Advances in Microbiology
Vol.2 No.4(2012), Article ID:25677,9 pages DOI:10.4236/aim.2012.24053
Transcriptional Expression Profile of Toll Like Receptor 1 - 10 mRNA in Bovine Peripheral Mononuclear Cells in Response to Foot and Mouth Disease Antigens
1Foot and Mouth Disease Research Laboratory, Indian Veterinary Research Institute, Bangalore Campus, Bangalore, India
2Foot and Mouth Disease Research Laboratory, Indian Veterinary Research Institute, Izatnagar, India
Email: *dechammahj@yahoo.com
Received April 9, 2012; revised May 11, 2012; accepted September 18, 2012
Keywords: Innate Immunity; FMDV; DNA Vaccine; TLR Genes
ABSTRACT
Toll like receptors are the primary component of innate immune response. The comparison of expression of innate immune genes TLR 1 - 10 was carried out by real time PCR by quantifying mRNA in bovine PBMCs. The bovine PBMCs in vitro stimulated with FMD infectious virus lead to up regulation of TLR 2, 3, 4 and down regulation of TLR 9. But stimulation with DNA vaccine carryingVP1 genes of FMDV leads to up regulation of TLR 2 only. Difference in the TLR gene expression by the virus and DNA vaccine, may be related to the outcome of the vaccine and carrier or persistent state of the animal which is seen in FMDV.
1. Introduction
Toll like receptors (TLR’s) are an important family of molecules which enable the immune cells to detect the presence of pathogens and activate innate immune defenses [1]. Innate immunity is either immediately available or rapidly activated and hinges on pattern recognition receptors (PRR) that recognizes conserved microbial signature molecules present on the pathogen and is relatively nonspecific. Foot and Mouth Disease is one of the economically important diseases of farm animals. The current conventional vaccine used for control of FMD induces protection due to induction of a combination of innate and adaptive immune response [2-4]. However doesn’t induce sterile immunity. Hence alternate DNA vaccine against FMD has been studied considerably [5-7]. But most of the DNA vaccine reports support protection against the disease in lab animals which unfortunately does not elicit full protection in cattle. Hence in-vitro studies involving homologous host immune cells become very important before in-vivo studies.
The importance of innate immune defenses for the control of FMDV, particularly early during infection has been reported [8]. FMDV can evade immune responses through its ability to shut down cellular protein synthesisincluding IFN type 1, in susceptible epithelial cells. Despite this, innate immune responses are probably induced in vivo, although detailed studies on this subject are lacking, accordingly, the interaction of FMDV with cells of the innate immune system is of particular interest. Moreover, innate response represents a promising strategy to develop improved emergency vaccine capable of rapidly establishing a protective status. Many studies on the interaction of FMDV with cells of innate immune system have been performed in porcine or mouse models, and it will be informative to verify these findings with ruminant cells. Hence in the current work expression of TLR genes in stimulated PBMCs with FMD virus and DNA vaccine plasmid is compared.
2. Materials and Methods
2.1. FMDV Multiserotype DNA Vaccine
Foot and mouth disease virus serotype O (Ind R2/75), A (Ind 17/77), Asia I (62/72) vaccine strains maintained in the FMD research Lab, IVRI, Bangalore, passaged in BHK-21 clone 13 monolayer cells and used as the source of virus for amplification of VP1 gene and stimulation of peripheral mononuclear cells. Total RNA was extracted by trizol method. VP1 gene of FMDV was amplified from each serotype separately by cDNA synthesis followed by PCR using serotype specific primers (Table 1). The amplicons were separately digested with restriction enzymes (“O” with Kpn1 & BamH1; “A” with BamH1 & EcoR1; “Asia 1” with EcoR1 & Not1) and ligated to pBSK+ and subcloned in pcDNA™3.1(+) vector digested with Kpn I and Not I sequentially (Figure 1(a)). Primers were designed with glycine and proline extra sequences as linkers between the FMDV VP1 genes of O, A and Asia. The final vaccine construct sequence was confirmed by sequence analysis (Bioserve India Ltd).
