Journal of Biosciences and Medicines
Vol.03 No.05(2015), Article ID:55956,11 pages
10.4236/jbm.2015.35002
DNA Marker Technologies in Plants and Applications for Crop Improvements
Djshwar Dhahir Lateef
Field Crop Departments, College of Agriculture, University of Sulaimani, Sulaimaniyah, Iraq
Email: djshwar.lateef@univsul.edu.iq
Copyright © 2015 by author and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 8 March 2015; accepted 22 April 2015; published 24 April 2015
ABSTRACT
Over the past several decades, especially through traditional breeding programme, intensive attempts have been made for the improvement of a large number of cereal varieties which adjusted to diverse agro-ecologies. However, increasing biotic and abiotic stresses, increasing populations, and sharply reducing natural resources especially water for agricultural purposes, push the breed- ers for organizing and developing improved cereal varieties with higher yield potential. In combination with developments in agricultural technology, plant breeding has made remarkable progress in increasing crop yields for over a century. Molecular markers are widely employed in plant breeding. DNA markers are being used for the acceleration of plant selection through marker-assist- ed selection (MAS). Genes of agronomic and scientific importance can be isolated especially on the basis of their position on the genetic map by using molecular markers technologies. In this review, the current status of marker development technologies for crop improvements will be discussed. It will also provide an outlook into the future approaches and most widely used applications in plant breeding in crop plants on the basis of present development.
Keywords:
RFLPs, RAPDs, SSRs, AFLPs, SNPs, KASPar, GBS

1. Introduction
The world’s most important sources of food are cereals. Cereals can be consumed directly as food by humans, or indirectly as inputs to improve animal production. Millions of consumers and farmers in both the developing and the developed world rely on cereals as their favoured staple food. The future of cereal production, affects not only the global food security, but also the source of revenue of small farmers worldwide [4] [5] . Over the past several decades, especially through traditional breeding programme, intensive attempts have been made for the improvement of a large number of cereal varieties which adjusted to diverse agro-ecologies. Nevertheless, increasing biotic and abiotic stresses, increasing populations, and sharply reducing natural resources especially water for agricultural purposes, push the breeders for organizing and developing improved cereal varieties with higher yield potential [4] . The genetics and breeding community found that there is an urgent need to introduce new technologies, including molecular marker-assisted breeding combined with high-throughput and precision phenotyping [6] . Before entering the next cycle of selection, molecular markers can offer genomic information for plant evaluation which is essential for successful breeding, and also help track polymorphisms with no clear phenotype [7] .
In general, DNA markers are a fragment of DNA indicating (mutations/variations), which can be used to detect polymorphism between alleles of a gene for a particular sequence of DNA or different genotypes. Such frag- ments are linked with a definite location within the genome and may be detected by using certain molecular technology [8] .
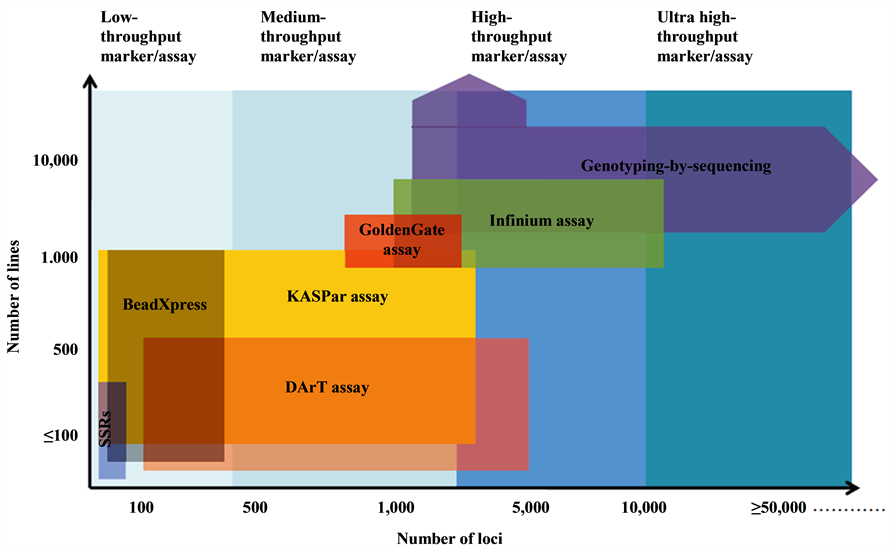
The marker systems that are now being progressively developed and also has shifted from the first and second generation marker systems including RFLPs, RAPDs, SSRs and AFLPs to the third and the fourth generation marker systems, which include SNPs, KASper, DArT assays, and Genotyping by Sequencing (GBS) [7] [9] (see Figure 1).
This review will address general principles and methodologies of different molecular markers in these categories with a major emphasis on emerging genotyping technologies in plants including SNPs and KASPer assays. Some issues related to applications of these methodologies in practical breeding will also be discussed.
2. Low-Throughput Marker Systems
Restriction Fragment Length Polymorphisms (RFLPs)
RFLP markers were mainly used in 1980s and 1990s in plant genetic studies, and are therefore, referred to as (first generation molecular markers) [10] . The polymorphisms detected by RFLPs are as a result of changes in nucleotide sequences in recognition sites of restriction enzymes, or due to mutation events (insertions ordeletions) of several nucleotides leading to obvious shift in fragment size [11] . The main advantages of RFLP markers are co-dominance, high reproducibility, no need of prior sequence information, and high locus-specificity.
