Open Journal of Applied Biosensor
Vol.2 No.3(2013), Article ID:35373,5 pages DOI:10.4236/ojab.2013.23010
Measurement of Temperature Induced Unfolding of DNA Hairpins by Microcantilever Sensors
1Laboratory for Structural Biology and the Department of Biological Science, University of Alabama, Huntsville, USA
2Department of Chemistry, University of Alabama, Huntsville, USA
3Department of Biology, University of Dayton, Dayton, USA
4Department of Chemical and Materials Engineering University of Alberta, Edmonton, Canada
Email: *georgem@uah.edu
Copyright © 2013 Joseph D. Ng et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received January 19, 2013; revised February 28, 2013; accepted March 5, 2013
Keywords: Hairpin DNA; Microcantilever; Thermal Melting; Unfolding
ABSTRACT
The technical feasibility of monitoring DNA melting and cooling transitions using a microcantilever substrate has been demonstrated and these results were compared with those measured in solution by circular dichroism. DNA hairpins have been immobilized on the surface of gold-coated microcantilever surfaces and their DNA melting and cooling transitions were monitored by nanomechanical deflections. The hairpins comprised of a 16 base-pair GACA repeat motif stem duplex with a 29 nucleotide variable region. Microcantilever deflection profiles, measured by the microcantilever response as a function of temperature, were unique to different hairpins indicative of the molecules’ general stability and denaturation characteristics. The major melting and cooling transition temperatures for all three immobilized oligonucleotides were between 41˚C - 52˚C. The composition and flexibility of the DNA stem loops were shown to influence the thermal transitions.
1. Introduction
We have examined the changes in microcantilever bending due to induced surface stress caused by the unfolding of DNA hairpin molecules with specific molecular binding [1,2]. Similar behavior has been observed in previous studies measuring macro-molecular interactions such as with protein [3,4] and DNA [5,6]. Microcantilever responses have also proven useful in label-free biodetection [6], quantitative binding assays [7,8] and proteomic analysis [9]. In this study, we immobilized DNA hairpin molecules on the surfaces of alkanethiol gold-coated microcantilevers and observed the response as the coated microcantilevers where systematically heated then cooled. The study indicates the feasibility of examining other similar biomolecules such as miRNA as a parallel study. Figure 1 depicts how the micromechanical motions of a microcantilever may be dependent upon the temperature-induced denaturation processes of DNA hairpin molecules.

Figure 1. Image depicting microcantilever deflection resulting from changes in surface stress as molecules unfold during melting. Hairpin molecules in (a) attached to microcantilever beam prior to heating. As heating begins (b), expansion of molecules lead to downward bending of beam. At (c) hairpins have expanded further and maximum deflection occurs. With continued heating, (d) hairpins open and this results in upward deflection of beam.
2. Discussion
Three large DNA hairpin molecules of the same size were synthesized (Phoenix BioTechnologies, Huntsville, AL) to examine the effects of loop flexibility on thermal denaturation and renaturation profiles. Each oligonucleotide contained 62 nucleotides with a 5’ C-6 thiol modifycation that can fold back on itself to form DNA hairpin structures. The thiol groups were used for immobilizing the hairpins on gold-coated microcantilevers. The hairpins were comprised of the same 16 base-pair stem duplex with a GACA repeat motif and a 5’ adenosine residue. The remaining 29 nucleotides were variable and contained sequences that were reverse compliment to genomic regions of the Satellite Tobacco Mosaic Virus, Borreliaburgdoferi, and Rickettsia rickettsiiin hairpins HpSt, HpBb, and HpRr, respectively. The theoretical secondary structures of the entire hairpin were predicted using nearest neighbor thermodynamic rules with the nucleic acid folding package, DNA MFOLD [10] (Figure 2).
The amount of hairpin DNA covalently bound on the cantilevers was estimated by a radiolabeling technique using P32-labeled thiolated DNA hairpins [1]. The surface density for all three hairpins was approximately 6 × 1010 molecules/cm [2]. The average distance between adjacent DNA molecules on the gold surface was calculated to be significantly bigger than its molecular gyration diameter.
Therefore, the thermal motion and conformational entropy force that contributes to the surface stress is not affected by the presence of neighboring molecules (calculation not shown). As such, the intramolecular forces arising from the DNA hairpin unfolding is the primary molecular process that contributes to the differential deflection observed as measured against a reference.