2.2. FMDV Marker Vaccine
FMDV marker vaccine was constructed by releasing EGFP 730 bp gene fragment from pCEGFP plasmid (available in lab) by EcoR1 digestion and ligated to EcoRI digested FMDV VP1 multiserotype vaccine. The marker vaccine had FMDV VP1 “O” at 5’ end followed by FMDV VP1 “A”, EGFP and FMDV VPI Asia I at the 3’ end. The full length linked gene with only one start codon at 5’ end and terminal sequence of the vector at 3’ end was constructed.
2.3. Functional Evaluations of Vaccine Construct
BHK 21 cell with 80% monolayer were transfected with FMDV multiserotype vaccine and FMDV EGFP marker vaccine separately. Cells transfected with FMDVVP1- EGFP was observed for green fluorescence in fluorescent microscope under 495 nm (Nikon). Cells transfected with FMDV VP1 multiserotype vaccine was collected after 48 h, lysed by freeze thawing and immunoblotted against
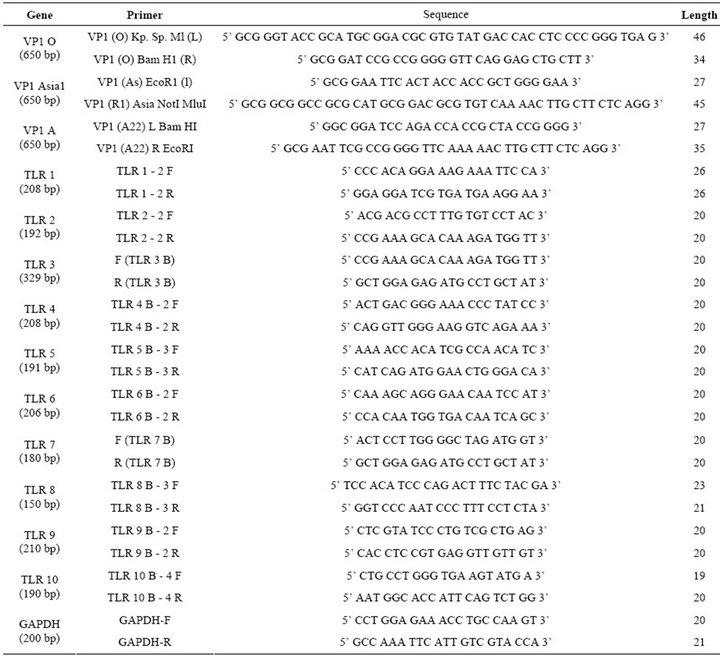
Table 1. List of primers showing the size and sequence used in the study.
FMDV O antibodies as per standard procedure [9].
2.4. Peripheral Mononuclear Cells Isolation and Stimulation
Growth of peripheral blood mononuclear cells: Animal ethics were followed as per the institute animal ethics committee guide lines. Freshly collected venous blood from Hallikar breed of cattle was used as source of PBMCs. Cells were isolated by a density gradient centrifugation using Histopaque1077 (Sigma, India) as reported [10] and cell number was counted by dye exclusion method [11]. Cells 2 × 106 in RPMI-1640 (Sigma) were electroporated with 5 µg of FMDV-VP1 multiserotype DNA vaccine in 0.4 cm gene pulse cuvette (Bio-Rad, India) with the following conditions 250 v, 950 µF capacitance and infinity resistance [12]. Immediately after the pulse cells wereresuspended in 1 mL RPMI 1640 containing 10% FBS and plated in 6 well tissue culture plate. For virus transfection 100 µL of pooled O, A, Asia1 FMDV of 3.9 pfu was used (strain “O” IndR2/75; “A” Ind 17/77; “Asia I” 62/72). Cells were incubated at 37˚C in 5% CO2 incubator for 48 h. Cells transfected with 5 µg of vector pcDNA™3.1(+) and without any plasmid DNA were used as control.
