American Journal of Plant Sciences
Vol.5 No.10(2014), Article ID:45661,7 pages DOI:10.4236/ajps.2014.510156
Genetic Variability among Genotypes of Physic Nut Regarding Seed Biometry
Leonardo Fardim Christo1, Tafarel Victor Colodetti1, Wagner Nunes Rodrigues1, Lima Deleon Martins1, Sebastião Batista Brinate1, José Francisco Teixeira do Amaral1, Bruno Galvêas Laviola2, Marcelo Antonio Tomaz1
1Centro de Ciências Agrárias, Universidade Federal do Espírito Santo (CCA/UFES), Alegre, Brazil
2Empresa Brasileira de Pesquisa Agropecuária, Embrapa Agroenergia, Brasília, Brazil
Email: wagnernunes@outlook.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 28 February 2014; revised 2 April 2014; accepted 12 April 2014
ABSTRACT
With probable center of origin in Brazil, the species Jatropha curcas L., known as physic nut, is a tropical oilseed with potential for cultivation aiming at the production of biodiesel. This study was conducted with the objective of investigating the genetic variability regarding the morphology of seeds of physic nut, using biometric analyses to identify variables that have potential for the study of the diversity of the species and the classification of genotypes. Seeds of 22 genotypes of physic nut, from the germplasm bank of Embrapa Agroenergia (Brazil) were evaluated regarding characteristics of size and mass. The genotypes of Jatropha curcas L. selected by the Brazilian breeding program presented high diversity for characteristics of the seeds, allowing the exploration of this genetic variability to classify genotypes in different groups. In addition, variables related to the biomass of the seeds present high relative contributions to the diversity observed in the genotypes.
Keywords:Jatropha curcas, Diversity, Size, Shape, Biomass

1. Introduction
The species Jatropha curcas L., known as physic nut, is a tropical plant from the Euphorbiaceae family, having Brazil as probable center of origin. It is reported that physic nut was introduced in the Archipelago of Cabo Verde and Guinea islands by Portuguese sailors before spreading to the African continent [1] .
Jatropha curcas L. is considered a species resistant to drought, pests and plant diseases, as well as adaptable to different soil and climatic conditions [2] . This species has wide geographical distribution and is actually being cultivated for different uses, such as fabrication of ink and paints, for polishing and varnishing furniture and in domestic medicine. In addition, the physic nut is considered an oilseed crop with great potential for the production of biodiesel [1] [3] [4] .
The concept of replacing part of petroleum diesel by renewable energy sources, such as the biodiesel, has gained widespread global attention in recent years [5] . Finding vegetal species with potential for the production of oil, which can be used to produce the biodiesel, is needed to achieve the objectives of the programs aiming at this partial substitution. The physic nut is one valuable option for this matter, due to this oilseed having high oil content on their seeds and presenting desirable chemical characteristics [6] .
The fruits of physic nut are ovoid capsules, with diameter normally ranging from 1.5 to 3.0 cm, having three locules with a seed each. However, great variability about weight and dimensions of fruits and seeds has been related between genotypes developed and cultivated in Brazil [7] , as well as for many other phenotypic characteristics [8] [9] .
Biometric analyses are an important tool to detect genetic variability within and between populations, also allowing studying the interaction between genetic and environmental factors, generating informations that can be used in the breeding programs [10] . Furthermore, morphological studies of biometrics seeds are essential for the development of efficient agricultural machinery and for adequate facilities to store production [7] .
The objective of this study was to investigate the genetic variability regarding the morphology of seeds of Jatropha curcas L., using biometric analyses to identify variables that have potential for the study of the diversity of the species and the classification of genotypes.
2. Materials and Methods
The experiment was conducted at the Laboratory of Plant Nutrition and Physiology located in the Centro de Ciências Agrárias of the Universidade Federal do Espírito Santo.
Seeds of genotypes of physic nut were provided by the breeding program of Embrapa Agroenergia, harvested from the in situ germplasm bank. The seeds from each genotype were benefited, eliminating immature and deteriorated seeds from the sample. The water content was kept at 10% - 12% and the seeds were packed and stored in refrigerator (3˚C) until the evaluations were performed.
