Open Journal of Marine Science
Vol.3 No.4(2013), Article ID:37961,8 pages DOI:10.4236/ojms.2013.34019
Mucus Glycoproteins Selectively Secreted from Bacteriocytes in Gill Filaments of the Deep-Sea Clam Calyptogena okutanii
1Institute of Biogeoscience (Biogeos), Japan Agency for Marine-Earth Science and Technology (JAMSTEC), Yokosuka, Japan
2Marine Works Japan, Ltd., Yokosuka, Japan
3School of Medicine, Yokohama City University, Yokohama, Japan
4Graduate School of Science and Engineering, Yamaguchi University, Yamaguchi, Japan
Email: #tyoshida@jamstec.go.jp
Copyright © 2013 Yoshimitsu Nakamura et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received July 3,, 2013; revised August 2, 2013; accepted August 14, 2013
Keywords: Mucus; Mucin; Bacteriocyte; Deep-Sea Clam; Symbiosis
ABSTRACT
The deep-sea clam Calyptogena okutanii possesses a large gill containing vertically transmitted symbiotic sulfur-oxidizing bacteria. It produces large amounts of highly viscoelastic mucus from the gill, which is thought to be a physical and chemical barrier. The mucus collected from the gill was shown to be composed of glycoproteins having the following sugar composition: Man (17.4%), GlcNAc (16.6%), GalNAc (15%), Glc (1.1%), Gal (29.9%), Xyl (3.0%), Fuc (14.4%), and unknown (2.6%), indicating that it contained mucin-like glycoproteins. In a monoclonal antibody library against the gill tissue, we found a monoclonal antibody (mAb), CokG-B3C10, reacting to the mucus. Western blot analysis using the mAb showed that it reacted to several glycoproteins in the mucus from the gill tissue, but not with those of other tissues such as the mantle, foot, and ovary, where mucus production has been reported in bivalves. Further, immunohistochemical analysis showed the CokG-B3C10 mAb reacting to glycoproteins was detected in the inner area of the gill, which was occupied by many bacteriocytes in the row of gill filaments. Strong mAb signals were found on the outer surface of the bacteriocytes facing the interfilamental space, and in the interfilamental spaces between filaments. Weaker signals were also observed in the bacteriocyte cells. These results indicate that the CokG-B3C10 mAbbinding mucus glycoproteins were produced from cells including bacteriocytes and nonbacteriocyte cells in the inner area of the gill filaments.
1. Introduction
In deep-sea hydrothermal vent and seep communities, a unique symbiotic partnership between chemoautotrophic bacteria and invertebrate hosts is found [1,2]. Vesicomyid clams, Calyptogena spp., are the dominant members in deep-sea chemosynthesis-based communities and harbor vertically transmitted symbiotic sulfur-oxidizing bacteria in the gills [3-7].
An outstanding anatomical feature of Calyptogena spp. is the large gill size. Calyptogena clams possess a pair of one or two plate-like demibranches, which are comprised of many rows of filaments [3,5,8]. Histological observation of the gill filaments has shown that their external area is composed of ciliated cells and some nonciliated cells, which have no symbiontic bacteria, which face seawater taken in through the inhalant siphon, and their inner area contains many bacteriocytes and smaller cells without symbiotic bacteria located between the bacteriocytes [3,5,8]. Calyptogena clams have a highly reduced gill ciliary groove, labial palps, and digestive tube and they lack mucus strings [3,5,8]. These observations suggested that the Calyptogena clams depend mainly on symbiont-mediated autotrophic nutrition but weakly on filter feeding [4,5,9]. The symbiotic bacteria of Calyptogena clams have been shown to have a reduced genome, indicating the mutual dependency of the symbiont on the host [10,11].
Calyptogena clams produce highly viscoelastic mucus. Mucus contains sugars covalently linked to lipid or protein, called mucin, and mucus secretion from the foot, mantle, gill, and digestive organs has been reported in bivalves [12,13]. The mucus-secreting cells, i.e., mucocytes, are distributed in the external area containing the ciliated cells of bivalve gills [8,14,15]. As the gill of suspension-feeding bivalves captures, processes, and transports particles to the mouth, the cilia and mucus are thought to function cooperatively [16-20]. In Calyptogena spp., mucocytes and ciliated cells are also distributed in the external area of the gill [8], although it is unlikely that they are involved in feeding. On the other hand, recent progress in research on innate immunity and the environment has focused on mucus as a physical and immunological barrier against environmental stressors and pathogens. As host-secreted mucus covers the surface of various tissues, it can provide a contact site for exogeneous microorganisms and host molecules. Despite the abundant mucus and the description of mucocytes in the gill of Calyptogena spp., neither the composition of the sugar nor the function of the mucus has been elucidated. In the present study, as the first step in understanding the mucus in the gill tissue of Calyptogena okuktanii, we isolated oligosaccharide conjugates from the mucus of the gill tissue and determined the structure of the oligosaccharides. Furthermore, we selected an oligosaccharide conjugate-specific monoclonal antibody (mAb) from an mAb library against the gill tissue cells and examined the localization of the mAb reacting to glycoproteins.
