Journal of Agricultural Chemistry and Environment
Vol.2 No.1(2013), Article ID:28189,8 pages DOI:10.4236/jacen.2013.21002
Application of PCR primer sets for detection of Pseudomonas sp. functional genes in the plant rhizosphere
![]()
1Department of Environmental Sciences, University of California, Riverside, USA; *Corresponding Author: jskim@gimb.or.kr
2Gyeongbuk Institute for Marine Bioindustry, Uljin, Republic of Korea
3Department of Primary Industries, Rutherglen, Australia
Received 23 November 2102; revised 28 December 2012; accepted 10 January 2013
Keywords: PGPR (Plant Growth-Promoting Rhizosphere); Pseudomonas; PCR; 16S rDNA; Plant-Microbe Interactions
ABSTRACT
Plant growth promoting pseudomonads play an important role in disease suppression and there is considerable interest in development of biomarker genes that can be used to monitor these bacteria in agricultural soils. Here, we report the application of a PCR primer sets targeting genes encoding the main antibiotic groups. Distribution of the genes was variably distributed across type strains of 28 species with no phylogenetic grouping for the detected antibiotics genes, phlD for 2,4-diacetylphloroglucinol (2,4-DAPG) and phzCD for phenazine-1-carboxylic acid or hcnBC for hydrogen cyanide production. Analysis of field soils showed that primer sets for phlD and phzCD detected these genes in a fallowed neutral pH soil following wheat production, but that the copy numbers were below the detection limits in bulk soils having an acidic pH. In contrast, PCR products for the phzCD, pltC, and hcnBC genes were detectable in mature root zones following planting with wheat. The ability to rapidly characterize populations of antibiotics producers using specific primer sets will improve our ability to assess the impacts of management practices on the functional traits of Pseudomonas spp. populations in agricultural soils.
1. INTRODUCTION
Plant growth promoting rhizosphere bacteria improve plant growth through the production of hormones and by suppression of root disease through the production of antibiotics and siderophores. Using culture based methods to identify these bacteria, PGPR have been isolated from plants in soils from diverse geographical regions [1-5]. One of the most important groups of PGPR are strains of antibiotic-producing fluorescent Pseudomonas spp. that contribute to suppression of fungal diseases including take-all of wheat, black root rot of tobacco, or fusarium wilt of tomato [6-9]. The genes that are responsible for production of antibiotics are variably dispersed among different genotypes, which vary in their efficacy for disease suppression [5,8,10-12]. Thus there is interest in development of PCR primer sets that can be used to detect the presence of these genes in environmental samples and to monitor the populations of bacteria that carry these genes [6,7,13-15].
Genes encoding the antibiotics pyoluteorin (pltC) [1], pyrrolnitrin (prnD) [1], phenazine-1-carboxylic acid (phzCD) [3], and 2,4-diacetylphloroglucinol (2,4-DAPG, phlD) [3] are a major focus of research in biological control of disease by pseudomonads. Other genes that are relevant to biocontrol include the gacA and hcnBC genes encoding hydrogen cyanide production. The gacA gene (global activator of cyanide production) is a response regulator that interacts with quorum regulated genes to control expression of small RNAs and antibiotics in various strains of PGPR pseudomonads [16]. In the PGPR bacterium P. chlororaphis, gacA also functions as a negative regulator of indole-3-acetic acid hormone production. The genes encoding aminocyclopropane carboxylic acid deaminase (acc) are still other potential targets, functioning for the destruction of ethylene [17,18].