The experimente followed a completelyrandomized design, with 22 genotypes of Jatropha curcas L. (Embrapa 248, Embrapa 184-I-2, Embrapa 249, Embrapa 283-I-1, Embrapa 144, Embrapa 245, Embrapa 246-I-4, Embrapa 308, Embrapa 273-I-2, Embrapa 301, Embrapa 201-I, Embrapa 167-II, Embrapa 149-I, Embrapa 192-I, Embrapa 127-I, Embrapa 304-I, Embrapa 180-I, Embrapa 275-I, Embrapa 255-I, Embrapa 211-I, Embrapa 171-I, Embrapa 110-II) studiedwithtenrepetitions. The experimental plot was composed by portions of 100 seeds.
According to the morphology of the seeds of physic nut, three dimensions were used to measure the main axis of their shape, using a digital caliper to determine length, larger diameter and smaller diameter of their middle section.
The weight of 1000 seeds was obtained in a precision weighing balance. The volume of the seeds was obtained in graduated cylinder, by the corresponding volume of water dislocated by samples of seeds, using two samples for each experimental plot. The seed samples were placed in paper bags and oven-dried in a drying oven, with forced air circulation at temperature of 65˚C, until constant weight, in order to determine their dry matter.
The data were subjected to analysis of variance by the F test (at 1% and 5% probability) and the genetic parameters were estimated based on the model using the model Yij = μ + gi + εij, with fixed genotypes.
The genetic divergence was assessed by means of multivariate cluster analysis. The Mahalanobis distance was used as dissimilarity measure, from which a dendrogram was constructed using the UPGMA criteria. The relative importance of each variable was calculated by the method proposed by Singh (1981). The analyses were performed with the statistical software GENES [11] . The template is used to format your paper and style the text. All margins, column widths, line spaces, and text fonts are prescribed; please do not alter them. You may note peculiarities. For example, the head margin in this template measures proportionately more than is customary. This measurement and others are deliberate, using specifications that anticipate your paper as one part of the entire journals, and not as an independent document. Please do not revise any of the current designations.
3. Results and Discussion
The results obtained for all the variables (Table 1) indicate the existence of significant variability in the shape
Table 1. Genotypic mean squares (MSg), means, estimate of phenotypic  genotypic
genotypic 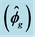 and environmental
and environmental 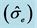 variances, coefficients of variation (CV), coefficients of genetic variation (CVg) and coefficients of genotypic determination (H2) of six characteristics of seeds of Jatropha curcas L.
variances, coefficients of variation (CV), coefficients of genetic variation (CVg) and coefficients of genotypic determination (H2) of six characteristics of seeds of Jatropha curcas L.
**Significant at 1%.
size and mass of the seeds among genotypes, making it possible to identify and classify them according to the characteristics of their seeds. Others author have also reported the existence of variability among genotypes of physic nut for many agronomic traits [7] -[9] .
The mean square related to the genotypes was statistically significant (p < 0.01) for all the variables, and the coefficients of variation (CV) were low and suitable for experimentation, with the highest value being 5.21% (Table 1). The range of dimensions for the size and the mass of seeds observed in the genotypes are in accordance with the described mean values for other Brazilian genotypes [1] [7] .
The estimate values of genotypic variances 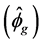 were higher than the values of environmental variance
were higher than the values of environmental variance  for all the characteristics of the seeds (Table 1). Therefore, the phenotypic variance
for all the characteristics of the seeds (Table 1). Therefore, the phenotypic variance  observed between the genotypes can be associated, in the most part, to the effect of their genetic differences, with smaller influence of environmental factors.
observed between the genotypes can be associated, in the most part, to the effect of their genetic differences, with smaller influence of environmental factors.
Considering the coefficient of genotypic determination, the characteristics of the seeds presented high values, ranging from 81.37% to 97.67% (Table 1), confirming that those characteristics; in special weight, dry matter and volume; were less influenced by environmental conditions.
The values of coefficients of genetic variation (CVg) compared to the low values of CV (Table 1), also indicate a favourable situation for the selection of genotypes aiming to change the overall size, shape and mass of physic nuts seeds. The weight of 1000 seeds, dry matter and volume of the seeds also presented a higher predominance of genetic factors over environmental factors, considering that the estimated CVg was superior to the CV (variation index over 1.00).
Figure 1 shows images from the seeds of some of the genotypes studied in this experiment, showing differences in the size and shape. Those differences, associate to the difference in the biomass accumulated in the seeds, allowed to classify the genotypes in different groups of homogeneous seed morphology through univariate and multivariate analyses.
The univariate study, following the Scott-Knott criteria (Table 2), allowed the separation of the genotypes in different homogenous groups for each variable. It was possible to identify a large number of different groups for the weight of 1000 seeds (seven groups), followed by the volume (six groups) and dry matter (five groups). For the dimensions of the seed (length and diameters), only three groups were formed.