2.5. RNA Isolation and RT PCR
Total cellular RNA was isolated using TRIZOL (Invitrogen, India). The total RNA was treated with Dnase I and followed by RNA clean up protocol according to the manufacturer protocol (Quiagen, India). RNA was stored at –80˚C until analyses. cDNA was synthesized by a reverse transcription PCR from 5 µg of total RNA using 200 units of superscript III (Invitorgen, India) using oligod (T) primer in a total reaction of 20 µL. The obtained cDNA was diluted 1/20 with water and 10 µL were used for amplification. TLR (1 - 10) specific primer sets were optimized for the real time PCR. The primer sequence and ampliconsize are shown in Table 1. The PCR was performed with the light cycler fast start DNA SYBR green (genetix) according to the protocol provided in the kit. The control for specificity of the amplification products, a melting curve analysis was performed. No ampli-
 (a)
(a) (b)
(b)
Figure 1. (a) Strategy of cloning FMDV VP1 of O; A and Asia1 in pBSK+ and subcloning the entire cassette in pCDNA3.1 vector. Pcpl3 (7.4 kbp) shows the final vaccine construct; (b) Nuceotide sequence of linked FMDV VP1 multiserotype vaccine (1993 bp).
fication of unspecific products was observed. All the primers efficiency was in the range of 1.0 to 1.25. Keeping the efficiency of primers as one the data obtained from each sample, detailed in methods was subjected to analysis using REST2008 programme. This programme uses boot strap randomization techniques to determine whether an observed up or down regulation in samples significant after normalization to housekeepers GADPH with integration of efficiency variation into hypothesis tests and the introduction of confidence intervals for expression. Amplification of GADPH was employed as an internal control. Reaction without cDNA was included as negative control. To ensure cDNA samples were not carrying genomic DNA, reactions were set up using 10 ng of RNA as negative control. Real time PCR reaction was run with ABI 7300 HT (Applied biosystems, India).
2.6. Statistical Analysis
For each of three samples gene expression was quantified by normalizing each target TLR against the reference gene, GADPH, through the statistical analysis package (REST2008). The near of three values was determined and the mean result is expressed in mean normalized expression ± standard error.
3. Results
3.1. Functional Evaluation of FMDV-VP1 Multiserotype Vaccine
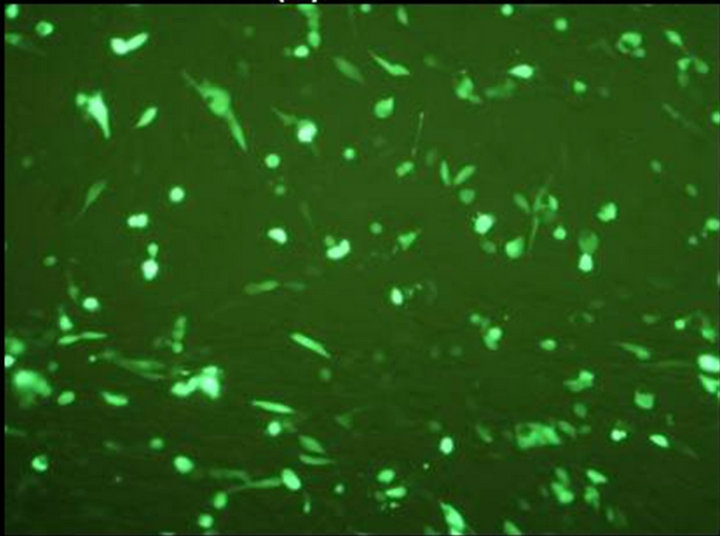
The final vaccine construct in pcDNA™3.1(+) (Figure 1(b)) was tested by transfecting the BHK-21 cells with marker vaccine which carried EGFP gene. Figure 2 shows the BHK-21 cells expressing green fluorescent protein under the green filter (495 nm). The cells transfected with EGFP marker vaccine show the green fluorescence and control cells transfected with pcDNA™3.1(+) vector look healthy.
