Open Journal of Soil Science
Vol.3 No.1(2013), Article ID:28719,6 pages DOI:10.4236/ojss.2013.31002
Diversity of Cultivable Actinomycetes in Tropical Rainy Forest of Xishuangbanna, China
![]()
1Yunnan Institute of Microbiology, Yunnan University, Kunming, China; 2Kunming University, Kunming, China.
Email: *jiangyi@ynu.edu.cn
Received November 19th, 2012; revised December 22nd, 2012; accepted January 5th, 2013
Keywords: Actinomycetes; Diversity; Tropical Rainy Forest; Xishuangbanna
ABSTRACT
In order to obtain much more un-known actinomycetes for discovering new drug lead, one hundred soil samples were collected from five national natural protection areas of tropical rain forests, Mengla, Menglun, Mandian, Xiaomengyang and Guanping, in Xishuangbanna, Yunnan, China. 1652 purified cultures of actinobacteria were isolated from these samples by using 5 media. The 16S rRNA gene sequences of 388 selected strains were analyzed, and the phylogenetic analysis was carried out. 35 genera which belong to 8 orders and 14 families of the Class actinobacteria were identified. It is showed from research results that actinomycete diversity in tropical rain forest of Xishuangbanna is the highest comparing with all areas studied in our laboratories before. Selective isolation methods for un-known actinomycetes from soil samples, including medium and inhibitors are discussed in this paper.
1. Introduction
Actinomycetes (Actinobacteria) have been paid a great attention owing to their production of various natural drugs and other bioactive metabolites including antibiotics, enzyme inhibitors and enzymes. Over 22,000 bioactive secondary metabolites (including antibiotics) were published on the scientific and patent literature, and about a half of them were produced by actinomycetes. About 150 antibiotics have being applied in human therapy and agriculture now, 100 - 120 of them were produced by actinomycetes [1]. Actinomycete is still an important source for new natural drugs development. So Baltz showed a proposition of “Renaissance in antibacterial discovery from actinomycetes” [2]. However, to develop new drugs from actinomycetes is more and more difficult [3]. In order to overcome these challenges, some new concepts based on genome was described, that is “new habitats, new methods, new species, new gene cluster, new products and new use” [3-6]. In other words, novel microbial kind should contain new gene cluster synthesizing new secondary metabolites, so far as getting new kind is an important premise for obtaining new compounds [3]. Many companies and laboratories focused on new actinomycete resources from new habitats, such as oceans, extreme environment and plants, for development of new drugs. In our view, making the uncultured to pure cultured actinomycetes is one new hope for getting new drug leads.
Mekong River (Ménam Khong, or Khong, or Mae Nam) is an international river. It originates from Yushu, Qinghai Province, China. It flows over Laos, Burma, Thailand, Cambodia and Vietnam, and flows into South China Sea. Total length of it is 4880 km, and it is the sixth river in length in the world.
The tropical rain forests are distributed widely on the river line of Mekong River, and one area of highest biodiversity in the world. Xishuangbanna is located at middle reaches of Mekong River. One hundred soil samples were collected from five national natural protection areas of tropical rain forests in Xishuangbanna. Diversity of cultivable actinomycetes was studied. Some results are reported in this paper.
Xishuangbanna is included in the Indo-Burma biodiversity hotspots and contains over 5000 species of vascular plants, comprising 16 percent of China’s total plant diversity [7-9]. The forests of Xishuangbanna biodiversity that is important both globally and nationally. Typical vegetation is mainly consisted of Family Sapindaceae, Annonaceae, Meliaceae, Euphobiaceaea, Moraceae, Lauracear, Datiscaceae, Rubiaceae, Binnoniaceae, and Orichdaceae etc. Despite Xishuangbanna’s high biodiversity, general ecological information about the tropical rain forests of the region has rarely been published for an international audience. The fauna of Xishuangbanna are no less diverse, as 36.2%, 21.7%, and 14.6% of China’s birds, mammals, and reptiles and amphibians occur in this region, respectively [10-13]. Five sampling areas, Mengla, Menglun, Mandian, Xiaomengyang and Guanping are located within a radius of N 21˚ and E 101˚, covering about 100 km2, altitude 450 to 900 m. Mean annual temperature is 21˚C, and accumulated temperature over 18˚C is more than 200 days. Annual precipitation is about 1500 mm. Annual sunshine is about 2000 hours. Soil belongs to brick-red soil, and pH 5.3 template, created in MS Word 2003 and saved as “Word 97-2003 & 6.0/95-RTF” for the PC, provides authors with most of the formatting specifications needed for preparing electronic versions of their papers. All standard paper components have been specified for three reasons: 1) ease of use when formatting individual papers, 2) automatic compliance to electronic requirements that facilitate the concurrent or later production of electronic products, and 3) conformity of style throughout a journal paper. Margins, column widths, line spacing, and type styles are built-in; examples of the type styles are provided throughout this document and are identified in italic type, within parentheses, following the example. Some components, such as multi-leveled equations, graphics, and tables are not prescribed, although the various table text styles are provided. The formatter will need to create these components, incorporating the applicable criteria that follow.