2. Materials and Methods
2.1. Sampling
The clams C. okutanii and Calyptogena soyoae, which are morphologically very similar to each other, were collected at depth of 856 m in the Off Hatsushima Island seep site in Sagami Bay (35˚00.952'N, 139˚13.32'E) using the ROV HyperDolphin during R/V Natsushima cruises (April 24 to May 6 in 2009, January 12 to 19, and May 11 to 18 in 2010). C. okutanii clams were selected after analyzing their partial cytochrome oxidase subunit 1 gene sequence [21]. After dissection, mucus (approximately 20 ml) was collected from their gill tissues. Then their tissues, gill, mantle, foot, and ovary were separately stored at −80˚C. Small pieces of the respective tissues were also immediately fixed with 4% paraformaldehyde at 4˚C in filtered seawater and stored until use.
2.2. Isolation of Oligosaccharide Samples
Oligosaccharide samples in the mucus were isolated following the method in a previous report [22]. Contaminated hematocytes and debris in the collected mucus were removed by centrifugation at 200 × g for 15 min. For precipitating the proteins, trichloroacetic acid (TCA, 10%, w/v) was added to the supernatant. The samples were then incubated at room temperature for 30 min and centrifuged at 9100 × g for 20 min. The precipitate was resuspended and dialyzed against 100 mM potassium phosphate buffer (pH 6.8) overnight and resolubilized. Resolubilized samples were digested at 37˚C overnight by a protease reagent of Streptomyces griseus, 1000 U of PRONASE (Roche Applied Science Japan, Tokyo, Japan). Proteins in the digested samples were removed by TCA treatment as described above. Then, oligosaccharide samples from which the protein had been removed were obtained by ethanol precipitation. The supernatant after TCA treatment was transferred into a new tube, diluted with a three-fold volume of ice-chilled ethanol, incubated at −30˚C overnight, and centrifuged at 9100 × g for 30 min. The precipitated oligosaccharide samples were solved in 1 ml of pure water, freeze-dried, and stored at −80˚C until use.
2.3. Protein Quantification
The freeze-dried oligosaccharide samples (1 mg) were solved in 1 ml of deionized water. The protein concentration in the aqueous solution was quantified with a BioRad protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA) using bovine albumin as a standard.
2.4. Molecular Mass Estimation
For molecular mass analysis, a high-pressure liquid chromatography (HPLC) system (Waters e2796 system, Waters Co., Milford, MA, USA) equipped with a gel filtration column (TSK-Gel GMPW, 7.5 × 300 mm, Tosoh, Tokyo, Japan) was used. One milligram of the freezedried oligosaccharide samples was dissolved in 100 μl of 200 mM sodium phosphate buffer (pH 6.8). Two hundred micromoles of sodium phosphate buffer were used as the running buffer at a flow rate of 0.5 ml/min. A refractive index detector (Waters e2796, Waters Co.) was used for the detection. A commercial molecular standard kit (P-82, Showa Denko K.K., Tokyo, Japan) was used for calibration.
2.5. Sugar Composition Analysis
The sugar composition of the oligosaccharide samples was analyzed using the 1-phenyl-3-methyl-5-pyrazolone (PMP) derivative method as described previously [23], with minor modification. One milligram of freeze-dried oligosaccharide samples was added to 1 ml of methanol containing 5% HCl, and the resulting mixture was heated at 100˚C for 2 h. After evaporation, 1 ml of 4 M trifluoroacetic acid was added and heated at 100˚C for 1 h. The resulting hydrolysate was evaporated, and 0.5 ml of deionized water was added. After solubilization, this solution (200 μl) was mixed with 200 μl of methanol containing 0.5 M PMP and 100 μl of 0.6 M NaOH, respectively, and then incubated at 70˚C for 30 min. After incubation, 0.1 M HCl was added and excess PMP was removed by chloroform extraction. PMP-labeled monomers were analyzed using the HPLC system (Waters e2796 system) equipped with a reverse-phase column (YMC pack ODS-AQ column, 4.6 × 150 mm, YMC Co. Ltd., Kyoto, Japan) and a Waters 2998 photodiode array detector (Waters Co.) at 245 nm. The hydrolysates were eluted with a gradient of mobile phase A (84:16 mixture of 100 mM sodium phosphate buffer, pH 7.0, and acetonitrile) and mobile phase B (acetonitrile), which used a gradient program (B%: 0% from 0 to 10 min, linear gradient from 0% to 30% between 10 and 30 min). The buffer flow rate was set at 1 ml/min.