Considering the dimensions of the seed, the genotypes 201-I, 149-I, 127-I, 304-I, 180-I, 255-I, 171-I and 110-II presented seeds with larger length, while the genotype 283-I-1 alone formed the group of smaller mean for this variable. The remaining genotypes were classified in a group of intermediate values, ranging from 16.70 to 17.27 mm (Table 2).
For the diameter of the rounder section of the seed (larger diameter), the genotypes 144, 308, 149-I, 127-I, 304-I, 180-I and 110-II formed the group with higher means, superior to 10.69 mm. The genotype 283-I-1 also returned the smaller mean for this variable (Table 2).
The diameters of the flatter section of the seed (smaller diameter) were bigger in the group formed by the genotypes 308, 201-I, 149-I, 127-I, 304-I and 180-I, with values over 8.57 mm. The smaller means were observed in the genotypes 283-I-1, 273-I-2, 167-II and 275-I (Table 2).
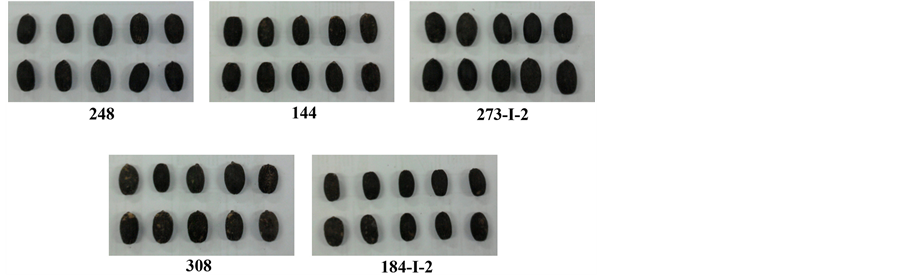
Figure 1. Seeds of genotypes of Jatropha curcas L. from the germplasm bank of the Brazilian breeding program developed by Embrapa Agroenergia.
Table 2. Means of six characteristics of seeds of 22 genotypes of Jatropha curcas L.
Means followed by the same letter in each column do not differ by the Scott-Knott test at 5% of probability.
Seeds from the genotype 110-II were heavier than all the others, presenting mean for weight of 1000 seeds of 764.99 g. There was high differentiation of genotypes for this variable, with formation of a larger number of homogeneous groups. The genotypes 283-I-1, 308 and 273-I-2 produced lighter seeds, with means under 554.40 g (Table 2).
Considering the dry matter accumulated in the seeds, the genotype 110-II also presented the higher mean values, with seeds weighting 0.66 g. After losing water content, the seeds with smaller mass were from the genotypes 283-I-1, 144, 273-I-2, 201-I and 149-I; presenting less than 0.52 g of dry matter (Table 2).
Seeds from the genotype 304-I had larger volume than all the others did, occupying 10.17 cm3, while the genotype 283-I-1 had the most compact seeds, with volume less than 33% smaller than the genotype 304-I (Table 2).
The genotypes 149-I, 127-I, 304-I and 180-I presented larger seeds in all dimensions, but their seeds didn’t have the higher means for weight of 1000 seeds or dry matter. This fact is associated to a lower density of their seeds, indicating that a larger space for storage may be required to store the same mass of seeds from those genotypes.
Thought the univariate analyses, two genotypes are highlighted for being part of the groups of superior and inferior means for most variables of dimension and mass: 283-I-1 and 110-II.
The dimensions of the seeds from the genotype 283-I-1 were smaller and with less accumulated biomass among the genotypes studied in this experiment. This genotype was included in the group of lowest means for all the variables in the univariate analyses.
The genotype 110-II presented seeds with larger length and diameter, but with a slightly flatter surface. In addition, this genotype alone presented the higher means for weight and dry matter, therefore, the bigger seeds of this genotype also shown an increased accumulated biomass.
Such characterization of the germplasm, allowing the study of the variability among the genotypes, is a preliminary activity that is fundamental for plant breeding. Studies of phenotypic variability have been developed for many vegetal species for centuries and are important to improve plant breeding programs, allowing preliminary selections of features [12] .
Table 3 presents the relative contributions of each variable of the seeds for the differentiation of genotypes. The variables related to the biomass of the seeds, weight of processed seeds (52.56%) and dry matter accumulated (41.88%), contributed more to the variability among the genotypes than their volume and dimensions. This result reaffirms the possibility of exploring the biomass of the seeds to identify differences between the genotypes.