The bound DNA hairpins were separately heated in a sin-
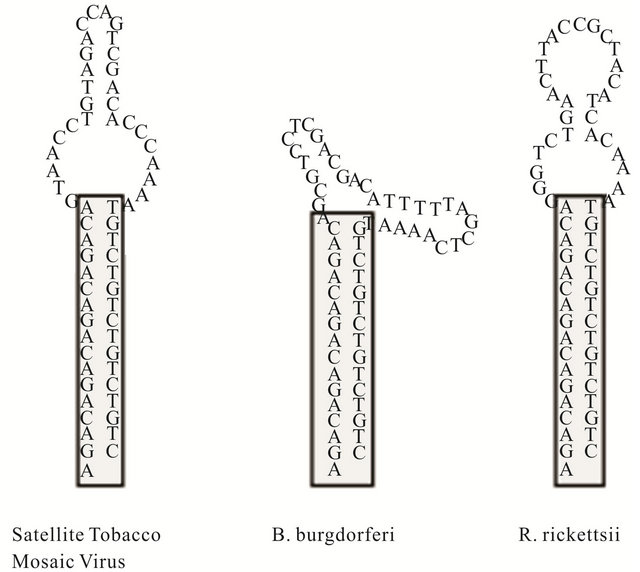
Figure 2. Secondary structureprediction of large DNA hairpins containing sequences that reverse complimentary to genomic sequences of Satellit Tobacco Mosaiv Virus (HpSt), Borrelinahurgdorferi (HpBp) and Rickettsiirickettsii (HpRr). These molecules will be represented in following figures by A, B and C respectively.
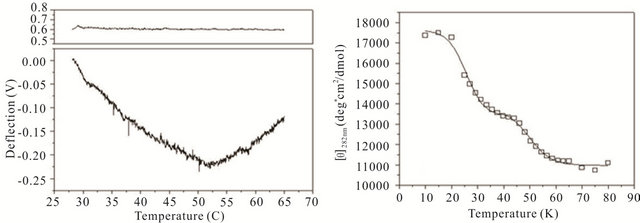
gle cantilever system. Microcantilever motion was generated from the interplay between changes in configurational entropy and intermolecular energetics as described by Alvarez et al. [1] and Wu et al. [2] Measuring the deflection differences between a blank and DNA coated cantilever as a function of temperature can produce a thermal denaturation (or melting) profile that reflects the molecular unfolding of a DNA hairpin. Figures 3(a)-(c) shows the changes in cantilever deflection over a range of increasing temperatures for the three DNA hairpins. Denaturation and renaturation profiles were obtained at heating and cooling rates at 1˚C/min. Each experimental profile of the DNA functionalized cantilever represents the average difference of nine reference and nine experimental deflection curves.
The hairpins were observed to undergo a multiple-state transition that represents an equilibrium process between the open random coil, partially opened hairpin, and completely closed hairpin states. The downward bending revealed by the negative deflection of the cantilever may be attributed to the unzipping process of internal stem structures within the hairpin loops caused by base pairing and stacking disruption. When the hairpins are completely opened, single-stranded DNA prevails having a variety of molecular conformations. Consequently the open state is characterized by high entropy and contributes to an upward cantilever bend resulting in a positive deflection. In the case of hairpin HpSt, minor transitions in the melting profile can be observed near 30˚C and 40˚C. The inflection point at 52˚C from downward to upward bending can be considered to be the principal melting temperature, Tm, where there exists an equal population of the opened and closed hairpin base stem region. A small transition temperature at 58˚C is finally noticed near the highest temperature examined. Similarly, the heating profile of HpBb had two minor temperature transitions near 34˚C and 55˚C and one major transition at 45˚C. Hairpin HpRr had only one major and one minor transition at 41˚C and 56˚C, respectively.
The heat denaturation behavior of the DNA hairpins was also observed by circular dichroism (CD) measurements to determine if oligonucleotides in solution have similar transition profiles to those immobilized on a microcantilever. Ellipticity measurements were collected between 200 and 320 nm wavelengths over a range of temperatures from 10˚C - 80˚C (Figures 3(d)-(f)). The characteristic wavelength for the DNA hairpins was empirically determined to be 282 nm and this data was plotted and fit using a non-linear squares fitting algorithm (Figures 3(d)-(f)). All three hairpins exhibited biphasic profiles, characterized by a possible duplex-hairpin transition at low temperature and a hairpin-coil at high temperature. We cannot conclude, at this point, any correlation to detailed hairpin structural changes with the temperature transitions observed. However, in the case of
 (a)
(a)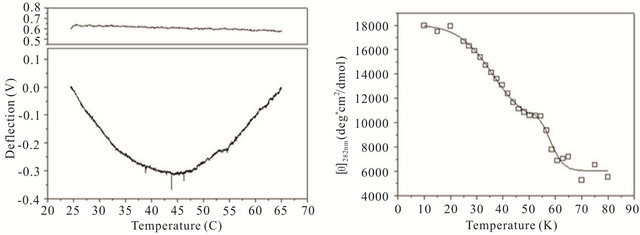 (d)
(d) (b)
(b)
Figure 3. Thermal denaturation profiles measured by differential deflection of microcantilever immobilized with hairpin DNA ((a), (b) and (c)) and optical ellipticity ((d), (e) and (f)) as a function of temperature. All experiments were compared to a reference deflection curve obtained from averaging measurements of nine blank cantilevers heated between 15.0˚C to 70.0˚C at an average rate of 1˚C/min. The upper panels on ((a), (b) and (c)) are the microcantilevers prior to attachment of hairpin molecules over the same heating range.