Molecular methods for the detection of these genes have been described, but are often utilized in combination with culture based methods to quantify the population sizes of Pseudomonas strains carrying specific genes [19]. The threshold population density at which 2,4- DAPG producing strains are effective for disease suppression is 105 CFU g−1 root for protection against takeall disease caused by Gaeumannomyces gramininis. 2,4-DAPG producing bacteria collected from various locations from around the world can be further grouped into at least 18 BOX-PCR genotypes that reflect the selection of local genotypes in different soils [20]. In general, the population densities of antibiotic producing Pseudomonas are highly dynamic, and generally increase by at least two orders of magnitude in the plant rhizosphere [21] and higher in the vicinity of plant roots than in unplanted soil [22]. The population size of different genotypes is further influenced by intrastrain competition as well as interactions with other rhizosphere bacterial species [23].
With the advent of rapid screening methods for detection of marker genes that are associated with plant health and growth in agricultural soils, we were interested in determining the utility of PCR based methods to detect a suite of genes for plant growth promoting pseudomonads in wheat soils in S.E. Australia where there is still little information on the ecology and importance of these bacteria in soil health. The research reported here examined selected PCR primer sets from the literature for detection of specific marker genes, and the association of antibiotic genes among different type strains of Pseudomonas spp. The primer sets were evaluated for three different soil classes representing the major soils used for wheat cultivation in Victoria, Australia. We examined both the field soil and adjacent remnant areas that had not been cultivated and further compared soils that were collected during the fallow period and in the rhizosphere after cultivation with wheat. The results demonstrate the utility of molecular methods to compare soils, but also illustrate the highly dynamic changes that occur in managed soils.
2. MATERIALS AND METHODS
2.1. Soil Description
Soil samples were collected in autumn from three soil classes according to the Australian soil classification system (Isbell, R.F., 2002. The Australian Soil Classification. CSIRO Publishing, Melbourne), including a dermosol (NE Victoria, Rutherglen), a calcarosol (NW Victoria, Birchip) and a chromosol (SW Victoria, Hamilton) collected in southeastern Australia. Within each of these regions, soils were collected from paired sites representing two contrasting land-uses, a “remnant” uncultivated site where indigenous plant species were present and a ‘managed’ site where the soils were subjected to normal agricultural practices with fertilizer and herbicide input regimes, grazing and cropping activities. Soil chemical and physical characteristics for the six sites are summarized in Table 1. Soils were collected during the wheat fallow period and were air-dried and shipped to the University of California, Riverside, for the experimental study. Subsamples of the six soils were planted with wheat (Triticum aestivum L. cv. Yecora rojo) in 1 cm thick root boxes, with a detachable side plate, and were watered daily to maintain constant 30% moisture content by weight. The plants were cultivated in a glasshouse under ambient conditions. Soil was collected from the root boxes after 3 weeks and included the bulk soil fraction, and the base of older plant roots and the root tips. Soil samples were passed through a 2-mm sieve to homogenize
Table 1. Land-use and soil chemical and physical characteristics.

the soil fractions, after which the soil and root were processed for DNA extraction.
2.2. Chemical and Physical Analysis
Soil pH and salinity (electrical conductivity, EC) were measured in soil extracts using 1:5 soil water and 1:1 soil water: 1 mM CaCl2 extraction solutions. Exchangeable cations, and heavy metals (iron, zinc, copper, and manganese) were quantified by atomic absorption spectrophotometer (Perkin-Elmer AAnalyst 800). Carbon and nitrogen was analyzed using a C-N-S analyzer (NA 1500 series 2, Carlo Erba). The soil particle size was analyzed using the hydrometer method. Nitrate, nitrite, ammonia and soluble reactive phosphorus were quantified with a Technicon TRAACS 800 Autoanalyzer (Tarrytown, NY).
2.3. DNA Extraction
DNA was extracted from 0.5 g samples of each soil and root using the FastPrepTM Soil DNA Extraction kit (Qbiogene, Carlsbad, CA) according to the manufacturers’ recommendations. Five replicates for each soil and root sample were prepared from each soil and were stored at −80˚C until use for subsequent analyses.