Figure 1. Low to ultra-high-throughput cost-effective marker assay platforms for genotyping. Horizontal axis shows number of loci that can be assayed in a single experiment, whereas the vertical axis specifies the number of lines per samples that could be genotyped in high-throughput manner at low cost [3] .
By using RFLP markers, genetic maps have been established in several crop species including rice maize, wheat [12] - [14] . However, since the last decade fewer direct uses of RFLP markers in genetic research and plant breeding have been stated. Most plant breeders would think that RFLP is too time consuming procedure and it requires relatively large amounts of pure DNA, tedious experimental procedure. Additionally, each point muta- tion has to be analysed individually [15] - [17] .
In the late 1980s, Colinearity across genomes was first reported between the three diploid genomes of hexaploid wheat (Chao et al., 1989) and between potato and tomato (Bonierbale et al., 1988). Soon after couple of years, cross-hybridization of RFLP markers which obtained from bread wheat with barley and rye revealed a few translocations of chromosome arms in the rye genome when compared to the wheat genomes, whereas most probes indicated that the order of the loci was preserved between those three species (Moore et al., 1995).
3. Medium-Throughput Marker Systems
3.1. Random Amplified Polymorphic DNA’s (RAPDs)
RAPDs are based on the PCR amplification of random DNA segments with primers of random nucleotide sequences that were inexpensive and easy to use. The primers bind to complementary DNA sequences and where two primers bind to the DNA sample in close enough for successful PCR reaction. The amplified of DNA products can then be visualized by gel electrophoresis [18] [19] .
RAPD markers have been widely used in diverse plant species for assessment of genetic variation in populations and species, fingerprinting and study of phylogenetic relationships among species and subspecies [20] . Nevertheless, disadvantages of RAPD markers are the fact that it predominantly provides dominant markers, and incapability to detect allelic differences in heterozygotes. Polymorphisms are detected only as the presence or absence of a band of a certain molecular weight, with no information on heterozygosity [21] . Additionally, because of their random nature of amplification and short primer length, they are not ideal for genome mapping. Moreover, these markers do not exhibit dependable amplification patterns and differ with the experimental conditions [16] .
3.2. Simple Sequence Repeats (SSRs)
During 1990s, Simple sequence repeats (SSRs) which is also known as microsatellites were established and provided a choice for many genetic researches since they are amenable to low, medium and high-throughput approaches. They are randomly tandem repeats of short nucleotide motifs (2 - 6 bp) [21] . SSRs are frequently highly polymorphic sequences normally present in animal and plant species [1] , and can be used to study the relationship between inherited traits within a species [22] . Microsatellite markers are often derived from non- coding/anonymous genomic regions, such as bacterial artificial chromosomes (BACs) and genomic survey sequences (GSSs). Therefore, development of SSR markers used to be expensive and laborious [3] . This assay is easily detectable by gel electrophoresis for few to hundreds of samples, which could be inexpensive by researchers with limited resources. Polymorphism is based on the variation in the number of repeats in different genotypes [23] . Since polymorphisms in longer penta-nucleotide and tetra repeats are easier to make a distinction in a variety of detection systems and longer repeats may be more robust [24] .
In recent years, SSR markers can easily be developed in silico due to the availability of large-scale gene (expressed sequence tag) EST sequence information for many plant species. Since EST sequencing projects have provided sequence data that is available in online databases and can be scanned for identification of SSRs [25] . The high degree of polymorphism as compared to RFLPs and RAPDs, their co-dominant nature and locus specific make them the markers of choice for a diversity of purposes including practical plant breeding. Therefore, (SSRs) have become a marker of choice for an array of applications in plants due to extensive genome coverage and hyper variable nature (Figure 2) [1] .
3.3. Amplified Fragment Length Polymorphism’s (AFLPs)
AFLPs are PCR-based markers, simply RFLPs visualized by selective PCR amplification of DNA restriction fragments. Such a marker is a multi-locus marker technique that combines the techniques of selective PCR amplification of restriction and fragments restriction digestion and it is possible to be applied into DNA of any origin [26] .
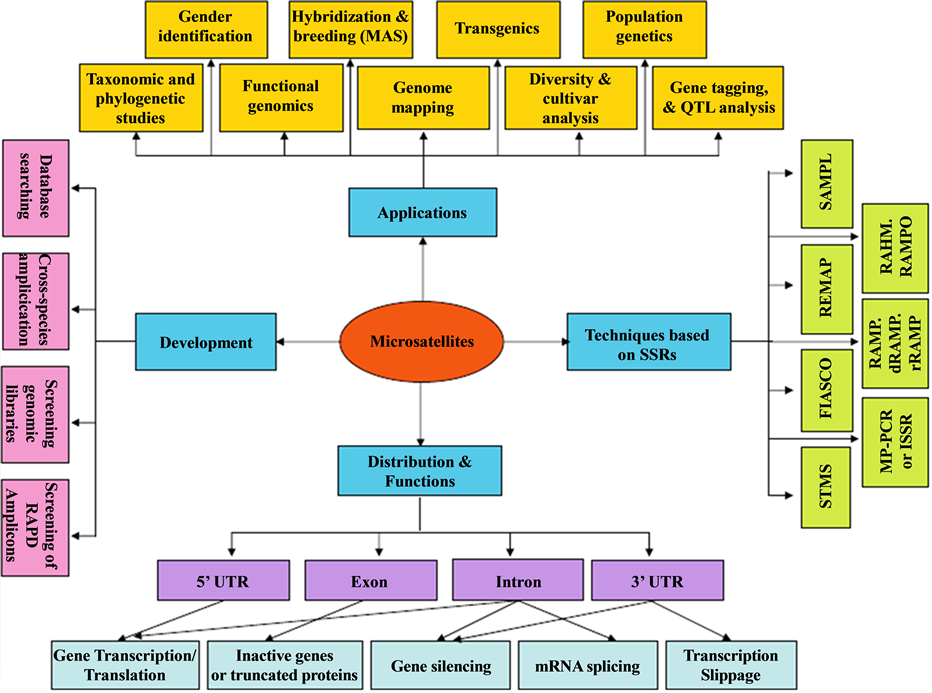
Figure 2. Microsatellites―a summary of development, distribution, functions and applications [1] .