The dissimilarity measures between the pairs of genotypes, estimated by the Mahalanobis generalized distance, are presented on Table 4. The largest distances were observed between the genotypes 308 and 110-II (D2 = 5089.47) and the lowest dissimilarity was observed between the genotypes 249 and 255-I (D2 = 4.55).
Additionally of presenting the highest value of D2, the genotype 110-II also participated in five of the ten largest dissimilarities between pairs of genotypes; followed by the genotype 184-I-2, which participated in three of the ten highest D2 values. The genotype 255-I presented similarities with 248, 249 and 192-I, resulting in three of the ten lowest measures of dissimilarities in this study.
Figure 2shows the clustering of the genotypes based on the Mahalanobis distance, using the value of 0.70 as cutoff for similarity in the dendrogram [13] , it is possible to distinguish five groups of genotypes: Group 1) 167-II, 171-I, 184-I-2, 301, 180-I; Group 2) 308, 149-I, 283-I-1, 304-I, 211-I; Group 3) 273-I-2, 127-I, 275-I; Group 4) 201-I, 255-I, 144, 246-I-4, 192-I; Group 5) 249, 245, 248, 110-II. Although the weight of 1000 seeds and the dry matter were the most important characteristics to explain the genetic variability between the 22 genotypes, those were not the only characteristics to group them together.
Grouping genotypes is an important tool to manage the selection of parents in breeding programs, allowing the establishment of combinations based on the dissimilarity between parental genotypes, exploring their potential to generate plants with better agronomic performance. This is especially useful for the selection individuals with different patterns of dissimilarity, evading the limitation of the genetic variability while also providing higher chance of gains through selection [13] .
Studies have been supporting the use of morphological characters of seeds as criteria for identification of species and genotypes, allowing to study the variability and to identify homogeneous groups between them [14] .
Table 3 . Relative contribution of six characteristics of seeds for the diversity observed between genotypes of Jatropha curcas L.
Contributions estimated by the method of Singh (1981), based on the Mahalanobis distance.
Table 4. Dissimilarity between pairs of genotypes of Jatropha curcas L., based on the Mahalanobis distance studied with six characteristics of seeds.
D2 maximum: 5089.47 (308 and 110-II); D2 minimum: 4.55 (249 and 255-I).
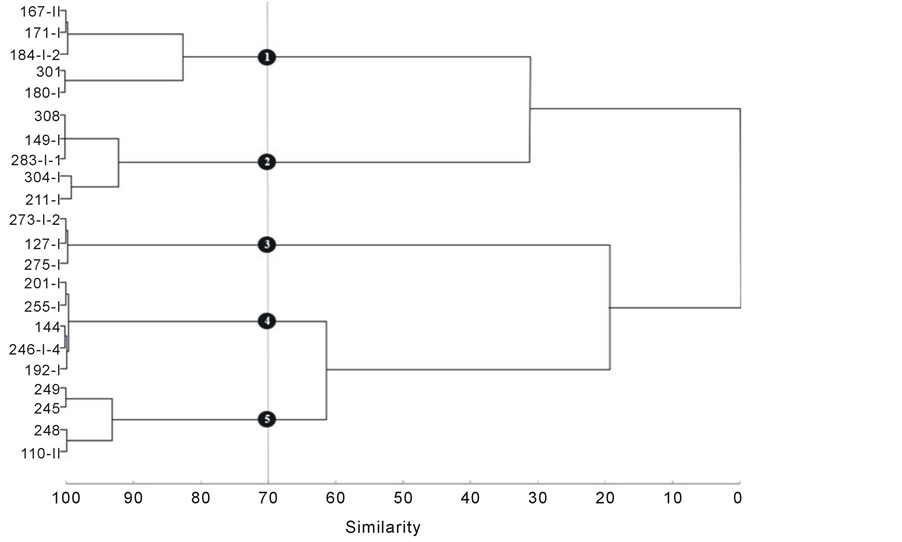
Figure 2. Dendrogram of similarity for 22 genotypes of Jatropha curcas L. using the UPGMA criteria, based on the Mahalanobis distance, obtained in the study of characteristics of seeds, classified in five groups.
4. Conclusions
The genotypes of Jatropha curcas L. selected by the Brazilian breeding program have high diversity for characteristics of the seeds, allowing the exploration of this genetic variability to classify genotypes in different groups.