2.4. Primers and PCR Reaction Conditions
Table 2 lists the gene targets and PCR primer sets that were used to detect genes encoding the selected antibiotics, regulatory genes, and markers for fluorescent pseudomonads. The 50 μL PCR reaction mixtures consisted of 1 μl of DNA template, 50 mM Tris (pH 8.3), 250 μg of bovine serum albumin (BSA) per ml, 2.5 mM MgCl2, 200 nM deoxynucleoside triphosphates, 200 nM each SSU rDNA primer and 3 U of Taq DNA polymerase. All primers were obtained from Invitrogen (Carlsbad, CA.). PCR was performed on a PTC 200 (Bio-Rad Laboratories, USA). The PCR program was 5 min at 95˚C; 30 cycles of 30 s at 94˚C, 30 s at 50 or 55˚C, and 1 min at 72˚C; and a final extension for 10 min at 72˚C. Optimal annealing temperatures were decided after gradient PCR or testing two different temperatures, 48˚C and 55˚C. Detection of PCR products was determined for ethidium bromide (EtBr) stained agarose gels using Gel-doc (BioRad) and image analysis using Quantity One (Bio-Rad).
2.5. Phylogenetic Analysis
PCR was conducted with 10 μl reaction volumes containing: 1 μl of DNA, 2 μl of Terminator Ready Reaction Mix, and 200 nM each of T7 and SP6 primers. The reagents were combined and heated to 96˚C for 1 min. Twenty-five cycles of PCR were run at 96˚C for 10 s, 50˚C for 10 s, and 60˚C for 4 min. Approximately 1 kb (position 289-1258 [24] were sequenced using the ABI 377 sequencer (Applied Biosystems, Foster, CA, USA)).
16S rDNA sequences were aligned with greengenes [25,26]. Evolutionary distances were calculated by the method of Kimura 2-parameter and a phylogenetic tree was constructed by the neighbor-joining method [27] with MEGA4 for Windows, including bootstrap analysis [28].
The nucleotide sequence data reported in this paper will appear in the GenBank/EMBL/DDBJ nucleotide sequence databases with the accession number(s) AY860450-AY860454.
3. RESULTS
3.1. Distribution of Antibiotic Genes among Pseudomonas Type Strains
PCR primer sets for conserved sequences of genes involved in expression and regulation of six antibiotics were targeted against 28 type strains of Pseudomonas spp. Of the six sequences evaluated, antibiotic gene sequences for 2,4-DAPG (phlD) and phenazine (phzCD) were detected in 3 and 5 of the type strains, respectively (Figure 1). In all cases where a positive signal was obtained, the PCR products were of the correct predicted size. Where no signals were obtained, the PCR reactions were rerun at less stringent conditions, but did not produce signals, indicating that the failure to obtain PCR products for those targets were most likely due to low copy numbers rather than base substitutions in the target sequence. Of the antibiotics detected, none occurred simultaneously in the same types strains, nor were there apparent relationships between distributions of the genes with taxonomic groupings by 16S rRNA gene.
PCR products were obtained in all of the type strains with primers for 16S rRNA sequences, and in 25 and 23 of the strains using primers for the proposed phylogenetic classification markers gyrB and rpoD. The hcnBC genes for cyanide production and gacA for global activator cyanide, which are involved in a regulatory circuit affecting antibiotic production, were widely dispersed in 8 and 14 of the 28 type strains. In this case, the gacA gene was detected in 5 of the 8 type strains having positive signals for hcnBC. Marker gene sequences for aminocyclopropane deaminase (acc) and indole acetic acid production (ipdc) were not detected in any of the type strains tested here.
3.2. Detection of PCR Products for Antibiotic Genes of Pseudomonas in Soil and Root
The fully set of gene targets for Pseudomonas sp. and functional genes were tested in bulk soils and in root samples from wheat for managed and remnant soils
Table 2. Target genes and primers used in PCR analysis of soil DNA.
