not all of them. Again more in vivo experimental evidence is needed for eventually proving such a possibility.
3.5. Lack of Specificity among CesA Interactions via Zinc-Binding Domains
The zinc-binding domains were thought to be the best candidate interaction regions between CesAs, as they were able to form both homodimers and heterodimers in the study of cotton fiber CesAs [20]. It was also shown that a mutation in the zinc-binding domain would inhibit the interaction between two CesA N-termini. In addition, Jacob-Wilk et al. [21] showed that the reduced CesA monomers were degraded rapidly while oxidized dimers were resistant to degradation during both in vivo and in vitro conditions, indicating the importance of CesA dimerization via zinc-binding domains for the stability of the CesA complexes. It seems that zinc-binding domains should play some specific roles in protein-protein interactions. However, our Y2H results showed that the interactions between CesA proteins via zinc-binding domains are insufficient to confer specificity of interactions among primary wall or secondary wall CesAs from aspen or Arabidopsis. CesAs are able to interact with each other either within primary and secondary wall specific groups or between these two groups. Moreover, the interactions even occurred between the inter-species CesAs, suggesting the structural capability of CesA zinc-binding domains for protein-protein interactions. Our results are consistent with the recent report [22], which showed that zinc-binding domains are not essential for the interactions between different CesAs. Using domain swapping techniques, Wang et al. [38] demonstrated that the catalytic or C-terminal domains were more important than other regions for the specific assembling in the CesA complex, which supports our results of lacking specificity of interactions between zinc-binding domains from different sources. While we cannot exclude the role of zinc-binding domains in the protein-protein interactions between CesAs, the other more specific interaction regions or other regulatory mechanisms need to be further discovered.
4. CONCLUSIONS
Here, we have demonstrated that all interactions among secondary wall-specific CesAs in both Arabidopsis and aspen, and intergeneric interactions between these two plant species are possible. We identified both qualitatively and quantitatively that intraspecies CesA interactions are stronger than the intergeneric CesA interactions. We also showed the interactions among primary CesAs, and the ones between primary and secondary cell wall-specific CesAs in aspen. These results suggest that two possibilities exist about CesA interactions. First, dynamic interactions might exist between the CesAs from two distinct wall-specific groups in the same species at least during the transition stage from primary to secondary cell wall formation. However, currently there is no supporting evidence for such a possibility. Secondly, no strong specificity among N-terminal domains of these CesAs was found in this study, indicating the possible existence of other more specific regions in other domains of the CesAs that are required for the proteinprotein interactions.
The genetic improvement of the cellulose biosynthetic process in crop plants or trees via overexpression of a single CesA has so far not been successful although such experiments are in progress in several laboratories including ours. We have hypothesized, therefore, that upregulation of CesA trios would improve cellulose production in cell walls ADDIN EN.CITE ADDIN EN.CITE.DATA [17-18] . ZnBD-mediated interactions of CesAs are difficult to demonstrate in vivo. Therefore, a systematic analysis of various combinations of CesA overexpressions in transgenic plants may be one of the methods available by which the CesA genes responsible for cellulose biosynthesis in economically important plants may be identified and manipulated for biofuel production.
5. ACKNOWLEDGEMENTS
This work was partially supported by the National Science Foundation’s CAREER award (IBN- 0236492) to CPJ. This work was also partially supported by the World Class University project of the Ministry of Science and Technology of Korea (R31-2009-000-20025-0).
REFERENCES
- Somerville, C. (2006) Cellulose synthesis in higher plants. Annual Review of Cell and Developmental Biology, 22(1), 53-78.
- Joshi, C.P. and Mansfield, S.D. (2007) The cellulose paradox—simple molecule, complex biosynthesis. Current Opinion in Plant Biology, 10(3), 220-226.
- Delmer, D.P. (1999) Cellulose biosynthesis: Exciting times for a difficult field of study. Annual Review of Plant Physiology and Plant Molecular Biology, 50, 245- 276.
- Doblin, M.S., Kurek, I., Jacob-Wilk, D. and Delmer, D.P. (2002) Cellulose biosynthesis in plants: From genes to rosettes. Plant Cell Physiology, 43(12), 1407-1420.
- Kimura, S., Laosinchai, W., Itoh, T., Cui, X., Linder, C.R. and Brown, R.M. Jr. (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant vigna angularis. Plant Cell, 11(11), 2075- 2086.
- Itoh, T., Kimura, S. and Brown, R.M. (2004) Theoretical considerations of immunogold labeling of cellulose synthesizing terminal complexes. Cellulose, 11(3-4), 385-394.
- Arioli, T., Peng, L.C., Betzner, A.S., Burn, J., Wittke, W., Herth, W., Camilleri, C., Hofte, H., Plazinski, J., Birch, R., Cork, A., Glover, J., Redmond, J. and Williamson, R. E. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science, 279(5351), 717-720.
- Scheible, W.R., Eshed, R., Richmond, T., Delmer, D. and Somerville, C. (2001) Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proceedings of the National Academy of Sciences of the United States of America, 98(18), 10079-10084.
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R. and Turner, S.R. (1999) The irregular xylem3 locus of arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell, 11(5), 769-779.
- Taylor, N.G., Howells, R.M., Huttly, A.K., Vickers, K. and Turner, S.R. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proceedings of the National Academy of Sciences of the United States of America, 100(3), 1450-1455.
- Taylor, N.G., Laurie, S. and Turner, S.R. (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell, 12(12), 2529-2540.
- Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., McCann, M., Rayon, C., Vernhettes, S. and Hofte, H. (2000) ROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell, 12(12), 2409-2424.