The specificity of the protein encoded by the gene
 (a)
(a) (b)
(b)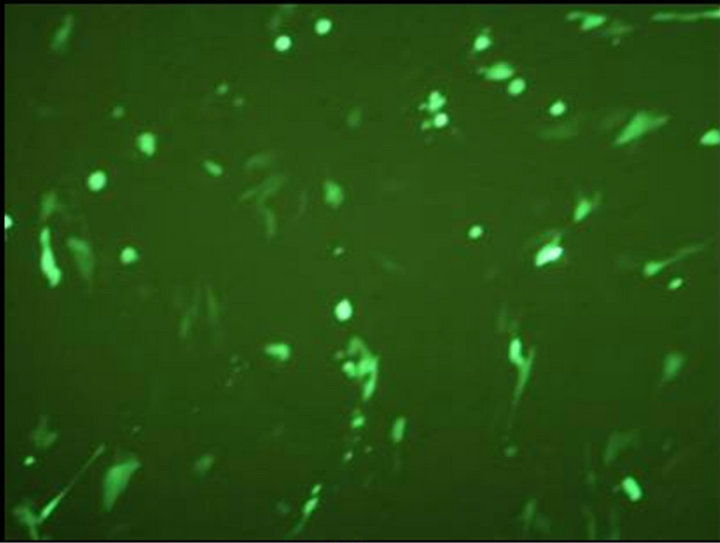 (c)
(c) (d)
(d)
Figure 2. Evaluation of FMDV VP1 multiserotype vaccine in BHK-21 Cells. (a) BHK21 cells transfected with marker vaccine carrying EGFP gene showing green fluorescence under 495 nm filter; (b) Same cells under white light; (c) BHK21 cells transfected with pcDNA™3.1(+) EGFP clone DNA (Positive control) showing green fluorescence under 495 nm filter; and (d) Under white light.
construct was evaluated by immunoblotting with FMDV “O” hyper immune sera. The cell lysate from FMDVVP1 multiserotype transfected BHK-21 cells reacted with FMDV “O” antibodies showing reaction band of 72 kDa protein which is encoded by all the three VP1 genes of FMDV (Figure 3).
3.2. TLR mRNA Expression in PBMCs Induced by FMDV
Report of expression of TLR 1, TLR 2, TLR 3, TLR 4, and TLR 7 was observed in viral diseases [13,14]. Here we studied the effect FMDV infection on TLR mRNA level in bovine PBMCs which are essential for innate immune response against viruses. Three independent experiments on bovine PBMCs were carried out from different individuals collected blood and real time RT-PCRs were run in parallel and mRNA quantifications were normalized relative to non infected PBMCs. In all cases similar Ct values were observed for internal gene control GAPDH suggesting cultures are treated under uniform conditions. Hence, the observed differences were due to the experimental conditions created. In the first condition of infecting the PBMCs with FMD virus, lead to up regulation of TLR 2, TLR 3, TLR 4 and down regulation of TLR 9 (Table 2). In the second condition of transfecting the PBMCs with FMDV VPI multi serotype DNA vaccine lead to up regulation of TLR 2 (Table 3). In the third condition of transfecting PBMCs with pcDNA™3.1(+) vector DNA lead to up regulation of TLR 9 (Table 4). The remaining TLR gene expression was not affected by transfection of FMDV or FMDV VPI multiserotype DNA vaccine.
4. Disscussion
The interaction of foot and mouth disease virus (FMDV) with the innate immune system is not much studied, partly because of restrictions on handling of this highly contagious virus and partly because of complexicity of the disease. More knowledge about the innate immune response to FMDV is urgently required to aid the devel-

Figure 3. Western blot analysis of FMDV VP1 multiserotype vaccine transfected in BHK21 cells. Lane 1: BHK21 control cell lysate. Lane 2: Cell lysate from pcDNA™3.1(+) vector transfected BHK21 cells. Lane 3: Cell lysate from FMDV VP1 multiserotype vaccine transfected BHK21 cells. Lane M: Prestained protein marker.
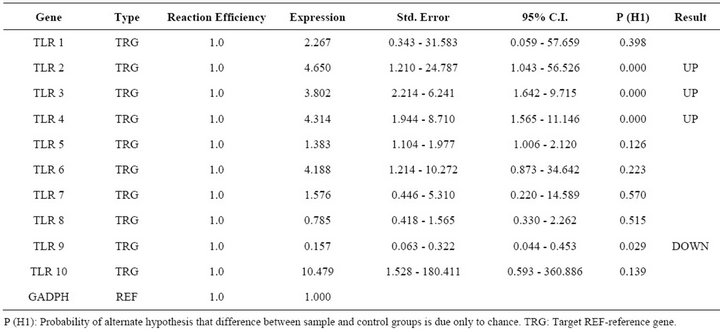
Table 2. Relative expression of TLR 1 - 10 genes in FMDV infected PBMCs.
opment of effective vaccines and therapeutics that prevent FMDV dissemination. In the current study attempts have been made to identify the TLRs (1 to 10) in response to FMDV infection and FMDV DNA vaccine in Bovine PBMCs in in-vitro. The FMDV VP1 antigen of all the serotypes carried by DNA vaccine was compared with infectious FMDV to understand at mechanism for failure of DNA vaccines in primary host cattle which show protective immune response in guinea pigs.