The technique involves three steps: Initially, oligonucleotide adapters are ligated to both ends of the resulting restriction fragments and genomic DNA is digested. Subsequently, the fragments are selectively amplified, using the adapter and restriction site sequences as primer binding sites for following PCR reactions. As the 3’ ends of the primers extend into the restriction fragments by (1 to 4 bp), only those fragments are amplified, whose ends are absolutely complementary to the 3’ ends of the selective primers. Therefore, only a certain amount of the restriction fragments is amplified. Finally, the amplified fragments are resolved by gel electrophoresis and visualized by either silver staining, autoradiography or fluorescence, resulting in a unique reproducible fingerprint for each individual [26] [27] .
The advantages of using this method are that it is cost-efficient, since a single assay allows detection of a large number of co-amplified restriction fragments and it requires moderate quantities of DNA. Additionally, higher levels of polymorphisms compared with RFLPs can be detected also AFLPs have much higher multiplex ratio (more markers per experiment) and better reproducibility than RAPDs. These features make this technology an attractive tool for saturating genomic region with low marker density and constricting genetic maps [16] [20] . However, AFLP assays have some limitations also. For instance, it requires the use of polyacrylamide gels for detection and it needs a greater technical skill. Moreover, most AFLP markers are dominant rather than co-dominant, because of the complex banding arrangements. In some cases the scoring of AFLP polymorphisms as co-dominant marker loci is possible, for the reason that diploid homozygous individuals cause more intense peak than heterozygous individuals [28] .
By group of researchers Colomba, Vischi [29] , genetic relatedness and identity of the durum wheat Graziella Ra, four Italian commercial durum cultivars (Grazia, Cappelli, Flaminio and Svevo and Kamut) were evaluated using (AFLPs). Their results revealed that the percentage of polymorphic loci within accession ranged from 6.57% to 19.71% (mean, 12.77%) and molecular variance was partitioned into 80% (variance among accessions) and 20% (within accession).
4. High-Throughput Marker Systems
4.1. Single Nucleotide Polymorphisms (SNPs)
SNP is a single nucleotide base difference between two DNA sequences or individuals. SNPs are typically bi- allelic and arise either due to substitutions/point mutations (transversion and transition) or as a result of deletion/insertion of nucleotides and are detectable when similar genomic regions from different genotypes of different or same species are aligned [30] .
SNPs provide the simplest and ultimate form of molecular markers as a single nucleotide base is the smallest unit of inheritance, and therefore they can provide a great marker density. SNPs happen frequently in animals and plants. The probability to find polymorphisms in a target gene are increases due to high density of SNP markers which provides a huge advantage over previous markers that are at best closely linked to a locus of interest and not within [31] . In the case of linkage it can easily happen that a linkage is lost when a marker is applied to other populations with different recombination patterns. Typically, SNP frequencies are in a range of one SNP every (100 - 300) bp in plants. SNPs may present within coding sequences of genes, non-coding regions of genes or in the intergenic regions between genes at different frequencies in different chromosome regions [17] [32] .
Several methods are currently available for SNP discovery, either following the database approach, where SNPs are detected by following the experimental approach, or mining sequence databases, where genome regions of interest are screened for SNPs with one of various techniques established for the detection of SNPs. Moreover, it can be categorized into four reaction chemistries or principles: hybridization with allele-specific oligonucleotide probes, oligonucleotide ligation, enzymatic cleavage, and single nucleotide primer extension [32] - [34] .
In principle, the SNP methods show differences between a probe of known sequence and a target DNA containing the SNP site. The target DNA sections are typically PCR products and mismatches with the probe reveal SNPs within the amplified target DNA segment. The mismatching DNA segments can be sequenced then as the most direct way to identify SNP polymorphisms [35] .
SNP markers are likely to become the marker of choice for breeding in the near future, especially as the full sequences of more plant genomes will become available with the advantage of next generation sequencing tech- nologies (NGS) [31] .
More recently, it has become very-cost effective and easier to quickly identify a large number of SNPs in short time in any plant species. This was due to the emergence of the third generation sequencing. The advantage of this new sequence technology are expected to further reduce sequencing costs extremely to levels below $1 per mega base compared to $60, $2, and $1 expected costs for sequences generated by next generation sequencing [36] .
SNPs in Wheat (Triticum aestivum)
The large size of wheat genome has led to various approaches to reduce the cost of data production. These include the targeted re-sequencing of captured exome fragments and the establishment of confederations to share the cost of genome sequence data generation [37] [38] . In a different study, by using illumina sequencing of cDNA libraries, 14,078 putative SNPs were recognized across representative samples of UK wheat germplasm, with a proportion of these SNPs validated using KASPar assays [39] . In addition, in developed countries such as UK, France and Australia many attempts for large-scale SNP development in bread wheat were undertaken, leading to the development of millions of SNPs. These SNPs will be widely used for molecular breeding in wheat [40] .