- Gardiner, J. C., Taylor, N.G. and Turner, S.R. (2003) Control of cellulose synthase complex localization in developing xylem. Plant Cell, 15(8), 1740-1748.
- Desprez, T., Juraniec, M., Crowell, E.F., Jouy, H., Pochylova, Z., Parcy, F., Hofte, H., Gonneau, M. and Vernhettes, S. (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 104(39), 15572-15577.
- Tanaka, K., Murata, K., Yamazaki, M., Onosato, K., Miyao, A. and Hirochika, H. (2003) Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiology, 133(1), 73-83.
- Burton, R.A., Shirley, N.J., King, B.J., Harvey, A.J. and Fincher, G.B. (2004) The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiology, 134(1), 224-236.
- Bhandari, S., Fujino, T., Thammanagowda, S., Zhang, D. Y., Xu, F.Y. and Joshi, C.P. (2006) Xylem-specific and tension stress-responsive coexpression of KORRIGAN endoglucanase and three secondary wall-associated cellulose synthase genes in aspen trees. Planta, 224(4), 828-837.
- Joshi, C.P., Bhandari, S., Ranjan, P., Kalluri, U.C., Liang, X., Fujino, T. and Samuga, A. (2004) Genomics of cellulose biosynthesis in poplars. New Phytologist, 164(1), 53-61.
- Ranik, M. and Myburg, A.A. (2006) Six new cellulose synthase genes from Eucalyptus are associated with primary and secondary cell wall biosynthesis. Tree Physiology, 26(5), 545-556.
- Kurek, I., Kawagoe, Y., Jacob-Wilk, D., Doblin, M. and Delmer, D. (2002) Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proceedings of the National Academy of Sciences of the United States of America, 99 (17), 11109-11114.
- Jacob-Wilk, D., Kurek, I., Hogan, P. and Delmer, D.P. (2006) The cotton fiber zinc-binding domain of cellulose synthase A1 from Gossypium hirsutum displays rapid turnover in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America, 103(32), 12191-12196.
- Timmers, J., Vernhettes, S., Desprez, T., Vincken, J.P., Visser, R.G. and Trindade, L.M. (2009) Interactions between membrane-bound cellulose synthases involved in the synthesis of the secondary cell wall. FEBS Letters, 583(6), 978-982.
- Atanassov, II, Pittman, J.K. and Turner, S.R. (2009) Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. Journal of Biological Chemistry, 284(6), 3833-3841.
- Persson, S., Wei, H.R., Milne, J., Page G.P. and Somerville, C.R. (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proceedings of the National Academy of Sciences of the United States of America, 102(24), 8633-8638.
- Brown, D.M., Zeef, L.A.H., Ellis, J., Goodacre, R. and Turner, S.R. (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell, 17(8), 2281-2295.
- Ding, S.Y. and Himmel, M.E. (2006) The maize primary cell wall microfibril: a new model derived from direct visualization. Journal of Agricultural and Food Chemistry, 54(3), 597-606.
- Samuga, A. and Joshi, C.P. (2004) Differential expression patterns of two new primary cell wall-related cellulose synthase cDNAs, PtrCesA6 and PtrCesA7 from aspen trees. Gene, 334, 73-82.
- Beeckman, T., Przemeck, G.K., Stamatiou, G., Lau, R., Terryn, N., De Rycke, R., Inze, D. and Berleth, T. (2002) Genetic complexity of cellulose synthase a gene function in Arabidopsis embryogenesis. Plant Physiology, 130(4), 1883-1893.
- Gillmor, C.S., Poindexter, P., Lorieau, J., Palcic M.M. and Somerville, C. (2002) Alpha-glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. Journal of Cell Biology, 156(6), 1003-1013.
- Williamson, R.E., Burn, J.E., Birch, R., Baskin, T.I., Arioli, T., Betzner A.S. and Cork, A. (2001) Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma, 215(1-4), 116-127.
- Ellis, C., Karafyllidis, I., Wasternack, C. and Turner, J.G. (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell, 14(7), 1557-1566.
- Kalluri, U.C. and Joshi, C.P. (2003) Isolation and characterization of a new, full-length cellulose synthase cDNA, PtrCesA5 from developing xylem of aspen trees. Journal of Experimental Botany, 54(390), 2187- 2188.
- Kalluri, U.C. and Joshi, C.P. (2004) Differential expression patterns of two cellulose synthase genes are associated with primary and secondary cell wall development in aspen trees. Planta, 220(1), 47-55.
- Pear, J.R., Kawagoe, Y., Schreckengost, W.E., Delmer, D.P. and Stalker, D.M. (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase," Proceedings of the National Academy of Sciences of the United States of America, 93(22), 12637-12642.
- Liang, X. and Joshi, C.P. (2004) Molecular cloning of ten distinct hypervariable regions from the cellulose synthase gene superfamily in aspen trees. Tree Physiology, 24(5), 543-550.
- Zhong, R., Morrison, W.H., 3rd, Freshour, G.D., Hahn, M.G. and Ye, Z.H. (2003) Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant physiology, 132 (2), 786-795.
- Bosca, S., Barton, C.J., Taylor, N.G., Ryden, P., Neumetzler, L., Pauly, M., Roberts, K. and Seifert, G.J. (2006) Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant physiology, 142(4), 1353-1363.
- Wang, J., Howles, P.A., Cork, A.H., Birch, R.J. and Williamson, R.E. (2006) Chimeric proteins suggest that the catalytic and/or C-terminal domains give CesA1 and CesA3 access to their specific sites in the cellulose synthase of primary walls. Plant Physiol, 142(2), 685- 695.