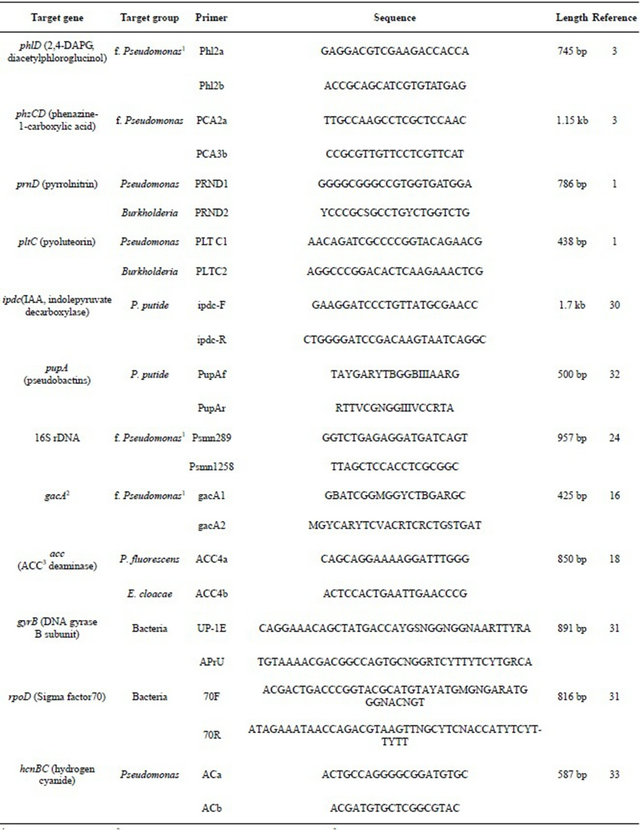
1fluoresecent Pseudomonas; 2global activator of cyanide and antibiotic production; 31-aminocyclopropane-1-carboxylic acid.
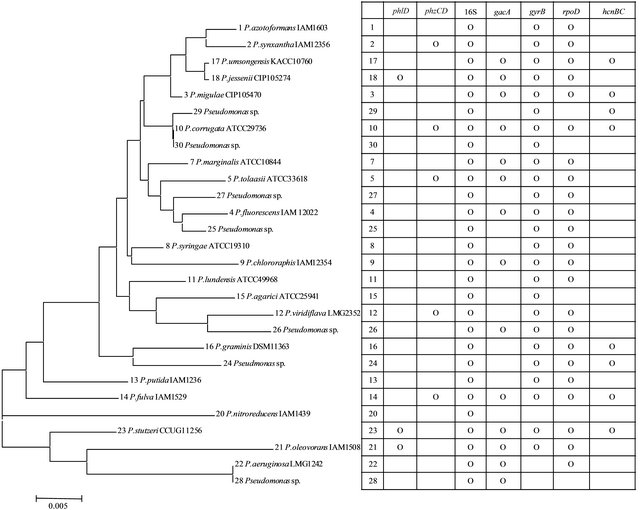
Figure 1. Phylogenetic analysis of type strains and PCR signals detected by amplification with primers for a range of target genes.
represented 3 soil geomorphic classes in Victoria, Australia. These soils were of similar texture, but varied in pH and organic carbon (Table 1). Among the antibiotic genes, PCR products were obtained for phlD and phzCD in bulk soils from Birchip (neutral pH soil) but were not detected in the two acid soils. In contrast, positive signals were obtained for these gene products in all of the soils following growth of wheat plants for 3 weeks, indicating that the copy number increased to detectable levels in the plant root. The phlD gene was detected in both mature root zones and in the root tip region. In contrast, the phzCD gene was detected only in mature root zones, and not at the root tip. This same pattern was observed for pltC. In this case, the pltC gene was not detected in any of the fallow bulk soil samples, but was detected in mature roots for all three soil classes. The prnD gene was an anomaly, producing, with the PCR primers for this gene producing multiple products of variable size in all of the soils tested, even under high stringency melting conditions (67˚C) during the PCR reaction.