Many factors influence the immunogenicity of DNA vaccines; particularly elements in the plasmid backbone play an important role [15-17]. Along with highly conserved FMDV sequence Arg-Gly-Asp (RGD) and highly variable antigenic VPI region [18-20] of O, A and AsiaI serotypes were included in the DNA vaccine. For the free mobility of the antigen of each serotype, Gly-Pro amino
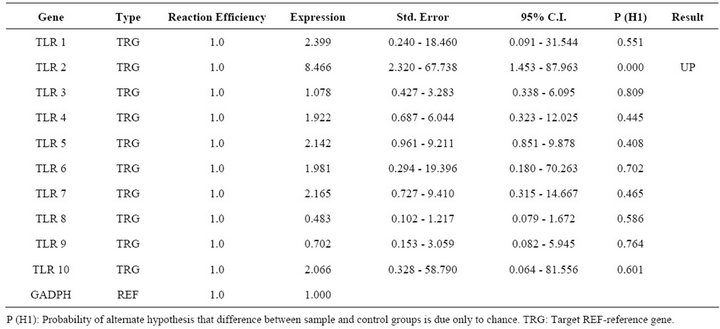
Table 3. Relative expression of TLR 1 - 10 genes in FMDV VP1 multiserotype vaccine DNA transfected PBMCs.
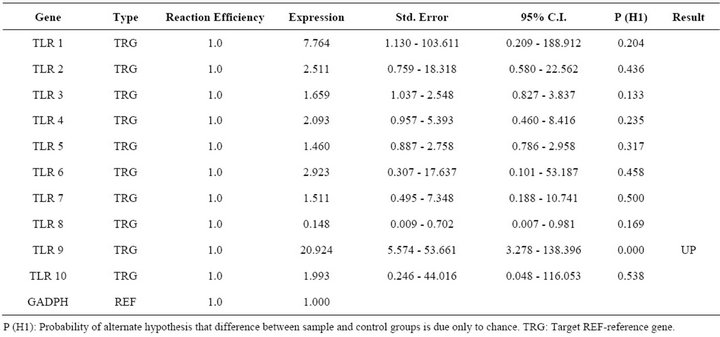
Table 4. Relative expression of TLR 1 - 10 genes in pCDNA3.1 vector transfected PBMCs.
acids were used as linkers which least affect the antigenicity of the protein [21]. FMDV VP1 multiserotype vaccine plasmid DNA encoded for 72 kDa protein, which is translated in single frame fusion protein. Also in linking 2A region of FMDV VP1 was excluded to avoid ribosomal skipping which is observed in FMDV. The expected single protein band was immuno detected with FMDV “O” hyper immune serum confirms the antigenicity of the expressed protein. Since FMDV serum shows cross reaction with other serotypes [22], reaction with A and O sera also reacted similarly (result not shown) wherein the size of the protein confirms the integrity of all the three serotypes.