4.2. The KBioscience Competitive Allele?Specific PCR (KASPar)
KASPar genotyping may be of specific interest to researchers and breeders who are interested in analysing a small number of targeted SNPs in a large number of samples. This makes KASPar a cost-effective, simpleand flexible genotyping system; since the assays can be modified with a range of DNA samples and it does notrequire a hybridization step; as an alternative it includes real-time detection of the product [41] .
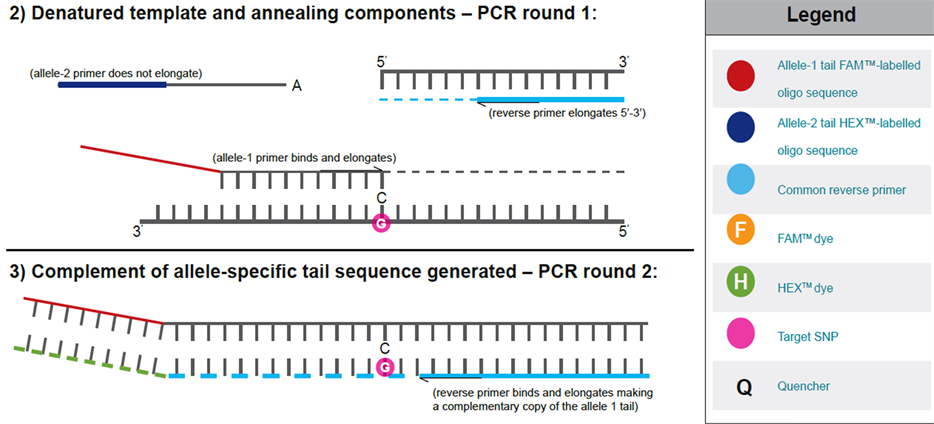
The chemistry of KASPar assyas involves one common reverse primer and two competitive allele specific tailed forward primers. To determine the alleles at a specific locus, this assay system relies on the discrimination power of a novel pattern of competitive allele specific PCR [3] . In such cases, this assay involves competitive allele-specific PCR for a given SNP, followed by SNP detection through Fluorescence Resonance Energy Trans- fer (FRET) [41] .
This assay for the target SNPs has been developed and used for genotyping commercially by Kbioscience UK (http://www.kbioscience.co.uk/). This company perfected this technique to improve the performance of the detection platform by incorporating a 5’ - 3’ exonuclease cleaved Taq DNA polymerase and a homogeneous Fluorescence Resonance Energy Transfer (FRET) detection system. The two allele-specific primers of a SNP are designed so that they incorporate with a unique (18) bp tail to the respective allele specific products, which in later cycles allow incorporation of allele specific fluorescent labels to the PCR products (with the help of corresponding labelled primers) (http://www.kbioscience.co.uk/). The mechanism of KASPar chemistry has been presented in (Figure 3) [42] .
In KASPar assays there is no need of sequencing to identify SNPs, instead SNP flanking sequences already known while developing different types of genotyping assays (for instance, illumina) can easily be used for primer design (one common and two allele-specific primers) for KASPar assays [34] .
Despite the fact that KASPar genotyping assays have come to the market very recently, they have started to be used for a large number of commercial species. In maize, a set of 695 highly polymorphic gene-based SNPs from a total of 13,882 GG-validated SNPs were selected and converted into KASPar genotyping assay with a success rate of 98% [43] . Additionally, in wheat, the technique has been used for constructing a linkage map containing several hundred SNPs [39] .
4.3. Genotyping-by-Sequencing (GBS)
With the increased throughput of NGS platforms, re-sequencing for genome-wide surveys of genetic diversity became reasonable [44] . However, this assay is bioinformatically challenging, impartial estimation of genetic diversity across the genome in both coding and non-coding regions could be determined. Additionally, it allows for the detection of various types of genetic variation; this detection capability contains not only SNPs and small indels, but also large mega-base scale indels [45] .
This assay involves the use of restriction enzymes for reducing the complexity of genomes followed by targeted sequencing of reduced proportions, in that way each marker can be sequenced at high coverage across many individuals at low cost and high accuracy [3] . A workflow of GBS has been presented in (Figure 4).
The main target for constructing GBS libraries was based on reducing genome complexity with restriction enzymes, which may reach important regions of the genome that are unreachable to sequence capture approaches [46] . The procedure has been demonstrated with barley (Oregon Wolfe Barley) at the recombinant inbred lines populations and maize (IBM) where about (25,000 to 200,000) sequence tags were mapped, respectively. With this method, species that lack a complete genome sequence can have a reference map settled around the restriction sites, which can be done in the process of sample genotyping. This system has been adjusted for reducing missing data points and improved SNP calls [2] .
5. Applications and Strategies of DNA Markers in Breeding Programs
5.1. Pyramiding Multiple Loci and Favourable Alleles
Gene pyramiding is defined as an assembly of multiple desirable genes which can be combined into a single genotype from multiple parents. This is referred as one of the major applications of marker assistant selection, as gene pyramiding via conventional plant breeding is difficult, if not impossible [47] .
The methods for pyramiding favourable alleles can be used in the same way to accumulate QTL controlling different traits. A main difference in the model is that alleles at different trait loci to be accumulated may have different favourable directions, for instance negative alleles are preferable for some traits but positive alleles are favourable for others. As a result, to meet breeding objectives one may need to combine the positive QTL alleles of some traits with the negative alleles of others [4] .