2.6. Construction of mAb against Glycoprotein in the Mucus
mAbs were raised against the homogenized gill tissues of C. okutanii using the conventional polyethylene glycol (PEG) method [24]. The tissue homogenate was injected three times at 2-week intervals into three BALB/c mice. On day 4 after the third immunization, the mice were killed and the spleen lymphocytes were collected and fused with the murine myeloma cell line NS-1 in the presence of 50% PEG. The fused hybridoma cells were selected by incubation in hypoxanthine-aminopterinthymidine selective medium (GIT medium, Wako Pure Chemical Industries, Osaka, Japan) supplemented with 100 μM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine at 37˚C for 10 days. Supernatants of the hybridoma cell cultures were screened for antibody production with the indirect immunofluorescence method described below. Cloning was performed for the immunofluoresence-positive hybridoma cells using the limited dilution method, which was repeated three times.
The antibody reacting with the oligosaccharides was selected by dot-blot analysis. The oligosaccharide samples obtained or N-glycosidase F-treated oligosaccharide samples were spotted on a polyvinyldifluoride (PVDF) membrane (Merck Milipore, Billerica, MA, USA). The membrane was reacted with each mAb at 37˚C for 1 h. After washing three times, the membrane was incubated at 37˚C for 1 h in anti-mouse IgG horseradish peroxidase (HRP)-linked goat antibody (Life Technologies Co., San Francisco, CA, USA) diluted at 1:1000. The HRP signal was detected using 3,3’-diaminobenxidine tetrahydrochloride (Wako Pure Chemical Industries). All animal experiments were conducted in accordance with the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan), and all protocols were approved by the institutional review board of the Animal Research Center, Yokohama City University School of Medicine.
2.7. SDS-PAGE and Western Blot Analyses
Immediately after collection of the Calyptogena clams, they were dissected and their tissues (gill, mantle, foot, and others) were separated. Approximately 30 mg of the tissues were respectively homogenized with 10 - 20 strokes by a Teflon homogenizer on ice in 1 ml of homogenization buffer (10 mM Tris-HCl, pH 7.8, 1% NP- 40, 0.15 M NaCl, 1 mM EDTA) with a protease inhibitor mix (GE Healthcare Bio-Science, Piscataway, NJ, USA). Homogenates were filtered through two-layered gauze to remove debris. The supernatants were sonicated at 10 pulses/2 sec with setting at 20% output with an UltraS homogenizer (Taitec, Koshigaya, Japan) in an ice-water bath. After centrifugation at 20,000 × g for 30 min, the supernatant was mixed with an equal volume of SDS sample buffer and boiled for 5 min. Then, 20 mg of each sample was subjected to 12.5% sodium dodecylsulfate polyacrylamide gel electrophoresis. For the SDS-PAGE analysis, the gel was stained by with Coomassie brilliant blue R-250 (CBB). After the electrophoresis, the separated proteins in the gel were electrophoretically transferred to a PVDF membrane. The membrane was incubated with the antibody (the supernatant of the hybridoma cells, which was positively selected by dot-blot analysis) and stained as described above for dot-blot analysis.
2.8. Indirect Immunofluorescence Staining
Gill tissues of C. okutanii were fixed with 4% paraformaldehyde in seawater for 18 h. They were then frozenembedded in OTC compound in liquid nitrogen. The frozen thin sections as 4-μm slices were prepared with cryomicrotome (Microm HM550, Thermo Fisher Scientific Inc., Tewksbury, MA, USA). Air-dried sections were washed three times with phosphate-buffered saline (PBS) for 10 min and then incubated with the supernatant of the hybridoma culture overnight at 4˚C. After washing three times, Alexa Fluor 488 (AF488)-conjugated goat antimouse IgG (Life Technologies Co.) diluted 1000-fold with PBS (135 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 8.0) was added and incubated for 3 h at 25˚C. In addition, the sections were reacted with anti-GroEL rabbit polyclonal antibody (SigmaAldrich Co., St. Louis, MO, USA) diluted at 1:1000, followed by incubation with Alexa Fluor 647-conjugated anti-rabbit IgG antibodies (Life Technologies Co.). After washing, the sections were stained with 6-diamidino-2- phenylindole dihydrochloride (DAPI, Sigma-Aldrich Co.) at a final concentration of 10 μg/ml for 15 min. A microscope equipped with phase-contrast and fluorescence optics (BZ-9000, Keyence, Osaka, Japan) was used for observation.
2.9. In Situ Hybridization
The sections were transferred to Teflon-coated slides and air-dried before dehydration by sequential washing in 50, 80, and 100% (v/v) ethanol for 3 min each. Hybridization using the FITC-labeled probe Cok 16S_1: antisense RNA fragment of the 16S rRNA gene of the symbiont of C. okutanii (5’-AGCTTCGCCACTAAAGGGTACCCCC-3’), was performed as described previously [25]. Microscopic observation was performed under fluorescence microscopy.