Quantitation of mRNA of TLR 1 - 10 in Bovine PBMCs during FMDV Infection and Transfection with DNA Vaccine in Vitro
The identification of combinatorial sets of TLRs which are associated with the recognition of FMDV molecules is critical to understand virus host interactions. The immunological repertoire elicited by FMDV comprises the effector functions of immune cells equipped with the corresponding set of TLR, as well as the regulatory role of these primary target cells on other immunocompetent cells. In general it is well established that four TLR members play a critical role in regulation of viral nucleic acids [14]. TLR 3 recognizes dsRNA, TLR 7 and 8 recognize ssRNA and TLR 9 responds to dsDNA viruses [23] recognizing non-methylated viral CpG containing DNA. Here we report the first study on TLRs mRNA expression in bovine PBMCs during FMDV infection. FMDV is ssRNA and it is expected that any of the above TLRs mRNA to be increased. However except for TLR 3 other two TLR 2 and TLR 4 are contradictory to previous results on other viruses [24]. Only one report on FMDV infected nasal associated lymphoid tissue reported increased expression of TLR 4 indicating a role in inducing expression of type I IFN mRNA [25]. Up regulation of TLR2 is observed in both virus infected cells as well as in DNA vaccine transfected cells in which FMDV antigen was a common factor. TLR 2 is a promiscuous receptor that generally recognizes bacterial lipotechic acid, peptidoglycans or lipoproteins and functions as a heterodimer with either TLR 1 or TLR 6 [26]. In FMDV infected or vaccinated animals an increased level of typeI IFN was reported [27]. So it could be due to complementary interaction of TLR 2 with TLR 3 or 4 followed with signal pathways which may induce antiviral agents. The vaccine DNA successfully reaching nucleus may express the encoded antigen which in turn processed as natural viral antigen in endosomes leading to increased expression of TLR 2.
Increased expression of TLR 3 and 4 only in case of FMDV infected cells may be due to viral replication in infected cells which result in generation of dsRNA intermediates, which are PAMP andnot a constituent of host cell [28]. Since unmethylated CpG motifs are present in the vector DNA, increased mRNA for TLR9 was observed. However, vaccine DNA transfected cells did not show increase in TLR 9 could be due to FMDV VP1 antigen if contributed for the down regulation of TLR 9 in FMDV infected cells, may be the same antigen carried by the DNA vaccine reduced expression of TLR 9 in cells but not significantly. Because in vivo replication of virus lead to more antigen expression in FMDV infected cells. Other possibility could be most RNA viruses have evolved strategies to sequester dsRNA by a variety of mechanisms to avoid activation of these antiviral pathways [29] and down regulation of TLRs might have important immunosuppressive effect [30] or viral persistence which is also observed in FMDV after the natural infection [25].
Our investigation showed very interesting results, showing TLR 2 is up regulated in FMDV infection and FMD DNA vaccine transfection conditions which contribute the antigen. However, major difference of up regulation of TLR 3 & 4, down regulation of TLR 9 observed in FMDV infection needs more studies to support the claim made in this report.
5. Acknowledgements
We are grateful to Department of Biotechnology for the financial support. The authors thank the Director, IVRI and the Joint Director for the facilities provided for carrying out this work.
REFERENCES
- D. Werling, J. Piercy and T. J. Coffey, “Expression of TollLike Receptors (TLR) by Bovine Antigen-Presenting CellsPotential Role in Pathogen Discrimination?” Veterinary Immunology and Immunopathology, Vol. 112, No. 1-2, 2006, pp. 2-11. doi:10.1016/j.vetimm.2006.03.007
- P. V. Barnett and H. Carabin, “A Review of Emergency Foot-and-Mouth Disease (FMD) Vaccines,” Vaccine, Vol. 20, No. 11-12, 2002, pp. 1505-1514. doi:10.1016/S0264-410X(01)00503-5