2. Materials and Methods
2.1. Collection and Preparation of Soil Samples
One hundred soil samples were collected from five national natural protection areas of tropical rain forests, Menngla, menglun, Mandian, Xiaomengyang and Guanping, in Xishuangbanna, Yunnan, China. 20 samples were collected in each area. The five protection areas cover about total 100 km2 range. Each sample was mixed by soil collected from 5 to 10 sampling holes and 5 to 20 cm depth. The samples were put in sterile glass dish immediately, and dried for 10 days at 28˚C. 2 g of each dried sample were pre-treated at 80˚C or 120˚C for 1 hour, and respectively put in 18 ml sterile water with 0.1% Na4P2O5, and shaken for 60 min at 220 rpm/min. The suspension was diluted from 10−1 to 10−5.
2.2. Isolation Medium of Actinobacteria
Following media were used for isolating actinobacteria in soil samples:
YIM 7 = HV medium [14].
YIM171 = Improvement Glycerol-Asparagine medium: 10 g Glycerol, 1 g asparagine, 1 g K2HPO4·H2O, 0.5 g MgSO4·7H2O, 0.3 g CaCO3, 3.7 mg Vit mixture of HV medium, 15 g agar, 1000 ml water, pH 7.2.
YIM212 = Mycose-Proline medium: 5 g Mycose, 1 g proline, 1 g (NH4)2SO4, 1 g NaCl, 2 g CaCl2, 1 g K2HPO4·H2O, 1 g MgSO4·7H2O, 3.7 mg Vit mixture of HV medium, 15 g agar, 1000 ml water, pH 7.2.
YIM213 = Raffinose-Histidine medium: 5 g Raffinose, 1 g histidine, 1 g K2HPO4·H2O, 0.5 g MgSO4·7H2O, 15g agar, 1000 ml water, pH 7.2 - 7.4.
Improvement HVG: 1 g Humic acid, 0.5 g Keratin, 0.3 g CaCl2, 10 mM MOPS, 0.1ml Trace salts, 7 g Gellan Gum, 1000 ml water, pH 7.2 - 7.4.
All of media were supplemented with filter sterilized mixture solution consisted of 50 or 100 mg cycloheximide, 50 or 100 mg nystatin and 20 or 40 mg nalidixic acid, or K2Cr2O7 50 or 75 mg for 1000 ml medium, as inhibitors against fungi and Gram negative bacteria.
Plate dilution method was used for selective isolating actinobacteria from the sample suspension. 0.2 ml of suspensions of each sample were spread on the medium plates, and cultivated for 7 to 35 days at 28˚C, then take count of colonies, and pick up actinobacteria to slant of the same isolation medium.
2.3. Identification of Pure Cultivated Actinobacteria
Total 1652 pure strains were isolated from the 100 soil samples, 388 strains of them were selected after throwing out many duplicates strains based on morphological and cultural characteristics. The DNA of pure strains was extracted for 16S rDNA analysis [15]. PCR amplifycation of the 16S rDNA, purification and sequence of the PCR products were done as described [16]. The forward primer F8 (8 ± 27), 5’-GAGAGTTTGATCCTGGCT-CAG-3’ and the reverse primer (1510 ± 1492), 5’-GGTTACCTTGTTACGACTT-3’ were used. The resultant sequences were manually aligned with available sequences from public databases. Phylogenetic trees (Not shown) were inferred by using the neighbour-joining [17] and maximum-likelihood methods [18]. All of pure cultivated strains were identified at a genus level.