Variables related to the biomass of the seeds present high relative contributions to the diversity observed in the genotypes.
References
- Arruda, F.P., Beltrão, N.E.M., Andrade, A.P., Pereira, W.E. and Severino, L.S. (2004) Physic Nut (Jatropha curca L.) Cultivation as an Alternative Crop for the Northeast Semi-Arid of Brazil. Revista Brasileira de Oleaginosas e Fibrosas, 8, 789-799.
- Drumond, M.A., Santos, C.A.F., Oliveira, V.R., Martins, J.C., Anjos, J.B. and Evangelista, M.R.V. (2010) Agronomic Performance of Different Genotypes of Physic Nut in the Semi-Arid Zone of Pernambuco State. Ciência Rural, 40, 44-47. http://dx.doi.org/10.1590/S0103-84782009005000229[in portuguese]
- Shrirame, H., Panwar, N. and Bamniya, B. (2011) Bio Diesel from Castor Oil—A Green Energy Option. Low Carbon Economy, 2, 1-6. http://dx.doi.org/10.4236/lce.2011.21001
- Nunes, C.F., Santos, D.N., Moacir, P. and Valente, T.C.T. (2009) External Morphology of Fruits, Seeds and Seedlings of Physic Nut. Pesquisa Agropecuária Brasileira, 44, 207-210. http://dx.doi.org/10.1590/S0100-204X2009000200014
- Francis, G., Edinger, R. and Becker, K. (2005) A Concept for Simultaneous Wasteland Reclamation, Fuel Production, and Socio-Economic Development in Degraded Areas in India: Need, Potential and Perspectives of Jatropha Plantations. Natural Resources Forum, 29, 12-24. http://dx.doi.org/10.1111/j.1477-8947.2005.00109.x
- Orhan, A.S., Dulger, Z., Kahraman, N. and Verizoglu, T.N. (2004) Internal Combustion Engines Fueled by Natural gas-Hydrogen Mixtures. International Journal of Hydrogen Energy, 29, 1527-1539. http://dx.doi.org/10.1016/j.ijhydene.2004.01.018
- Christo, L.F., Amaral, J.F.T., Laviola, B.G., Martins, L.D. and Amaral, C.F. (2012) Biometric Analysis of Seeds of Genotypes of Physic Nut (Jatropha curcas L.). Agropecuária Científica no Semi-Árido, 8, 1-6.
- Martins, L.D., Lopes, J.C., Laviola, B.G., Colodetti, T.V. and Rodrigues, W.N. (2013) Selection of Genotypes of Jatrophacurcas L. for Aluminium Tolerance Using the Solution-Paper Method. Idesia, 31, 81-86. http://dx.doi.org/10.4067/S0718-34292013000400011
- Amaral, J.F.T., Martins, L.D., Laviola, B.G., Christo, L.F., Tomaz, M.A. and Rodrigues, W.N. (2012) A Differential Response of Physic Nut Genotypes Regarding Phosphorus Absorption and Utilization Is Evidenced by a Comprehensive Nutrition Efficiency Analysis. Journal of Agricultural Science, 4, 164-173. http://dx.doi.org/10.5539/jas.v4n12p164
- Gusmão, E., Vieira, F.A. and Júnior, E.M.F. (2006) Fruits and Endocarps Biometry of Murici (Byrsonima verbascifolia Rich. Ex. A. Juss.). Revista Cerne, 12, 1.
- Cruz, C.D. (2013) GENES—A Software Package for Analysis in Experimental Statistics and Quantitative Genetics. Acta Scientiarum. Agronomy, 35, 271-276. http://dx.doi.org/10.4025/actasciagron.v35i3.21251
- Nunziata, A., Ruggieri, V., Greco, N., Frusciante, L. and Barone, A. (2010) Genetic Diversity within Wild Potato Species (Solanum spp.) Revealed by AFLP and SCAR Markers. American Journal of Plant Sciences, 1, 95-103. http://dx.doi.org/10.4236/ajps.2010.12012
- Cruz, C.D., Regazzi, A.J. and Carneiro, P.C.S. (2004) Biometric Models Applied to Genetic Improvement. Imprensa Universitária, Viçosa.
- Al-Ghamdi, F.A. (2011) Seed Morphology of Some Species of Indigofera (Fabaceae) from Saudi Arabia (Identification of Species and Systematic Significance). American Journal of Plant Sciences, 2, 484-495. http://dx.doi.org/10.4236/ajps.2011.23057.