HpSt, the high temperature transition measured by CD near 53˚C corresponded to the major inflection temperature at 52˚C measured with the microcantilever. It is speculated that in this case, this transition temperature corresponded to the putative hairpin-coil transition. The major melting temperatures for all three immobilized oligonucleotides were between 41˚C - 52˚C and their variation may be influenced by the structural differences in the duplex-hairpin. In contrast to immobilized HpSt, cantilever bound HpBr and HpRr do not have any coincident transition temperatures with their corresponding hairpins in solution. Immobilized hairpins may undergo different unfolding mechanisms compared to the same molecules in free sution.
The temperature transitions for all of the hairpins result from the dynamic equilibrium between closed and opened confirmations. HpRr appeared to be the most kinetically active hairpin in this respect, resulting in easier opening transitions. It also appears that the loop configuration may, as it unfolds, impose a mechanistic molecular drive for the overall denaturation. The large loop in HpSt may take a highly distorted globular shape with very high entropy. As a result, the compact bulge would prevent the initiation of stem opening of the closing base pair closest to the loops, consequently raising the overall denaturation temperature. In comparison, the loops of HpBb are more rigid than those found in HpSt and do not fluctuate as much as HpRr. Hence, the major transition temperatures increase in the order of HpRr, HpBb and HpSt, respectively.
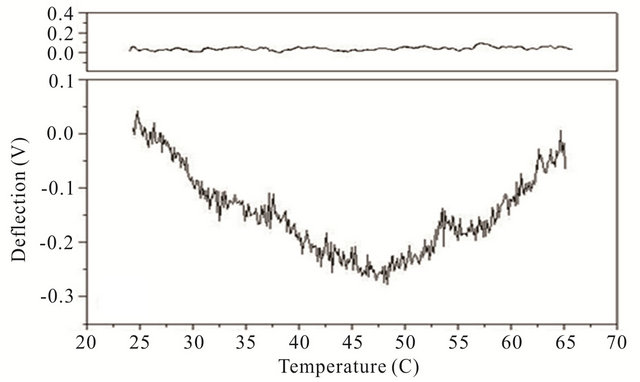
Cooling transitions of the DNA hairpin can also be observed using microcantilever deflection. Figure 4 shows the cooling profiles for the DNA hairpins. Microcantilever deflection profiles for denaturation and renaturation are different even though their starting and ending deflections are the same. Interestingly the major deflection, indicated by an upward cantilever bend, oc-
 (a)
(a)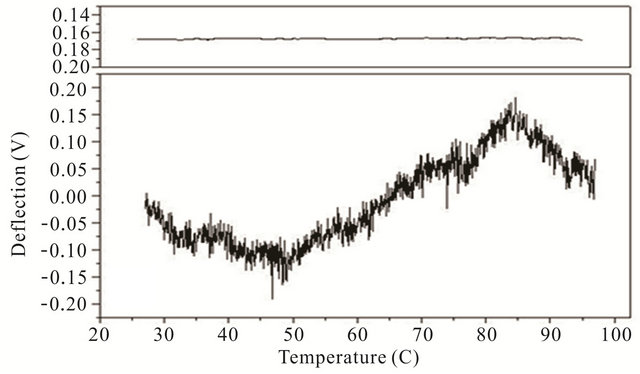 (b)
(b)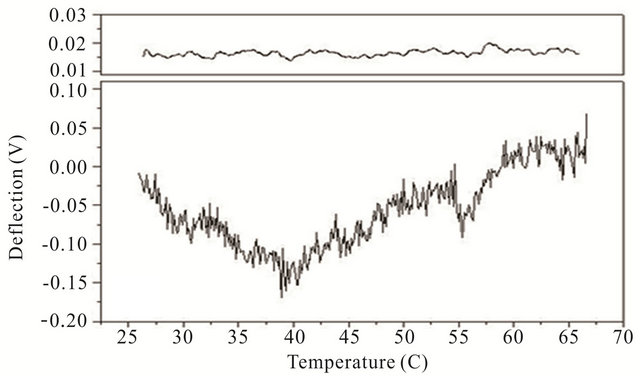 (c)
(c)
Figure 4. Cooling profiles for HpSt, HpBb and HpRr measured by microcantilever response. Top panels are heating profiles for the same microcantilevers prior to attachment of hairpin molecules.
cur-red near the same temperature as the major deflection point in the heating denaturation curves for all hairpins. This is not observed in cooling experiments with the same hairpins in solution measured by CD (data not shown). The opening transition depends on the unzipping energy of the hairpin while the closing transition depends on a more intricate pathway involving multiple interacttions of the stem stalks and base pairing events. Thus a stable closed hairpin configuration is only achieved after it undergoes a sequence of folded and unfolded states.