- R. C. Rigden, C. P. Carrasco, P. V. Barnett, A. Summerfield and K. C. McCullough, “Innate Immune Responses Following Emergency Vaccination against Foot-andMouth Disease Virus in Pigs,” Vaccine, Vol. 21, No. 13-14, 2003, pp. 1466-1477. doi:10.1016/S0264-410X(02)00663-1
- A. L. Barnard, A. Arriens, S. Cox, P. Barnett, B. Kristensen, A. Summerfield and K. C. McCullough, “Immune Response Characteristics Following Emergency Vaccination of Pigs against Foot-and-Mouth Disease,” Vaccine, Vol. 23, No. 8, 2005, pp. 1037-1047. doi:10.1016/j.vaccine.2004.07.034
- Q. Yao, P. Qian, Q. Huang, Y. Cao and H. Chen, “Comparison of Immune Responses to Different Foot-andMouth Disease Genetically Engineered Vaccines in Guinea Pigs,” Journal of Virological Methods, Vol. 147, No. 1, 2008, pp. 143-150. doi:10.1016/j.jviromet.2007.08.027
- D. Dory, M. Remond, V. Beven, R. Cariolet, S. Zientara and A. Jestin, “Foot-and-Mouth Disease Virus neutralizing Antibodies Production Induced by pcDNA3 and Sindbis Virus Based Plasmid Encoding FMDV P1-2A3C3D in Swine,” Antiviral Research, Vol. 83, No. 1, 2009, pp. 45- 52. doi:10.1016/j.antiviral.2009.03.004
- A. K. Chockalingam, S. Thiyagarajan, N. Govindasamy, R. Patnaikuni, S. Garlapati, R. R. Golla, D. H. Joyappa, P. Krishnamshetty and V. V. Veluvarti, “Study of a Chimeric Foot-and-Mouth Disease Virus DNA Vaccine Containing Structural Genes of Serotype O in a Genome Backbone of Serotype Asia 1 in Guinea pigs,” Acta Virologica, Vol. 54, No. 3, 2010, pp. 189-195. doi:10.4149/av_2010_03_189
- A. Summerfield, L. Guzylack-Piriou, L. Harwood and K. C. McCullough, “Innate Immune Responses against Footand-Mouth Disease Virus: Current Understanding and Future Directions,” Veterianry Immunology and Immunopathology, Vol. 128, No. 1-3, 2009, pp. 205-210. doi:10.1016/j.vetimm.2008.10.296
- J. Sambrook and D. W. Russell, “The Condensed Protocols from Molecular Cloning: A Laboratory Manual,” Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 2006.
- A. Boyum, “Isolation of Mononuclear Cells and Granulocytes from Human Blood. Isolation of Monuclear Cells by One Centrifugation, and of Granulocytes by Combining Centrifugation and Sedimentation at 1 g,” Scandinavian Journal of Clinical & Laboratory Investigation, Vol. 97, 1968, pp. 77-89.
- S. A. Altman, L. Randers and G. Rao, “Comparison of Trypan Blue Dye Exclusion and Fluorometric Assays for Mammalian Cell Viability Determinations,” Biotechnology Progress, Vol. 9, No. 6, 1993, pp. 671-674. doi:10.1021/bp00024a017
- P. Konstantinova, O. Brake, J. Haasnoot, P. De Haan and B. Berkhout, “Trans-Inhibition of HIV-1 by a Long Hairpin RNA Expressed within the Viral Genome,” Retrovirology, Vol. 4, 2007, p. 15. doi:10.1186/1742-4690-4-15
- K. W. Boehme, M. Guerrero and T. Compton, “Human Cytomegalovirus Envelope Glycoproteins B and H Are Necessary for TLR2 Activation in Permissive Cells,” Journal of Immunology, Vol. 177, No. 10, 2006, pp. 7094-7102.
- G. M. Barton, “Viral Recognition by Toll-Like Receptors,” Seminars in Immunology, Vol. 19, No. 1, 2007, pp. 33-40. doi:10.1016/j.smim.2007.01.003
- Y. Sato, M. Roman, H. Tighe, D. Lee, M. Corr, M. D. Nguyen, G. J. Silverman, M. Lotz, D. A. Carson and E. Raz, “Immunostimulatory DNA Sequences Necessary for Effective Intradermal Gene Immunization,” Science, Vol. 273, No. 5273, 1996, pp. 352-354. doi:10.1126/science.273.5273.352
- D. M. Klinman, G. Yamshchikov and Y. Ishigatsubo, “Contribution of CpG Motifs to the Immunogenicity of DNA Vaccines,” Journal of Immunology, Vol. 158, No. 8, 1997, pp. 3635-3639.
- W. W. Leitner, H. Ying and N. P. Restifo, “DNA and RNA-Based Vaccines: Principles, Progress and Prospects,” Vaccine, Vol. 18, No. 9-10, 1999, pp. 765-777. doi:10.1016/S0264-410X(99)00271-6
- E. Pfaff, H. J. Thiel, E. Beck, K. Strohmaier and H. Schaller, “Analysis of Neutralizing Epitopes on Foot-andMouth Disease Virus,” Journal of Virology, Vol. 62, No. 6, 1988, pp. 2033-2040.
- M. Leippert, E. Beck, F. Weiland and E. Pfaff, “Point Mutations within the Beta G - Beta H Loop of Foot-andMouth Disease Virus O1K Affect Virus Attachment to Target Cells,” Journal of Virology, Vol. 71, No. 2, 1997, pp. 1046-1051.