The hcnBC gene was detected in two of the six bulk soil samples, and in mature root zones in all three soil classes. The gacA gene was not detected in any of the bulk or root samples, nor were the two growth regulator genes, the ipdc and acc genes were not detected in any of the root samples. The phylogenetic group marker 16S rRNA gene target for Pseudomonas was detected in all soil samples, but the gyrB and rpoD genes were not detected in any of the soil samples.
4. DISCUSSION
The objective of this study was to evaluate a selected set of gene targets for detection of Pseudomonas sp. and functional genes for antibiotic production, regulation of cyanide and antibiotic production, and plant growth hormone production across different soil types representing a range of important soils used for wheat production in Australia. There is considerable interest in developing molecular methods to monitor these and other species of plant growth promoting bacteria as part of a strategy to monitor soil health and the impacts of various management practices on the population densities of PGPR bacteria. Previous research has identified a number of candidate genes that are potential markers for antibiotic-producing Pseudomonas spp. It has also been shown that there is considerable variation in genotypes of specific antibiotic-producing Pseudomonas that vary in their ability to colonize the rhizosphere (Bergsma-Vlami et al., 2005). In general, population densities of pseudomonads are highly dynamic in soils and are highest in the plant rhizosphere, but are also influence by the host plant species [6]. To characterize the abundance and distribution of these marker genes in the literature, we used PCRbased methods to screening for the antibiotics, 2,4- DAPG (phlD) [3], phenazine-1-carboxylic acid (phzCD) [3], pyoluteorin (pltC) [1], pyrrolnitrin (prnD) [1], and hydrogen cyanide (hcnBC) [29]. Other gene targets included indole-3-acetic acid (ipdc) [30] and ACC deaminase (acc) which degrades a precursor for ethylene [18]; gacA, the global activator of cyanide and antibiotic production [16], and phylogenetic group markers (gyrB) [31], RNA polymerase beta-subunit encoding gene (rpoB) [31], and 16S rRNA genes with Pseudomonas specific primers [24]. Our results showed that among the antibiotic genes targets that were evaluated, the two that were detected with pure cultures, gyrB and rpoD, were broadly distributed among types strains of the 25 and 23 different Pseudomonas species that were tested in pure culture. The broad distribution of these genes in different species illustrates the difficulty of monitoring PGPR based solely on tracking species using 16S rRNA genes or other phylogenetic markers. In this manner, monitoring of PGPR may be better achieved by tracking the presence and copy number of specific functional genes. The phlD and phzCD genes were readily detected all three of the field soils, but detection was enhanced by use of root samples and was further influenced by sample location in the mature roots or the root tips. The phlD gene was found in all locations in the root in all of the soils, whereas the phzCD, pltC and hcnBC genes were detected only in mature root zones (Table 3). Altogether, these data suggests that sampling procedures must be highly standardized for using the presence, absence, or copy number of specific marker genes as parameters for measuring soil health and the impact of management practices on PGPR bacteria. The method used here in which wheat plants were cultivated in the soil for a defined period of time provides a standardized condi
Table 3. PCR signals detected using plant growth promoting and population specific-primers.
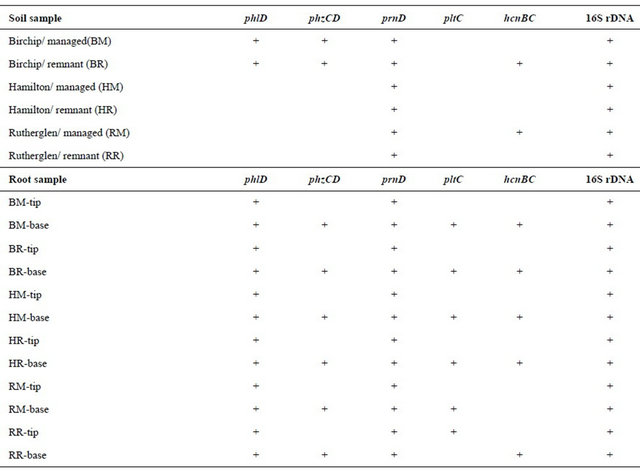
tion that may be preferable, allowing uniform conditions for establishment of a stable population size for comparative analysis of soils over time and across different soil types and management regimes. This is analogous to monitoring of chemical and physical variables used to describe soil quality, where specific reference locations, or “benchmarks”, are used for comparison of changes in chemical and physical variables such as bulk density and hydraulic conductivity. In the case of soil biological variables, the benchmark conditions employ a standardized test plant species, plant age, and rhizosphere location.