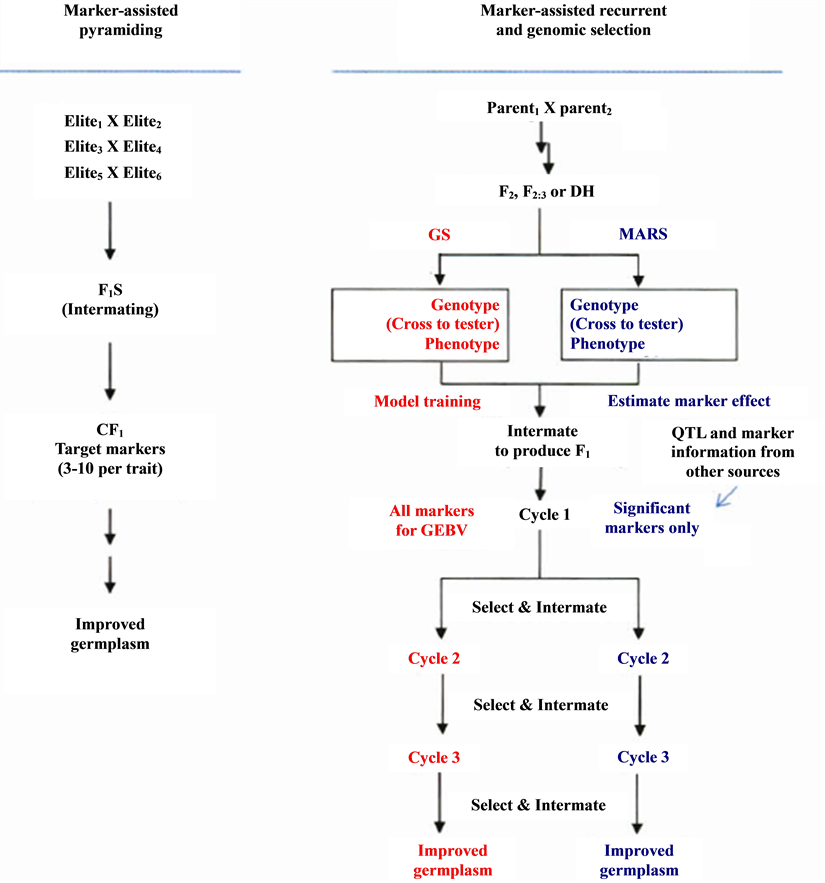
Selection for multiple traits may be completed in one cycle if the population size is large enough to allow desirable individuals to combine different traits (see Figure 5). Nevertheless, the number of trait loci that can be manipulated in one cycle is restricted because the population size required covering the recombinants increases exponentially with the increase of the number of traits/loci [4] .
Figure 3. The mechanism of KASPar assays.
Figure 4. A workflow for genotyping-by-sequencing (GBS) approach. A schematic representation of various steps involved in GBS approach has been shown (Adopted from Poland, Brown [2] ).
To overcome this limitation, a strategy was proposed by Bonnett, Rebetzke [48] . In this method, individuals has been selected by all target markers for both heterozygous and homozygous forms to gain a subset of population that contain higher frequencies of the target alleles so that to obtain the homozygotes at the target loci, a smaller population size is required in the following generation.
For Barley Yellow Mosaic Virus complex, a variety of markers have been developed for selection of the rym5 and rym4 resistance genes on chromosome 3H [49] . In another study, the regions of a typical Spanish barley line that have VRNH2 and VRNHl were introgressed into a winter variety. A set of 12 lines introgressed with all four possible combinations of VRNHl and VRNH2 has been assessed for frost tolerance and vernalization requirement [50] . In practical, what has to be taken into account when applying such strategies pyramiding has to be repeated after each crossing, since the pyramided resistance genes are segregating in the progeny [51] .
5.2. Marker-Assisted Recurrent Selection (MARS)
In the 1990s, marker-assisted recurrent selection was proposed, which uses markers at each generation to target all traits of importance and for which genetic information can be achieved. Parents contribute different favourable alleles when the QTL mapping is conducted based on a bi-parental population. Therefore, the perfect genotype is a mosaic of chromosomal segments produced by recombination between the two parents [47] .
MARS refers to the improvement of an (F2) population by one cycle of marker-assisted selection (for instance, based on data marker scores and phenotypic) followed usually by two or three cycles of marker-based
Figure 5. Methodologies for marker-assisted breeding. Genomic selection (CS, red) and markerassisted recurrent selected (MARS, blue) can start with the same type of population, F2, F2:3, BC, or OH. Crossing to tester can be included in the procedure for hybrid crops. For the current MARS, Marker/QTL information from other sources can be combined for selection with the significant markers identified at the beginning stage. Results from GS of breeding populations can be used to improve the prediction and model training for next cycles of selection or other GS projects [4] .
selection (for instance, based on marker scores only) [4] . It is possible today to define an ideal genotype as a pattern of QTLs, all QTLs carrying favourable alleles from various parents (Figure 5). After several successive generations of crossings, it might be possible to get close to the ideal genotype. In addition, this system can start without any QTL information, and selection can be based on significant marker-trait association established during the MARS process [6] .
Studies have revealed that, in accumulating favourable alleles, (MARS) was superior to phenotypic selection [52] . Moreover, through MARS in maize, the rates of genetic gain accomplished for complex traits were about twice those of phenotypic selection in some reference populations [53] . The usefulness of including prior knowl- edge of QTL under genetic models has been studied that included QTL number, gene effects, heritability, epistasis and linkage. It is concluded that with known QTL, MARS is most favourable for traits controlled by a large number of QTL [54] .