Our investigation demonstrates that a micromechanical cantilever is responsive to thermally induced local and global conformational changes in DNA hairpins. This technique may be useful in conducting, single molecule experiments on folding and denaturation characteristics of DNA hairpin formation. While the primary purpose of this study was to demonstrate the technical feasibility of monitoring DNA melting transitions using a microcantilever substrate, it was not our intention to explain the mechanism of sequence-dependent formation of multimembered loops or provide any thermodynamic or kinetic detail on structural hairpin formation. However, our results indicate that the composition and flexibility of large DNA stem loops influence thermal transitions of DNA hairpins, revealing general stability and denaturetion characteristics. Learning how DNA sequence affects the stability and conformational flexibility of hairpin structures is important in understanding regulation sites of genomic sequences as well as potential binding regions for exogenous drugs and proteins. We have observed similar results with RNA molecules using this technique and extensive studies are in progress.
3. Acknowledgements
This work was sponsored by NASA EPSCOR Grant Number NCC5-580, and the Office of Biological and Environmental Research (OBER), U.S. DOE. Oak Ridge National Laboratory (ORNL) managed by UT-Battelle, LLC for the U. S. Department of Energy under Contract No. DE-AC05-00OR22725.
REFERENCES
- M. Alvarez, L. G. Carrascosa, M. Moreno, A. Calle, A. Zaballos, L. M. Lechuga, A. C. Martinez and J. Tamayo, “Nanomechanics of the Formation of DNA Self-Assembled Monolayers and Hybridization on Micro-Cantilevers,” Langmuir, Vol. 20, No. 22, 2004, pp. 9663-9668. doi:10.1021/la0489559
- G. Wu, H. Ji, K. Hansen, T. Thundat, R. Datar, R. Cote, M. F. Hagan, A. K. Chakraborty and A. Majumdar, “Origin of Nanomechanical Cantilever Motion Generated from Biomolecular Interactions,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 98, No. 4, 2001, pp. 1560-1564. doi:10.1073/pnas.98.4.1560
- A. Kooser, K. Manygoats, M. P. Eastman and T. L. Porter, “Investigation of the Antigen Antibody Reaction between Anti-Bovine Serum Albumin (a-BSA) and Bovine Serum Albumin (BSA) Using Piezoresistive Microcantilever Based Sensors,” Biosensors and Bioelectronics, Vol. 19, No. 5, 2003, pp. 503-508.
- C. Grogan, R. Raiteri, G. M. O’Connor, T. J. Glynn, V. Cunningham, M. Kane, M. Charlton and D. Leech, “Characterisation of an Antibody Coated Micro-Cantilever as a Potential Immuno-Based Biosensor,” Biosensors and Bioelectronics, Vol. 17, No. 3, 2002, pp. 201-207. doi:10.1016/S0956-5663(01)00276-7
- F. Liu, Y. Zhang and Z. C. Ou-Yang, “Flexoelectric Origin of Nanomechanic Deflection in DNA-Microcantilever System,” Biosensors and Bioelectronics, Vol. 18, No. 5-6, 2003, pp. 655-660. doi:10.1016/S0956-5663(03)00047-2
- M. Hegner and Y. Arntz, “Advanced Biosensing Using Micromechanical Cantilever Arrays,” Methods in Molecular Biology, Vol. 242, 2004, pp. 39-49.
- J. Pei, F. Tian and T. Thundat, “Glucose Biosensor Based on the Microcantilever,” Analytical Chemistry, Vol. 76, No. 2, 2004, pp. 292-297. doi:10.1021/ac035048k
- R. McKendry, J. Zhang, Y. Arntz, T. Strunz, M. Hegner, H. P. Lang, M. K. Baller, U. Certa, E. Meyer, H. J. Guntherodt and C. Gerber, “Multiple Label-Free Biodetection and Quantitative DNA-Binding Assays on a Nanomechanical Cantilever Array,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 99, No. 15, 2002, pp. 9783-9788. doi:10.1073/pnas.152330199
- C. A. Savran, S. M. Knudsen, A. D. Ellington and S. R. Manalis, “Micromechanical Detection of Proteins Using Aptamer-Based Receptor Molecules,” Analytical Chemistry, Vol. 76, No. 11, 2004, pp. 3194-3198. doi:10.1021/ac049859f
- M. Zuker, “Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction,” Nucleic Acids Research, Vol. 31, No. 13, 2003, pp. 3406-3415. doi:10.1093/nar/gkg595
NOTES
*Corresponding author.