The gacA sequence was of particular interest here for its role in regulation of both cyanide and antibiotic production. However, our study of the type strains for 28 species shows that the hcnBC and gacA genes were coassociated only in 30% of the strains. This could be due to variations in the sequence that was targeted. The gacA gene, which was detected in 3 of the 14 strains was consistently co-associated with the phlD gene for 2,4-DAPG production, but was inconsistently associated with phenazine production for 3 of the 5 phzCD containing type strains.
Several species of soil bacteria contain the acc gene [17,18]. The gene appears to undergo horizontal transfer and has been found in various Pseudomonas sp. The sequence used here was not detected in any of the type strains, or in the soils that were screened. This was unexpected as the target sequence we used is conserved. It is possible of course that our inability to detect this gene and others in our primer set such as the pupA gene for siderophores production is due to low copy numbers or interferences from PCR inhibitors in the soil extracts that lowered the detection efficiency.
Molecular methods for detection of specific gene sequences for monitoring bacterial species population densities and copy numbers of functional genes are advanceing rapidly. Results of this research demonstrate that it will be necessary to empirically test many of the sequences that have been identified in the laboratory for their efficacy in actual field samples, and standardize the conditions that are to be used for screening soils. This may be best achieved by bringing soils to a standard reference state that reflects the conditions under which the PGPR perform their ecosystem function; in this case the presence of a plant rhizosphere of a particular plant species. The ability to rapidly characterize populations of antibiotics producers will greatly enhance our understanding of their role in enhancing plant health and yields, and the impacts of management practices on PGPR bacteria.
5. ACKNOWLEDGEMENTS
The authors gratefully acknowledge the support of the Department of Primary Industries Victoria as part of the ‘Our Rural Landscape’ initiative (sub-project 1.2 Our Soils: Understanding and Protecting the Soils of our Rural Landscapes) funded by the Victorian State Government (Department of Innovation, Industry and Regional Development).
![]()
![]()
REFERENCES
- de Souza, J.T. and Raaijmakers, J.M. (2003) Polymorphisms within the prnD, and pltC genes from pyrrolnitrin and pyoluteorin-producing Pseudomonas and Burkholderia spp. FEMS Microbiology Ecology, 43, 21-34. doi:10.1111/j.1574-6941.2003.tb01042.x
- Landa, B.B., Mavrodi, O.V., Raaijmakers, J.M., McSpadden Gardener, B.B., Thomashow, L.S. and Weller, D.M. (2002) Differential ability of genotypes of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens strains to colonize the roots of pea plants. Applied and Environmental Microbiology, 68, 3226-3237. doi:10.1128/AEM.68.7.3226-3237.2002
- Raaijmakers, J.M., Weller, D.M. and Thomashow, L.S. (1997) Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Applied and Environmental Microbiology, 63, 881-887.
- Raaijmakers, J.M., Vlami, M. and de Souza, J.T. (2002) Antibiotic production by bacterial biocontrol agents. Antonie van Leeuwenhoek, 81, 537-547. doi:10.1023/A:1020501420831
- Wang, C.X., Ramette, A., Punjasamarnwong, P., Zala, M., Natsch, A., Moenne-Loccoz, Y. and Defago, G. (2001) Cosmopolitan distribution of phlD-containing dicotyledonous crop-associated biocontrol pseudomonads of worldwide origin. FEMS Microbiology Ecology, 37, 105-116.