3. Results
3.1. Isolation of Oligosaccharides from Mucus and Determination of Components
Approximately 6 mg of mucus from which protein had been removed was obtained from the protease-digested TCA precipitate. No free oligosaccharide was detected either in the secreted mucus or in the mucus from which protein had been removed. A protein assay showed that mucus without protein contained 15.2% ± 1.2% peptides (15.2 ± 1.2 mg/mg dry weight), implying that the oligosaccharide samples contained small amounts of peptides. Gel-permeation chromatography showed that the molecular weight of the prepared oligosaccharide samples was approximately 14 kDa. The sugar composition of the oligosaccharide samples was analyzed by HPLC after the hydrolysis and PMP-labeling treatments (Table 1). Several minor peaks were unidentified (2.6% of the total peak area). The major sugar compound was galactose (Gal), which made up approximately 30% (Table 1). The contents of mannose (Man), fucose (Fuc), GlcNAc, and GalNAc were around 15% each.
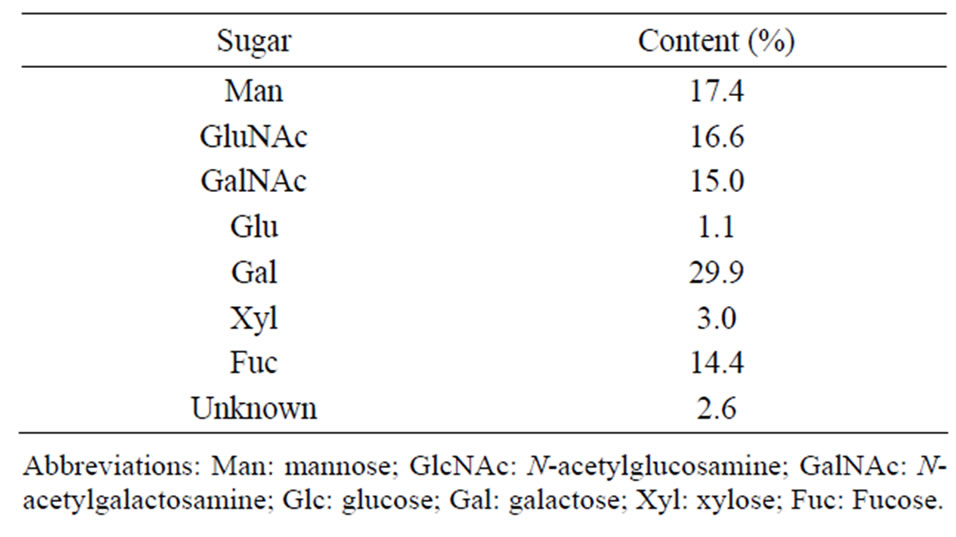
Table 1. Sugar composition of the mucus oligosaccharides from C. okutanii.
3.2. mAb Reacting to Mucus Glycopeptides
To determine the localization of the mucus glycoproteins in the gill tissue, the CokG-B3C10 mAb was selected from a mAb library prepared using homogenized gill tissue as the antigen. According to the results of dot-blot analysis, the mAb showed the strongest signal to the oligosaccharide samples in the approximately 500 tested antibodies (data not shown). The mAb reaction was lost after treatment with N-glycosidase F (data not shown). These results suggested that the CokG-B3C10 mAb reacted to the glycoproteins.
3.3. Localization of Mucus Glycoproteins in C. okutanii
To examine the localization of the CokG-B3C10 mAb antigen, tissues of the gill, ovary, foot, and mantle of C. okutanii were separately homogenized and their supernatants were subjected to Western blot analysis. The mAb reacted only to several bands from the gill homogenate, in the range of molecular weight between 25 and 250 kDa (Figure 1). This result indicated that the gill tissue contained several different antigens that reacted to the mAb, all of which were exclusively localized in the gill tissue.
 (a) (b)
(a) (b)
Figure 1. SDS-PAGE analysis of different tissues from C. okutanii with Coomassie brilliant blue (CBB) staining and western blotting analysis using the CokG-B3C10 mAb. (a) CBB staining gel; (b) Western blotting analysis. The samples were from the gill (lane 1), foot (lane 2), mantle (lane 3), and ovary (lane 4). M: molecular weight marker.