3. Results and Discussion
3.1. Effects of Isolating Actinomycetes with Five Media
100 soil samples from Xishuangbanna were isolated by using five media. It is showed from Table 1 and Figure 1 that total 1652 pure cultural actinomyces strains were isolated, and 238 (60%) of the 1652 strains were rare actinomycetes with YIM 212 medium; 220 strain were
Table 1. Comparison of five media for isolating actinomycetes (amount of pure cultivated strains).
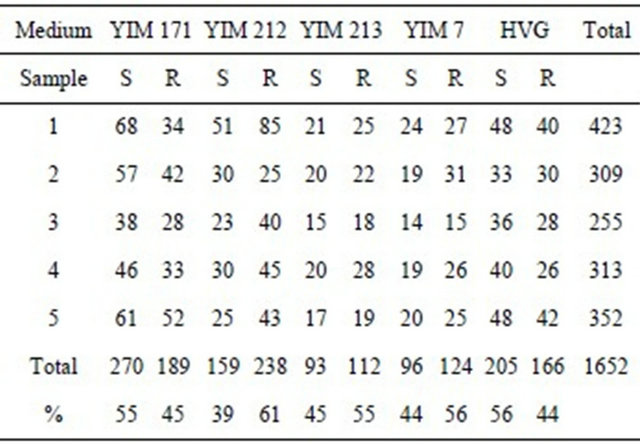
S = Streptomyces; R = rare actinomycetes; YIM 171 = Improvement Glycerol-Asparagine medium; YIM YIM 212 = Mycose-Proline medium; YIM 213 = Raffinose-Histidine medium; 7 = HV medium; HVG = Improvement HVG.

Figure 1. Amount of pure strains isolated with five media.
isolated, 124 (56%) of them were rare actinomycetes with YIM 7 (HV); 205 strains were isolated, and 112 (55%) were rare actinomycetes with YIM 213. 459 pure strains were isolated, and 270 strains (55%) of them were Streptomycetes with YIM 171; 371 pure strains were isolated, and 205 strains (56%) of them were Streptomycetes with HVG. Therefore we propose that the former three media can use to selection isolation of rare actionmycetes, and the last two media use to isolate streptomycetes from forest soil samples.
Kind and concentration of inhibitors for isolating actinobacteria from forest soil were tested many times in our laboratories, the optimum composition was 50 mg/L K2Cr2O7, or 100 mg/L mixture solution of nystatin, 50 mg/L cycloheximide and 20 mg/L nalidixic acid, most part of Gram negative bacteria were inhibited, and no fungi grown on all five medium plates.
3.2. Composition of Actinobacetria
16S rDNA sequences of 388 selected pure strains were determined. The phylogenetic analysis was carried out. Strains were identified at a genus level. Total 35 genera of actinobacteria were isolated and identified. These genera belong to 8 orders and 14 families (Table 2) [19]. Strepptomyces, Micromonospora, Pseudonocardia and Nocadia were widely distributed and the most in number in all of soil samples. But Actinomycetospora, Actinopolymorpha, Curtobacterium, Isoptericola, Kineicoccus, Lentzea, Planosporangium, Polymorphospora and Sphaerisporangium which are very rare in nature, were isolated from soil samples in Xishuangbanna.
Based on general regularization in taxonomic world, the similarity of 16S rRNA gene sequence of a strain with the closest valid species is below 98.5%, the strain should be possible novel species [12]. Table 3 is a phylogenetic analysis based on the 16S rRNA gene sequence of a part of actinomycete strains. Therefore 19 strains were possible new species, it took 6% of 588 sequencing strains.
Two new genera, Planosporangium [6] and Actinomycetospora [3], and five new species, Planosporangium flavigriseum [6], Planosporangium mesophilum [20], Actinomycetospora chiangmaiensis [3], Actinopolymorpha alba [21] and Agromyces aurantiacus [22] were isolated and identified in soil samples collected from the river line of Mekong river. All of them were valid published before.
In our previous studies of pure cultivable actinobacteria, 17 genera were isolated in 60 soil samples collected from primeval forest in Grand Shangri-La, southwest China [23]. 13 genera in 50 soil samples from primeval subtropical every green forest in Gulin, Sichuan Province, China [24]. Only five genera, Mycobacterium, Nocardia, Promocromonopora, Dactylosporangium and Streptomyces were isolated and identified in 100 soil samples collected from secondary forest, which located in Emei and Qingcheng Mountains, Sichuan [24]. 35 genera were isolated in soil samples from Xishuangbanna. Therefore actinomycete diversity in tropical rain forest of Xishuangbanna is the highest comparing with all areas studied in our laboratories before.