- Bergsma-Vlami, M., Prins, M.E. and Raaijmakers, J.M. (2005) Influence of plant species on population dynamics, genotypic diversity and antibiotic production in the rhizosphere by indigenous Pseudomonas spp. FEMS Microbiology Ecology, 52, 59-69. doi:10.1016/j.femsec.2004.10.007
- Bergsma-Vlami, M., Prins, M.E., Staats, M. and Raaijmakers, J.M. (2005) Assessment of genotypic diversity of antibiotic-producing Pseudomonas species in the rhizosphere by denaturing gradient gel electrophoresis. Applied and Environmental Microbiology, 71, 993-1003. doi:10.1128/AEM.71.2.993-1003.2005
- Haas, D. and Keel, C. (2003) Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annual Review of Phytopathology, 41, 117-153. doi:10.1146/annurev.phyto.41.052002.095656
- Mazzola, M. (2004) Assessment and management of soil microbial community structure for disease suppression. Annual Review of Phytopathology, 42, 35-59. doi:10.1146/annurev.phyto.42.040803.140408
- Chin-A-Woeng, T.F.C., Bloemberg, G.V. and Lugtenberg, B.J.J. (2003) Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytologist, 157, 503-523. doi:10.1046/j.1469-8137.2003.00686.x
- Weller, D.M., Raaijmakers, J.M., McSpadden Gardener, B.B. and Thomashow, L.S. (2002) Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annual Review of Phytopathology, 40, 309-348. doi:10.1146/annurev.phyto.40.030402.110010
- Weller, D.M., Landa, B.B., Mavrodi, O.V., Schroeder, K.L., De La Fuente, L., Bankhead, S.B., Allende Molar, R., Bonsall, R. F., Mavrodi, D. V. and Thomashow, L. S. (2007) Role of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant Biology, 9, 4-20.
- Costa, R., Gomes, N.C.M., Peixoto, R.S., Rumjanek, N., Berg, G., Mendonca-Hagler, L.C.S. and Smalla, K. (2006) Diversity and antagonistic potential of Pseudomonas spp. associated to the rhizosphere of maize grown in a subtropical organic farm. Soil Biology and Biochemistry, 38, 2434-2447. doi:10.1016/j.soilbio.2006.03.003
- Garbeva, P., Voesenek, K. and Van Elsas, J.D. (2004) Quantitative detection and diversity of the pyrrolnitrin biosynthetic locus in soil under different treatments. Soil Biology and Biochemistry, 36, 1453-1463. doi:10.1016/j.soilbio.2004.03.009
- Moynihan, J.A., Morrissey J.P., Coppoolse E.R., Stiekema W.J., O’Gara, F. and Boyd, E.F. (2009) Evolutionary history of the phl gene cluster in the plant-associated bacterium Pseudomonas fluorescens. Applied and Environmental Microbiology, 71, 4951-4959.
- De Souza, J.T., Mazzola, M. and Raaijmakers, J.M. (2003) Conservation of the response regulator gene gacA in Pseudomonas species. Environmental Microbiology, 5, 1328-1340. doi:10.1111/j.1462-2920.2003.00438.x
- Hontzeas, N., Richardson, A.O., Belimov, A., Safronova, V., Abu-Omar, M.M. and Glick, B.R. (2005) Evidence for horizontal transfer of 1-aminocyclopropane-1-carboxylate deaminase genes. Applied and Environmental Microbiology, 71, 7556-7558. doi:10.1128/AEM.71.11.7556-7558.2005
- Wang, C., Knill, E., Glick, B.R. and Defago, G. (2000) Effect of transferring 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase genes into Pseudomonas fluorescens strain CHA0 and its gacA derivative CHA96 on their growth-promoting and disease-suppressive capacities. Canadian Journal of Microbiology, 46, 898-907.