- S. B. Nagendrakumar, G. S. Reddy, D. Chandran, D. Thiagarajan, P. N. Rangarajan and V. A. Srinivasan, “Molecular Characterization of Foot-and-Mouth Disease Virus Type C of Indian Origin,” Journal of Clinical Microbiology, Vol. 43, No. 2, 2005, pp. 966-969. doi:10.1128/JCM.43.2.966-969.2005
- A. Rudensky, P. Preston-Hurlburt, S. C. Hong, A. Barlow and C. A. Janeway Jr., “Sequence Analysis of Peptides Bound to MHC Class II Molecules,” Nature, Vol. 353, No. 6345, 1991, pp. 622-627. doi:10.1038/353622a0
- A. A. Andersen, “Cross Reactions of Normal Bovine Sera with Foot-and-Mouth Disease Virus: Incidence, Duration, and Effect of Shipping Stress,” American Journal of Veterinary Research, Vol. 39, No. 4, 1978, pp. 603-606.
- K. Tabeta, P. Georgel, E. Janssen, X. Du, K. Hoebe, K. Crozat, S. Mudd, L. Shamel, S. Sovath, J. Goode, L. Alexopoulou, R. A. Flavell and B. Beutler, “Toll-Like Receptors 9 and 3 as Essential Components of Innate immune Defense against Mouse Cytomegalovirus Infection,” Proceedings of the National Academy of Sciences USA, Vol. 101, No. 10, 2004, pp. 3516-3521. doi:10.1073/pnas.0400525101
- K. Crozat and B. Beutler, “TLR7: A New Sensor of Viral Infection,” Proceedings of the National Academy of Sciences USA, Vol. 101, No. 18, 2004, pp. 6835-6836. doi:10.1073/pnas.0401347101
- Z. Zhang, J. B. Bashiruddin, C. Doel, J. Horsington, S. Durand and S. Alexandersen, “Cytokine and Toll-Like Receptor mRNAs in the Nasal-Associated Lymphoid Tissues of Cattle during Foot-and-Mouth Disease Virus Infection,” Journal of Comparitive Pathology, Vol. 134, No. 1, 2006, pp. 56-62. doi:10.1016/j.jcpa.2005.06.011
- U. Zahringer, B. Lindner, S. Inamura, H. Heine and C. Alexander, “TLR2-Promiscuous or Specific? A Critical Re-Evaluation of a Receptor Expressing Apparent Broad Specificity,” Immunobiology, Vol. 213, No. 3-4, 2008, pp. 205-224. doi:10.1016/j.imbio.2008.02.005
- G. Cheng, X. Zhao, W. Yan, W. Wang, X. Zuo, K. Huang, Y. Liu, J. Chen, J. Wang, W. Cong, M. Liu, H. Gao, Y. Lu and Z. Zheng, “Alpha Interferon Is a Powerful Adjuvant for a Recombinant Protein Vaccine against Footand-Mouth Disease Virus in Swine, and an Effective Stimulus of in Vivo Immune Response,” Vaccine, Vol. 25, No. 28, 2007, pp. 5199-5208. doi:10.1016/j.vaccine.2007.04.089
- E. Peterhans, T. W. Jungi and M. Schweizer, “BVDV and Innate Immunity,” Biologicals, Vol. 31, No. 2, 2003, pp. 107-112. doi:10.1016/S1045-1056(03)00024-1
- M. Schroder and A. G. Bowie, “An Arms Race: Innate Antiviral Responses and Counteracting Viral Strategies,” Biochemical Society Transactions, Vol. 35, No. 6, 2007, pp. 1512-1514. doi:10.1042/BST0351512
- M. V. Borca, I. Gudmundsdottir, I. J. Fernandez-Sainz, L. G. Holinka and G. R. Risatti, “Patterns of Cellular Gene Expression in Swine Macrophages Infected with Highly Virulent Classical Swine Fever Virus Strain Brescia,” Virus Research, Vol. 138, No. 1-2, 2008, pp. 89-96. doi:10.1016/j.virusres.2008.08.009
NOTES
*Corresponding author.