To determine more detailed localization of the mAb antigen in the gill tissue, histochemical analysis of frozen transverse sections of the gill tissue was performed. Figure 2 shows the frontal zone of the external asymbiotic area and inner symbiotic area of the gill filament. In the differential interference contrast micrograph of the inner area of gill filaments, the symbiotic bacteria in bacteriocytes were observed as dark spots (arrows in Figure 3(a)). Green fluorescence signals of the CokG-B3C10 mAb were observed only in the inner symbiotic area of gill filament, but not in the frontal zone of the external asymbiotic area of gill filament (Figure 3(b)). No signals were detected in the negative control micrograph without mAb treatment. To visualize the location of the symbiotic bacteria, immunostaining with antibacterial GroEL antibody was performed (red fluorescence in Figures 3(c), (f) and (g)). The enlarged images showed that strong green signals appeared on the apical surface of the gill filament cells facing seawater and in the interfilamental spaces, and weaker signals were also detected in the bacteriocyte cells (Figures 3(d) and (e)). The density of the CokG-B3C10 mAb-reactive antigen in the secretions was higher near the external area (A in Figures 3(a)-(c)) in the gill filaments than in the central region of the inner area (B in Figures 3(a)-(c)). Symbionts were detected in the cytoplasm of bacteriocytes in the inner area of the gill filaments (Figure 3(c), (f) and (g)). The localization of symbionts was furher confirmed by FISH analysis using a C. okutanii symbiont-specific probe targeting its 16S ribosomal RNA. This probe was shown to be specific to the symbionts of C. okutanii and did not hybridize with symbionts of Bathymodiolus septemdierum (data not shown). The symbionts were detected only in the bacteriocytes but not in the asymbiotic cells in the inner area in the frontal zone (Figure 4). The green signals for the mAb antigen seemed to appear on both bacteriocytes and asymbiotic cells in the inner area (Figure 3(g), arrows).
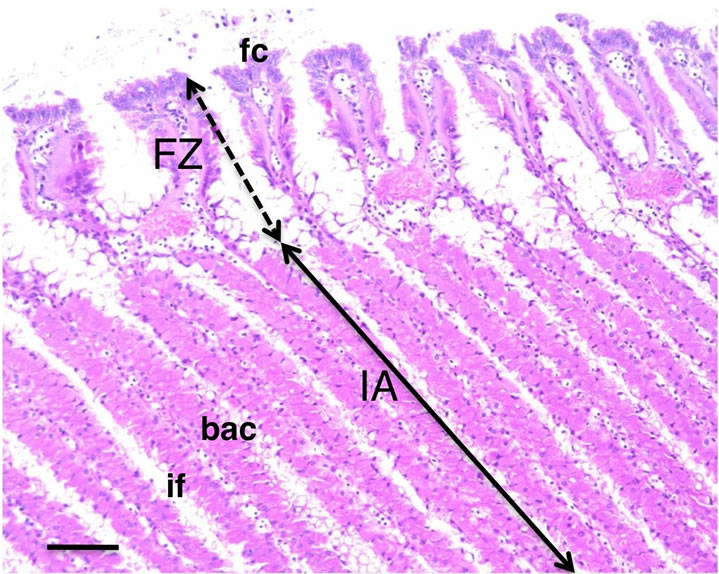
Figure 2. Transverse sections of C. okutanii gill. Frontal zone (FZ) of the external area and inner area (IA) with filament, stained with hematoxylin and eosin. Scale bar: 100 μm. Abbreviations: bac: bacteriocyte; if: interfilamental space; fc: frontal ciliated cells.

Figure 3. Immunostaining images of the frontal zone of the external and internal areas of transverse sections from the C. okutanii gill. (a) Phase difference microscopy image; (b), (d) and (e) Staining with the CokG-B3C10 mAb (green); (c), (f) and (g) Merged with the mAb (green), anti-GroEL antibody (red), and DAPI staining; (d) and (f) Enlarged images of part A in Figures 3(a) and (e); (g) Enlarged images of part B in Figure 3(a). Scale bar: (a)-(c): 50 μm; (d)-(g): 5 μm. Arrows: bacteriocytes. Abbreviations: FZ: frontal zone; bac: bacteriocyte; if: interfilamental space.

Figure 4. FISH image analysis of gill filament. (a) Phase difference microscopy image of the gill filament; (b) Merged image of DAPI staining (blue) and FISH image using a C. okutanii symbiont-specific oligo-DNA probe targeting 16S RNA (green); (c) Enlarged merged image of the boxed area. Scale bar: (a) and (b): 50 μm; (c): 5 μm. Abbreviations: FZ: frontal zone; IA: inner area.
4. Discussion
4.1. Sugar Composition of Mucus Secreted from Gill Tissue of C. okutanii
In human mucin, 50% - 90% of O-linked glucans are attached to the protein backbone (apomucin), and molecular weights of the complexes are often many million Daltons [26]. Seventeen human mucin genes have been assigned to the MUC gene family according to the Human Genome Project. The analyzed more than 100 different O-linked glucans shown to contain two to 12 monosaccharides, of which the major saccharides are Gal, GalNAc, GlcAc, Fuc, and sialic acid [26]. As the mucins of C. okutanii had not been characterized yet, we examined the saccharide composition of the gill mucus and showed that the mucus was comprised of Gal, Man, GlcNAc, GalNAc, and Fuc (Table 1). This composition is similar to that of human mucus, except for the high amount of Man. It was also consistent with the results of the analysis of Mytilus edulis mucus glycoprotein showing that it contained Man, Fuc, Gal, GlcNAc, and GalNAc as major sugar components [27]. Thus, the present study confirmed that mucins from bivalves have similar sugar components to those of humans. This may indicate that mucin in vertebrates and invertebrate has a similar function. Furthermore, we obtained the gill tissue-specific CokG-B3C10 mAb bound to the oligosaccharide samples from mucus. The analysis of the mAb-binding epitope is important for a better understanding of the function of mucin from clams.