- Landa, B.B., Mavrodi, O.V., Schroeder, K.L., AllendeMolar, R. and Weller, D.M. (2006) Enrichment and genotypic diversity of phlD-containing fluorescent Pseudomonas spp. in two soils after a century of wheat and flax monoculture. FEMS Microbiology Ecology, 55, 351-368. doi:10.1111/j.1574-6941.2005.00038.x
- De La Fuente, L., Mavrodi, D.V., Landa, B.B., Thomashow, L.S. and Weller, D.M. (2006) phlD-based genetic diversity and detection of genotypes of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens. FEMS Microbiology Ecology, 56, 64-78. doi:10.1111/j.1574-6941.2006.00074.x
- Bankhead, S.B., Landa, B.B., Lutton, E., Weller, D.M. and McSpadden Gardener, B.B. (2004) Minimal changes in rhizobacterial population structure following root colonization by wild type and transgenic biocontrol strains. FEMS Microbiology Ecology, 49, 307-318. doi:10.1016/j.femsec.2004.04.005
- Haas, D. and Defago, G. (2005) Biological control of soilborne pathogens by fluorescent pseudomonads. Nature Reviews Microbiology, 3, 307-319. doi:10.1038/nrmicro1129
- Landa, B.B., Mavrodi, D.M., Thomashow, L.S. and Weller, D.M. (2003) Interactions between strains of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens in the rhizosphere of wheat. Phytopathology, 93, 982-994. doi:10.1094/PHYTO.2003.93.8.982
- Widmer, F., Seidler, R.J., Gillevet, P.M., Watrud, L.S. and Di Giovanni, G.D. (1998) A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Applied and Environmental Microbiology, 64, 2545-2553.
- DeSantis, T.Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E.L., Keller, K., Huber, T., Dalevi, D., Hu, P. and Andersen, G. L. (2006a) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology, 72, 5069- 5072. doi:10.1128/AEM.03006-05
- DeSantis, T.Z., Jr., Hugenholtz, P., Keller, K., Brodie, E.L., Larsen, N., Piceno, Y.M. Phan, R. and Andersen, G. L. (2006b) NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Research, 34, W394-399. doi:10.1093/nar/gkl244
- Saitou, N. and Nei, M. (1987) The Neighbor-joining method a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406-425.
- Tamura, K., Dudley, J., Nei, M. and Kumar, S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596-1599. doi:10.1093/molbev/msm092
- Ramette, A., Moenne-Loccoz, Y. and Defago, G. (2003) Prevalence of fluorescent pseudomonads producing antifungal phloroglucinols and/or hydrogen cyanide in soils naturally suppressive or conducive to tobacco black root rot. FEMS Microbiology Ecology, 44, 35-43. doi:10.1111/j.1574-6941.2003.tb01088.x
- Patten, C.L. and Glick, B.R. (2002) Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Applied and Environmental Microbiology, 68, 3795-3801. doi:10.1128/AEM.68.8.3795-3801.2002
- Yamamoto, S., Kasai, H., Arnold, D.L., Jackson, R.W., Vivian, A. and Harayama, S. (2000) Phylogeny of the genus Pseudomonas: Intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology, 146, 2385-2394.
- Koster, M., Ovaa W., Bitter W. and Weisbeek P. (1995) Multiple outer membrane receptors for uptake of ferric pseudobactins in Pseudomonas putida WCS358. Molecular & General Genetics, 248, 735-743. doi:10.1007/BF02191714
- Ramette, A., Frapolli, M., Defago, G. and Moenne-Loccoz, Y. (2003) Phylogeny of HCN synthase-encoding hcnBC genes in biocontrol fluorescent pseudomonads and its relationship with host plant species and HCN synthesis ability. Molecular Plant-Microbe Inreactions, 16, 525- 535. doi:10.1094/MPMI.2003.16.6.525