Recent 20 years, un-cultivable microorganisms in various habitats, soil, ocean, extreme environments, animal and plant were analyzed with DGGE, TGGE and 454 sequencing in many laboratories [25-31]. But in our view, in order to develop new drug and other industry products, only knowing the existence of un-cultivable microbes is not enough completely, we should make the un-cultivable to pure cultured actinomycetes by using every means. This is an important presupposition and new hope for development and utilization of actinomycete resources.
3.3. Key of Selective Isolating Actinobacteria from Soil Samples
Selective isolation methods for actinobacteria, especially
Table 2. Composition of actinobacteria in tropical rain forest in Xishuangbanna.
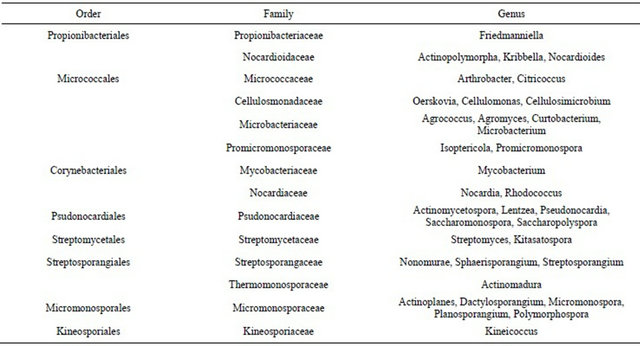
Table 3. A phylogenetic analysis based on the 16S rRNA gene sequence of a part of actinomycete strains.

un-known actinomycetes, are very important, and itself as a research project should be studied, improved and replaced constantly. A large number of Gram negative bacteria, fungi and even known actinomycetes in soil samples is a main problem for selective isolation of un-known actinobacteria. In order to eliminate the appearance of the former, and obtain much more un-known actinobacteria for discovering novel drug leader and other products, sampling and isolation methods should be put as the key point.
Based on many test results for a long term in our laboratory, first, it is best to collect test samples from primeval habitats which have never been disturbed by action of human, such as primeval forests and extreme environments; second, soil samples have to been dried at 25˚C - 28˚C for 7 to 10 days; third, pre-treatment of dried samples at 80˚C to 100˚C for 60 min has to be carried out before isolation; fourth, potassium bichromate 50 mg, or mixture solution of nystatin 100 mg, cycloheximide 50 mg and nalidixic acid 20 mg for 1000 ml medium, as inhibitors, have to be added in the isolation medium for inhibiting fungi and Gram negative bacteria; fifth, improvement Glycerol-Asparagine medium, Mycose-Proline medium and HV medium were better for isolation of actinobacteria from soil samples.
4. Acknowledgements
This work is supported by The GMSTEC (The Greater Mekong Subregion Tertiary Education Consortium Trust), the National Natural Science Foundation of China (No. 30900002 and No. 21062028), National Major scientific and technology special projects (2009ZX09302- 003), and National Institutes of Health USA (1P41GM0- 86184-01A1).
REFERENCES
- J. Berdy, “Bioactive Microbial Metabolites,” The Journal of Antibiotics, Vol. 58, No. 1, 2005, pp. 1-26. doi:10.1038/ja.2005.1
- R. H. Baltz, “Renaissance in Antibacterial Discovery from Actinomycetes,” Current Opinion in Pharmacology, Vol. 8, No. 5, 2008, pp. 1-7. doi:10.1016/j.coph.2008.04.008
- Y. Jiang, J. Wiese, S. K. Tang, L. H. Xu, J. F. Imhoff and C. L. Jiang, “Actinomycetospora Chiangmaiensis gen. nov., sp. nov., a New Member of the Family Pseudonocardiaceae,” International Journal of Systematic and Evolutionary Microbiology, Vol. 58, No. 2, 2008, pp. 408-413. doi:10.1099/ijs.0.64976-0
- Z. T. Kou, H. Zhang, “A Herpetological Report of Xishuangbanna,” In: Y. C. Xu, H. Q. Jiang and F. Quan, (Eds.), Proceedings of Synthetical Investigation of Xishuangbanna Nature Reserves, Yunnan Science and Technology Press, Kunming, 1987, pp. 350-368.