4.2. Glycoproteins in Mucus Localized in Gill Tissue and Possible Function
In bivalves, mucocytes have been morphologically observed in epithelial cells of mucus-producing organs such as the gill, mantle, foot, and digestive organs [3,5,8,12, 13]. In a variety of bivalves, including Calyptogena magnifica and Mytilus galloprovincialis, mucocytes were morphologically identified in the frontal and abfrontal zones of the gill external area [8,14,15]. The mucocytes of M. galloprovincialis were stained by periodic acid Schiff-alcian blue, indicating mucus production [15]. Those previous reports suggested that the mucus of the gill is produced from the external asymbiotic area of gill filaments in C. okutanii. The co-localization of mucocytes and ciliated cells in gill filaments is reasonable for feeding; the mucus captures plankton or particles, and the ciliated cells generate the flow to transport them to the digestive organs in filter-feeding asymbiotic bivalves. We did not show the distribution of the mucus in the gill tissue, although we determined that of the glycoproteins contained in the mucus using the CokG-B3C10 mAb. The glycoproteins were specifically produced from the inner asymbiotic area of gill tissues. Both the bacteriocytes and the adjacent asymbiotic cells appeared to produce them (Figures 3(f) and (g)).
Although the function of the glycoprotein antigens remains to be elucidated, it is not likely for feeding because the digestive tract is vestigial, but may act as a defense barrier and/or for maintenance of the symbiotic bacteria. In the human intestinal extracellular microbiota system, mucin not only plays a role in protection against pathogens but also in cross-talk and maintenance of the microbiota [28-31]. Recently, in invertebrates, factors binding with sugars on the surface of the symbionts have been identified in the host mucus in some horizontal transmission-type symbiotic systems: a squid (Euprymna scolopes); tropical clam (Codakia orbicularis); and marine nematode (Laxus oneistus) [32-34]. They are a novel lipopolysaccharide-binding protein, mannose-binding protein, and C-type lectins, respectively, and contribute to the acquision of symbionts. The CokG-B3C10 mAb appeared to bind to several glycoproteins (Figure 1). It is not clear whether they have different functions. However, they are produced in bacteriocytes and adjacent asymbiotic cells in the inner area of the gill filaments, and this localization suggests that they have some role in the maintenance of symbiosis.
In symbioses between invertebrates and microorganisms, glycoproteins of the host invertebrates and of the symbionts may play important roles in the recognition of their partners [35] and in the maintenance of their stable interactions. Further identification of the innate immune factors in mucus will be useful for understanding the mucosal immunity of deep-sea clams and of the maintenance of their symbiotic partnerships. While the function of the CokG-B3C10 mAb antigen is still not clear, this antibody will be useful in revealing the function of the antigen in gill mucosal immunity and symbiosis.
5. Acknowledgements
We thank the crews of the R/V Natsushima, and the operation team of the ROV HyperDolphin for helping to collect deep-sea biological samples. We also thank Mr. S. Shimamura for assisting in the FISH probe design, and Ms. E. Seo for the identification of Calyptogena clams.
REFERENCES
- C. M. Cavanaugh, S. L. Gardiner, M. L. Jones, H. W. Jannasch and J. B. Waterbury, “Prokaryotic Cells in the Hydrothermal Vent Tube Worm Riftia pachyptila Jones: Possible Chemoautotrophic Symbionts,” Science, Vol. 213, No. 4505, 1981, pp. 340-342. http://dx.doi.org/10.1126/science.213.4505.340
- H. Felbeck, J. J. Childress and G. N. Somero, “CalvinBenson Cycle and Sulphide Oxidation Enzymes in Animals from Sulphide-Rich Habitats,” Nature, Vol. 293, No. 5830, 1981, pp. 291-293. http://dx.doi.org/10.1038/293291a0
- K. J. Boss and R. D. Turner, “The Giant White Clam from the Galapagos Rift, Calyptogena magnifica, Species Novum,” Malacologia, Vol. 20, No. 1, 1980, pp. 161- 194.