6. Conclusions
As it is evident from the discussion above, that different levels of throughput are available. Therefore, an appropriate marker system can be selected based on the need. The earlier types of molecular markers include neutral markers. For instance, (RFLPs), and which were later followed by based on the (PCR) reaction, a faster and less expensive technology. A PCR-based DNA marker includes (RAPDs), (AFLPs).
RFLPs offer the best marker type for many purposes. The main disadvantages of using RFLPs are low through- put and high cost of genotyping. RAPDs and AFLPs have also been widely used in genetic diversity studies and gene mapping. Both technologies are particularly useful when there is a necessity to assay loci across the entire genome. However, their lack of reproducibility, dominant nature of RAPDs compared with AFLPs and the lack of specificity in both cases, are limiting factors for their application in precise MAS breeding approaches [16] .
Recently, the availability of whole genome sequences of a few selected crops and the sequence information has also led to the development of a new generation of markers, such as KASpar, SNP assays. SNP markers which are transferable through different genotyping chemistries will offer as flexible selection tools for plant breeders in (MAS). In addition, the focus should be placed in the identification of SNPs in as many genes as possible and the parallel analysis of many different lines [43] . It is very likely that improvement of complex traits will depend on the ability to manipulate genes, which have minor effects, and show interaction with each other.
As shown by some simulation studies, genotype by sequencing seems to be the best approach for improvement of complex traits. However, the parameters and conditions included in simulations may not fully reflect the complex situations of diverse plant breeding programs, the genetic gain per unit time and cost that has been achieved in simulations needs to be supported by the long-term selection response by comparing with other breeding approaches. In addition, this approach can technically lead to the discovery of thousands of SNPs in one single experiment. Moreover; it can be used of those plants that do not have the reference genome available [2] [4] .
Acknowledgements
I would like to express my most sincere gratitude to Dr. Ahmad Hama Amin. Thank you for your trust, support and encouragement. I would like to extend my appreciation to all lecturers in our department especially Dr. Nawroz Abdul Razzak for providing wise advice. Last but not least, I devote this review to my parents and my lovely wife for their love, trust and support during writing this paper.
References
- Kalia, R.K., et al. (2011) Microsatellite Markers: An Overview of the Recent Progress in Plants. Euphytica, 177, 309- 334. http://dx.doi.org/10.1007/s10681-010-0286-9
- Poland, J.A., et al. (2012) Development of High-Density Genetic Maps for Barley and Wheat Using a Novel Two- Enzyme Genotyping-by-Sequencing Approach. PloS One, 7, e32253. http://dx.doi.org/10.1371/journal.pone.0032253
- Mir, R.R., et al. (2013) Evolving Molecular Marker Technologies in Plants: From RFLPs to GBS. In: Lübberstedt, T. and Varshney, R.K., Eds., Diagnostics in Plant Breeding, Springer, Berlin, 229-247. http://dx.doi.org/10.1007/978-94-007-5687-8_11
- Xu, Y., et al. (2013) Marker-Assisted Selection in Cereals: Platforms, Strategies and Examples. In: Cereal Genomics II, Springer, Berlin, 375-411.
- Conway, G.R. and Barbier, E.B. (2013) After the Green Revolution: Sustainable Agriculture for Development. Rout- ledge, London.
- Gupta, P.K., Langridge, P. and Mir, R.R. (2010) Marker-Assisted Wheat Breeding: Present Status and Future Possibilities. Molecular Breeding, 26, 145-161. http://dx.doi.org/10.1007/s11032-009-9359-7
- Paux, E., et al. (2010) Insertion Site―Based Polymorphism Markers Open New Perspectives for Genome Saturation and Marker-Assisted Selection in Wheat. Plant Biotechnology Journal, 8, 196-210. http://dx.doi.org/10.1111/j.1467-7652.2009.00477.x
- Henry, R.J. (2012) Molecular Markers in Plants. Wiley. http://dx.doi.org/10.1002/9781118473023
- Paux, E., et al. (2012) Sequence-Based Marker Development in Wheat: Advances and Applications to Breeding. Biotechnology Advances, 30, 1071-1088. http://dx.doi.org/10.1016/j.biotechadv.2011.09.015
- Jones, N., et al. (2009) Markers and Mapping Revisited: Finding Your Gene. New Phytologist, 183, 935-966. http://dx.doi.org/10.1111/j.1469-8137.2009.02933.x
- Tanksley, S., et al. (1989) RFLP Mapping in Plant Breeding: New Tools for an Old Science. Nature Biotechnology, 7, 257-264. http://dx.doi.org/10.1038/nbt0389-257
- Cho, Y., et al. (1998) Integrated Map of AFLP, SSLP and RFLP Markers Using a Recombinant Inbred Population of Rice (Oryza sativa L.). Theoretical and Applied Genetics, 97, 370-380. http://dx.doi.org/10.1007/s001220050907
- Smith, O., et al. (1990) Similarities among a Group of Elite Maize Inbreds as Measured by Pedigree, F1 Grain Yield, Grain Yield, Heterosis, and RFLPs. Theoretical and Applied Genetics, 80, 833-840. http://dx.doi.org/10.1007/BF00224201
- Nagaoka, T. and Ogihara, Y. (1997) Applicability of Inter-Simple Sequence Repeat Polymorphisms in Wheat for Use as DNA Markers in Comparison to RFLP and RAPD Markers. Theoretical and Applied Genetics, 94, 597-602. http://dx.doi.org/10.1007/s001220050456
- Wong, L.-J.C. (2013) Next Generation Molecular Diagnosis of Mitochondrial Disorders. Mitochondrion, 13, 379-387. http://dx.doi.org/10.1016/j.mito.2013.02.001
- Edwards, J.D. and McCouch, S.R. (2007) Molecular Markers for Use in Plant Molecular Breeding and Germplasm Evaluation. Marker-Assisted Selection-Current Status and Future Perspectives in Crops, Livestock, Forestry and Fish, Food and Agriculture Organization of the United Nations (FAO), Rome, 29-49.