- P. R. Jensen, “Linking Species Concepts to Natural Product Discovery in the Post-Genomic Era,” Journal of Industrial Microbiology and Biotechnology, Vol. 37, No. 3, 2010, pp. 219-224.
- J. Wiese, Y. Jiang, S. K. Tang, V. Thiel, R. Schmaljohann, L. H. Xu, C. L. Jiang and J. F. Imhoff, “A New Member of the Family Micromonosporaceae, Planosporangium Flavigriseum gen. nov., sp. Nov.,” International Journal of Systematic and Evolutionary Microbiology, Vol. 58, No. 6, 2007, pp. 1324-1331. doi:10.1099/ijs.0.65211-0
- M. Cao and J. H. Zhang, “Tree Species Diversity of Tropical Forest Vegetation in Xishuangbanna, SW China,” Biodiversity & Conservation, Vol. 6, No. 7, 1997, pp. 995-1006. doi:10.1023/A:1018367630923
- Y. Li, S. J. Pei and Z. F. Xu, “List of Plants in Xishuangbanna,” Yunnan Nationality Press, Kunming, 1996.
- N. Myers, R. A. Mittermeier, C. G. Mittermeier, G. A. B. Dafonseca and J. Kent, “Biodiversity Hotspots and Conservation Priorities,” Nature, Vol. 403, 2000, pp. 853-858. doi:10.1038/35002501
- Y. Jiang, Y. R. Cao, J. Wiese, K. Lou, L. X. Zhao, J. F. Imhoff and C. L. Jiang, “A New Approach of Research and Development on Pharmaceuticls from Actinomycetes,” Journal of Life Sciences, Vol. 3, No. 7, 2009, pp. 52-56.
- Y. X. Wang and B. Jin, “Mammals in Xishuangbanna Area and a Brief Survey of Its Fauna,” In: Y. C. Xu, H. Q. Jiang and F. Quan, Eds., Proceedings of Synthetical Investigation of Xishuangbanna Nature Reserves, Yunnan Science and Technology Press, Kunming, 1987, pp. 289- 304.
- L. H. Xu, W. J. Li, Z. H. Liu and C. L. Jiang, “Actinomycete taxonomy,” Acedemic Press, Beijing, 2007, pp. 202- 208.
- Y. Yang, T. Xie, Y. Duan, W. Xu and D. H. Zhu, “On Birds from Xishuangbanna,” In: Y. C. Xu, H. Q. Jiang and F. Quan, Eds., Proceedings of Synthetical Investigation of Xishuangbanna Nature Reserves, Yunnan Science and Technology Press, Kunming, 1987, pp. 326-330.
- M. Hayakawa and H. Nonomura, “Humic Acid-Vitamin Agar, a New Medium for the Selective Isolation of Soil Actinomycetes,” Journal of Fermentation Technology, Vol. 65, No. 5, 1987, pp. 501-509. doi:10.1016/0385-6380(87)90108-7
- M. Orsini and V. Romano-Spica, “A Micrwave-Based Method for Nucleic Acid Isolation from Environmental Samples,” Letters in Applied Microbiology, Vol. 33, No. 2, 2001, pp. 17-20. doi:10.1046/j.1472-765X.2001.00938.x
- X. L. Cui, P. H. Mao, M. Zeng, L. H. Xu and C. L. Jiang, “Streptomonospora Salina gen. nov., sp. nov., a New Member of the Family Nocardiopsaceae,” International Journal of Systematic and Evolutionary Microbiology, Vol. 51, No. 2, 2001, pp. 357-363.
- N. Saitou and M. Nei, “The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees,” Molecular Biology and Evolution, Vol. 4, No. 4, 1987, pp. 406-425.
- J. Felsenstein, “Evolutionary Trees from DNA Sequences: A Maximum Likelihood Approach,” International Journal of Systematic and Evolutionary Microbiology, Vol. 17, 1981, pp. 368-376. doi:10.1007/BF01734359
- W. B. Whitman, M. Goodfellow, P. Kämpfer, H. J. Busse, M. E. Trujillo, W. Ludwig, K. I. Suzuki and A. Parte, “Bergey’s Manual of Systematic Bacteriology: Volume 5: The Actinobacteria (Bergey’s Manual/Systemic Bacteriology,” 2nd Edition, Springer, New York, 2012.