- C. M. Cavanaugh, “Symbiotic Chemoautotrophic Bacteria in Marine Invertebrates from Sulphide-Rich Habitats,” Nature, Vol. 302, No. 5903, 1983, pp. 58-61. http://dx.doi.org/10.1038/302058a0
- A. Fiala-Medioni and C. Metivier, “Ultrastructure of the Gill of the Hydrothermal Vent Bivalve Calyptogena magnifica, with a Discussion of Its Nutrition,” Marine Biology, Vol. 90, No. 2, 1986, pp. 215-222. http://dx.doi.org/10.1007/BF00569130
- S. C. Cary and S. J. Giovannoni, “Transovarial Inheritance of Endosymbiotic Bacteria in Clams Inhabiting Deep-Sea Hydrothermal Vents and Cold Seeps,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 90, No. 12, 1993, pp. 5695- 5699. http://dx.doi.org/10.1073/pnas.90.12.5695
- K. Endow and S. Ohta, “Occurrence of Bacteria in the Primary Oocytes of Vesicomyid Clam Calyptogena soyoae,” Marine Ecology Progress Series, Vol. 64, No. 3, 1990, pp. 309-311. http://dx.doi.org/10.3354/meps064309
- B. Morton, “The Functional Morphology of the Organs of Feeding and Digestion of the Hydrothermal Vent Bivalve Calyptogena magnifica (Vesicomyidae),” Journal of Zoology, Vol. 208, No. 1, 1986, pp. 83-98. http://dx.doi.org/10.1111/j.1469-7998.1986.tb04711.x
- M. J. Kennish and R. A. Lutz, “The Hydrothermal Vent Clam, Calyptogena magnifica (Boss and Turner, 1980): A Review of Existing Literature,” Reviews in Aquatic Science, Vol. 6, No. 1, 1992, pp. 29-66.
- H. Kuwahara, T. Yoshida, Y. Takaki, S. Shimamura, S. Nishi, M. Harada, K. Matsuyama, K. Takishita, M. Kawato, K. Uematsu, Y. Fujiwara, T. Sato, C. Kato, M. Kitagawa, I. Kato and T. Maruyama, “Reduced Genome of the Thioautotrophic Intracellular Symbiont in a Deep-Sea Clam, Calyptogena okutanii,” Current Biology, Vol. 17, No. 10, 2007, pp. 881-886. http://dx.doi.org/10.1016/j.cub.2007.04.039
- I. L. G. Newton, T. Woyke, T. A. Auchtung, G. F. Dilly, R. J. Dutton, M. C. Fisher, K. M. Fontanez, E. Lau, F. J. Stewart, P. M. Richardson, K. W. Barry, E. Saunders, J. C. Detter, D. Wu, J. A. Eisen and C. M. Cavanaugh, “The Calyptogena magnifica Chemoautotrophic Symbiont Genome,” Science, Vol. 315, No. 5814, 2007, pp. 998-1000. http://dx.doi.org/10.1126/science.1138438
- M. S. Davies and S. J. Hawkins, “Mucus from Marine Molluscs,” In: J. H. S. Blaxter, A. J. Southward and P. A. Tyler, Eds., Advances in Marine Biology, Academic Press Ltd.-Elsevier Science Ltd., London, 1998, pp. 1-71.
- C. Calabro, M. P. Albanese, S. Martella, P. Licata, E. R. Lauriano, C. Bertuccio and A. Licata, “Glycoconjugate Histochemistry and nNOS Immunolocalization in the Mantle and Foot Epithelia of Tapes philippinarum (Bivalve Mollusc),” Folia Histochemica et Cytobiologica, Vol. 43, No. 3, 2005, pp. 151-156.
- S. C. Dufour and P. G. Beniger, “A Functional Interpretation of Cilia and Mucocyte Distributions on the Abfrontal Surface of Bivalve Gills,” Marine Biology, Vol. 138, No. 2, 2001, pp. 295-309. http://dx.doi.org/10.1007/s002270000466
- A. Gomez-Mendikute, M. Elizondo, P. Venier and M. P. Cajaraville, “Characterization of Mussel Gill Cells in Vivo and in Vitro,” Cell and Tissue Research, Vol. 321, No. 1, 2005, pp. 131-140. http://dx.doi.org/10.1007/s00441-005-1093-9
- M. A. Sleigh, “Adaptations of Ciliary Systems for the Propulsion of Water and Mucus,” Comparative Biochemistry and Physiology Part A: Physiology, Vol. 94, No. 2, 1989, pp. 359-364. http://dx.doi.org/10.1016/0300-9629(89)90559-8
- P. G. Beninger and S. C. Dufour, “Mucocyte Distribution and Relationship to Particle Transport on the Pseudolamellibranch Gill of Crossostrea virginica (Bivalve: Ostreidae),” Marine Ecology Progress Series, Vol. 137, No. 1-3, pp. 133-138.