- Edwards, D. and Batley, J. (2009) Plant Genome Sequencing: Applications for Crop Improvement. Plant Biotechnology Journal, 8, 2-9. http://dx.doi.org/10.1111/j.1467-7652.2009.00459.x
- Williams, J.G., et al. (1990) DNA Polymorphisms Amplified by Arbitrary Primers Are Useful as Genetic Markers. Nucleic Acids Research, 18, 6531-6535. http://dx.doi.org/10.1093/nar/18.22.6531
- Gupta, P.K. and Varshney, R.K. (2013) Cereal Genomics II. Springer-Verlag GmbH.
- Gupta, P., et al. (1999) Molecular Markers and Their Applications in Wheat Breeding. Plant Breeding, 118, 369-390. http://dx.doi.org/10.1046/j.1439-0523.1999.00401.x
- Jiang, G.-L. (2013) Molecular Markers and Marker-Assisted Breeding in Plants. Plant Breeding from Laboratories to Fields.
- Dunn, G., et al. (2005) Microsatellites versus Single-Nucleotide Polymorphisms in Linkage Analysis for Quantitative and Qualitative Measures. BMC Genetics, 6, S122. http://dx.doi.org/10.1186/1471-2156-6-S1-S122
- Ellegren, H. (2000) Microsatellite Mutations in the Germline: Implications for Evolutionary Inference. Trends in Genetics, 16, 551-558. http://dx.doi.org/10.1016/S0168-9525(00)02139-9
- Koelling, J., et al. (2012) Development of New Microsatellite Markers (SSRs) for Humulus Lupulus. Molecular Breed- ing, 30, 479-484. http://dx.doi.org/10.1007/s11032-011-9637-z
- Varshney, R.K., Graner, A. and Sorrells, M.E. (2005) Genic Microsatellite Markers in Plants: Features and Applications. Trends in Biotechnology, 23, 48-55. http://dx.doi.org/10.1016/j.tibtech.2004.11.005
- Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., et al. (1995) AFLP: A New Technique for DNA Fingerprinting. Nucleic Acids Research, 23, 4407-4414. http://dx.doi.org/10.1093/nar/23.21.4407
- Nicod, J.C. and Largiadèr, C.R. (2003) SNPs by AFLP (SBA): A Rapid SNP Isolation Strategy for Non-Model Organisms. Nucleic Acids Research, 31, e19. http://dx.doi.org/10.1093/nar/gng019
- Meudt, H.M. and Clarke, A.C. (2007) Almost Forgotten or Latest Practice? AFLP Applications, Analyses and Advances. Trends in Plant Science, 12, 106-117. http://dx.doi.org/10.1016/j.tplants.2007.02.001
- Colomba, M., Vischi, M. and Gregorini, A. (2012) Molecular Characterization and Comparative Analysis of Six Durum Wheat Accessions Including Graziella Ra. Plant Molecular Biology Reporter, 30, 168-175. http://dx.doi.org/10.1007/s11105-011-0328-z
- Xu, Y. (2010) Molecular Plant Breeding. CABI International, Wallingford, Oxfordshire.
- Ganal, M.W., Altmann, T. and Röder, M.S. (2009) SNP Identification in Crop Plants. Current Opinion in Plant Biology, 12, 211-217. http://dx.doi.org/10.1016/j.pbi.2008.12.009
- Edwards, D., Forster, J.W., Chagné, D., Batley, J., et al. (2007) What Are SNPs? In: Oraguzie, N., et al., Eds., Association Mapping in Plants, Springer, New York, 41-52. http://dx.doi.org/10.1007/978-0-387-36011-9_3
- Syvänen, A.-C. (2005) Toward Genome-Wide SNP Genotyping. Nature Genetics, 37, S5-S10. http://dx.doi.org/10.1038/ng1558
- Gupta, P.K., Rustgi, S. and Mir, R.R. (2008) Array-Based High-Throughput DNA Markers for Crop Improvement. Heredity, 101, 5-18. http://dx.doi.org/10.1038/hdy.2008.35
- Weising, K., Nybom, H., Wolff, K. and Kahl, G. (2005) DNA Fingerprinting in Plants: Principles, Methods, and Applications. 2nd Edition, Taylor & Francis, UK. http://dx.doi.org/10.1201/9781420040043
- Thudi, M., Li, Y., Jackson, S.A., May, G.D. and Varshney, R.K. (2012) Current State-of-Art of Sequencing Technologies for Plant Genomics Research. Briefings in Functional Genomics, 11, 3-11. http://dx.doi.org/10.1093/bfgp/elr045
- Winfield, M.O., Wilkinson, P.A., Allen, A.M., Barker, G.L.A., Coghill, J.A., Burridge, A., et al. (2012) Targeted Re- Sequencing of the Allohexaploid Wheat Exome. Plant Biotechnology Journal, 10, 733-742. http://dx.doi.org/10.1111/j.1467-7652.2012.00713.x
- Edwards, D., Wilcox, S., Barrero, R.A., Fleury, D., Cavanagh, C.R., Forrest, K.L., et al. (2012) Bread Matters: A National Initiative to Profile the Genetic Diversity of Australian Wheat. Plant Biotechnology Journal, 10, 703-708. http://dx.doi.org/10.1111/j.1467-7652.2012.00717.x