- Y. R. Cao, Q. Wang, R. X. Jin, Y. Jiang, H. X. Lai, W. X. He, L. H. Xu and C. L. Jiang, “Planosporangium Mesophilum sp. nov., Isolated from Rhizosphere Soil of Bletilla Striata,” International Journal of Systematic and Evolutionary Microbiology, Vol. 61, No. 6, 2011, pp. 1330- 1333. doi:10.1099/ijs.0.026096-0
- Y. R. Cao, Y. Jiang, J. Y. Wu, L. H. Xu and C. L. Jiang, “Actinopolymorpha Alba sp. nov., Isolated from a Rhizosphere Soil,” International Journal of Systematic and Evolutionary Microbiology, Vol. 59, No. 9, 2009, pp. 2200-2203. doi:10.1099/ijs.0.010280-0
- W. J. Li, L. P. Zhang, P. Xu, X. L. Cui, L. H. Xu, Z. Zhang, P. Schumann, E. Stackebrandt and C. L. Jiang, “Agromyces Aurantiacus sp. nov., Isolated from a Chinese Primeval Forest,” International Journal of Systematic and Evolutionary Microbiology, Vol. 53, No. 1, 2003, pp. 303-307. doi:10.1099/ijs.0.02350-0
- Y. R. Cao, Y. Jiang, L. H. Xu, “Actinomycete Composition and Bioactivities in Grand Shangri-La,” Acta Microbiol Sinica, Vol. 49, No. 1, 2009, pp. 105-109.
- Y. R. Cao, Y. Jiang, Q. Wang, L. X. Zhao, R. X. Jin and C. L. Jiang, “Diversity and Some Bioactivities of Cultured Actinomycetes in Four Areas in Sichuan and Yunnan,” Acta Microbiol Sinica, Vol. 50, No. 8, 2010, pp. 995-1000.
- J. B. Hughes, J. J. Hellmann and H. Ricketts, “Counting the Uncountable: Statistical Approaches to Estimating Microbial Diversity,” Applied and Environmental Microbiology, Vol. 67, No. 10, 2001, pp. 4399-4406. doi:10.1128/AEM.67.10.4399-4406.2001
- R. C. Gao, Z. X. Huang, C. F. Wu, B. Xu and X. Y. Wang, “Culture-Independent Analysis of Microflora in Gayals (Bos frontalis) Feces,” African Journal of Biotechnology, Vol. 9, No. 19, 2010, pp. 2774-2788.
- S. J. Joseph, P. Hugenholtz and P. Sangwan, “Laboratory Cultivation of Widespread and Previously Uncultured Soil Bacteria,” Applied and Environmental Microbiology, Vol. 69, No. 12, 2003, pp. 7210-7215. doi:10.1128/AEM.69.12.7210-7215.2003
- I, S. Middelbos, B. M. V. Boler, A. Qu, B. A. White, K. S. Swanson and G. C. F. Jr, “Phylogenetic Characterization of Fecal Microbial Communities of Dogs Fed Diets with or without Supplemental Dietary Fober Using 454 Pyrosequencing,” PloS One, Vol. 5, No. 3, 2010, pp. 1-9.
- S. Qin, X. Ke, J. H. Jiang, L. H. Xu and W. J. Li, “Biodiversity, Bioactive Natural Products and Biotechnological Potential of Plant Associated Endophytic Actinobacteria,” Applied Microbiology and Biotechnology, Vol. 89, No. 3, 2011, pp. 457-473. doi:10.1007/s00253-010-2923-6
- H. H. Zhang and L. Chen, “Phylogenetic Analysis of 16S rRNA Gene Sequences Reveals Distal Gut Bacterial Diversity in Wild Wolves (Canis lupus),” Molecular Biology Reports, Vol. 37, No. 8, 2010, pp. 4013-4023. doi:10.1007/s11033-010-0060-z
- K. Zengler, G. Toledo and M. Rappé, “Cultivating the Uncultured,” Proceedings of the National Academy of Sciences, Vol. 99, No. 24, 2002, pp. 15681-15686. doi:10.1073/pnas.252630999
NOTES
*Corresponding author.