- Allen, A.M., Barker, G.L.A., Berry, S.T., Coghill, J.A., Gwilliam, R., Kirby, S., et al. (2011) Transcript-Specific, Single-Nucleotide Polymorphism Discovery and Linkage Analysis in Hexaploid Bread Wheat (Triticum aestivum L.). Plant Biotechnology Journal, 9, 1086-1099. http://dx.doi.org/10.1111/j.1467-7652.2011.00628.x
- Lorenc, M.T., Hayashi, S., Stiller, J., Lee, H., Manoli, S., Ruperao, P., et al. (2012) Discovery of Single Nucleotide Polymorphisms in Complex Genomes Using SGSautoSNP. Biology, 1, 370-382. http://dx.doi.org/10.3390/biology1020370
- McCouch, S.R., Zhao, K.Y., Wright, M., Tung, C.-W., Ebana, K., Thomson, M., et al. (2010) Development of Genome- Wide SNP Assays for Rice. Breeding Science, 60, 524-535. http://dx.doi.org/10.1270/jsbbs.60.524
- LGC Genomics (2013) KASP™ Genotyping Chemistry User Guide and Manual. http://www.lgcgenomics.com/genotyping/kasp-genotyping-reagents/?download_file=22_1_kasp_manual.pdf&download_cat=downloads
- Mammadov, J., Chen, W., Mingus, J., Thompson, S. and Kumpatla, S. (2012) Development of Versatile Gene-Based SNP Assays in Maize (Zea mays L.). Molecular Breeding, 29, 779-790. http://dx.doi.org/10.1007/s11032-011-9589-3
- Wheeler, D.A., Srinivasan, M., Egholm, M., Shen, Y.F., Chen, L., McGuire, A., et al. (2008) The Complete Genome of an Individual by Massively Parallel DNA Sequencing. Nature, 452, 872-876. http://dx.doi.org/10.1038/nature06884
- Kiani, S., Akhunova, A. and Akhunov, E. (2013) Application of Next-Generation Sequencing Technologies for Genetic Diversity Analysis in Cereals. In: Gupta, P.K. and Varshney, R.K., Eds., Cereal Genomics II, Springer, Dordrecht, 77-99.
- van Oeveren, J., de Ruiter, M., Jesse, T., van der Poel, H., Tang, J., Yalcin, F., et al. (2011) Sequence-Based Physical Mapping of Complex Genomes by Whole Genome Profiling. Genome Research, 21, 618-625. http://dx.doi.org/10.1101/gr.112094.110
- Gupta, P.K., Kumar, J., Mir, R.R. and Kumar, A. (2010) Marker-Assisted Selection as a Component of Conventional Plant Breeding. Plant Breeding Reviews, 33, 145-217. http://dx.doi.org/10.1002/9780470535486.ch4
- Bonnett, D.G., Rebetzke, G.J. and Spielmeyer, W. (2005) Strategies for Efficient Implementation of Molecular Markers in Wheat Breeding. Molecular Breeding, 15, 75-85. http://dx.doi.org/10.1007/s11032-004-2734-5
- Rae, S.J., Macaulay, M., Ramsay, L., Leigh, F., Matthews, D., O’Sullivan, D.M., et al. (2007) Molecular Barley Breed- ing. Euphytica, 158, 295-303. http://dx.doi.org/10.1007/s10681-006-9166-8
- Gracia, M.P. and Casas, A.M. (2012) Barley Adaptation: Teachings from Landraces Will Help to Respond to Climate Change. In: Zhang, G.P., Li, C.D. and Liu, X., Eds., Advance in Barley Sciences: Proceedings of 11th International Barley Genetics Symposium, Springer, Dordrecht.
- Werner, K., Friedt, W. and Ordon, F. (2005) Strategies for Pyramiding Resistance Genes against the Barley Yellow Mosaic Virus Complex (BaMMV, BaYMV, BaYMV-2). Molecular Breeding, 16, 45-55. http://dx.doi.org/10.1007/s11032-005-3445-2
- Wang, J.K., Chapman, S.C., Bonnett, D.G. and Rebetzke, G.J. (2009) Simultaneous Selection of Major and Minor Genes: Use of QTL to Increase Selection Efficiency of Coleoptile Length of Wheat (Triticum aestivum L.). Theoretical and Applied Genetics, 119, 65-74. http://dx.doi.org/10.1007/s00122-009-1017-2
- Crosbie, T.M., Eathington, S.R., Johnson, G.R., Edwards, M., Reiter, R., Stark, S., et al. (2006) Plant Breeding: Past, Present, and Future. In: Lamkey, K.R. and Lee, M., Eds., Plant Breeding: The Arnel R. Hallauer International Symposium, Wiley-Blackwell, Hoboken. http://dx.doi.org/10.1002/9780470752708.ch1
- Bernardo, R. and Charcosset, A. (2006) Usefulness of Gene Information in Marker-Assisted Recurrent Selection: A Simulation Appraisal. Crop Science, 46, 614-621. http://dx.doi.org/10.2135/cropsci2005.05-0088