- P. G. Beninger, S. St. Jean, Y. Poussart and J. E. Ward, “Gill Function and Mucocyte Distribution in Placopecten magellanicus and Mytilus edulis (Mollusca: Bivalvia): The Role of Mucus in Particle Transport,” Marine Ecology Progress Series, Vol. 98, No. 3, 1993, pp. 275-282. http://dx.doi.org/10.3354/meps098275
- J. E. Ward, J. S. Levinton, S. E. Shumway and T. Cucci, “Particle Sorting in Bivalves: In Vivo Determination of the Pallial Organs of Selection,” Marine Biology, Vol. 131, No. 2, 1998, pp. 283-292. http://dx.doi.org/10.1007/s002270050321
- H. Silverman, J. W. Lynn, P. G. Beninger and T. H. Dietz, “The Role of Latero-Frontal Cirri in Particle Capture by the Gills of Mytilus edulis,” The Biological Bulletin, Vol. 197, No. 3, 1999, pp. 368-376. http://dx.doi.org/10.2307/1542791
- M. Harada, T. Yoshida, H. Kuwahara, S. Shimamura, Y. Takaki, C. Kato, T. Miwa, H. Miyake and T. Maruyama, “Expression of Genes for Sulfur Oxidation in the Intercellular Chemoautotrophic Symbiont of the Deep-Sea Bivalve Calyptogena okutanii,” Extremophiles, Vol. 13, No. 6, 2009, pp. 895-903. http://dx.doi.org/10.1007/s00792-009-0277-8
- N. Seno, K. Anno, Y. Yaegashi and T. Okuyama, “Microheterogeneity of Chondroitin Sulfates from Various Cartilages,” Connective Tissue Research, Vol. 3, No. 1, 1975, pp. 87-96. http://dx.doi.org/10.3109/03008207509152345
- N. Izawa, T. Hanamizu, R. Iizuka, T. Sone, H. Mizukoshi, K. Kimura and K Chiba, “Streptococcus thermophilus Produces Exopolysaccharides including Hyaluronic Acid,” Journal of Bioscience and Bioengineering, Vol. 107, No. 2, 2009, pp. 119-123. http://dx.doi.org/10.1016/j.jbiosc.2008.11.007
- G. Galfre and C. Milstein, “Preparation of Monoclonal Antibodies: Strategies and Procedures,” Methods in Enzymology, Vol. 73, 1981, pp. 3-46. http://dx.doi.org/10.1016/0076-6879(81)73054-4
- Y. Sekiguchi, Y. Kamagata, K. Sytsubo, A. Ohashi, H. Harada and K. Nakamura, “Phylogenetic Diversity of Mesophilic and Thermophilic Granular Sludges Determined by 16S rRNA Gene Analysis,” Microbiology, Vol. 144, No. 9, 1998, pp. 2655-2665. http://dx.doi.org/10.1099/00221287-144-9-2655
- M. Derrien, M. W. van Passel, J. H. van de Bovenkamp, R. G. Schipper, W. M. de Vos and J. Dekker, “MucinBacterial Interactions in the Human Oral Cavity and Digestive Tract,” Gut Microbes, Vol. 1, No. 4, 2010, pp. 254-268. http://dx.doi.org/10.4161/gmic.1.4.12778
- H. Y. Ahn, L. F. Sue, J. K. Ma, C. A. Pinkstaff, R. S. Pore, D. O. Overman and C. J. Malanga, “Synthesis and Secretion of Mucus Glycoprotein by the Gill of Mytilus edulis. I. Histochemical and Chromatographic Analysis of [14C] Glucosamine Bioincorporation,” Biochimica et Biophysica Acta (BBA)-General Subjects, Vol. 966, No. 1, 1988, pp. 122-132. http://dx.doi.org/10.1016/0304-4165(88)90136-5
- S. Salminen, M. Laine, A. Vonwright, J. Vuopio-Varkila, T. Korhonen and T. Mattila-Sandholm, “Development of Selection Criteria for Probiotic Strains to Access their Potential in Functional Food: A Nordic and European Approach,” Bioscience and Mircoflora, Vol. 15, No. 2, 1996, pp. 61-67. http://dx.doi.org/10.1111/j.1574-6968.1998.tb13226.x
- P. V. Kirjavainen, A. C. Ouwehand, E. Isolauri and S. J. Salminen, “The Ability of Probiotic Bacteria to Bind to Human Intestinal Mucus,” FEMS Microbiology Letters, Vol. 167, No. 2, 1998, pp. 185-189.
- V. Lievin-Le Moal and A. L. Servin, “The Front Line of Enteric Host Defense against Unwelcome Intrusion of Harmful Microorganisms: Mucins, Antimicrobial Peptides, and Microbiota,” Clinical Microbiology Reviews, Vol. 19, No. 2, 2006, pp. 315-337. http://dx.doi.org/10.1128/CMR.19.2.315-337.2006
- S. K. Linde, P. Sutton, N. G. Karlsson, V. Korolik and M. A. McGuckin, “Mucins in the Mucosal Barrier to Infection,” Mucosal Immunology, Vol. 1, No. 3, 2008, pp. 183- 197. http://dx.doi.org/10.1038/mi.2008.5
- C. K. Chun, J. V. Troll, I. Koroleva, B. Brown, L. Manzella, E. Snir, H. Almabrazi, T. E. Scheetz, M. F. Bonaldo, T. L. Casavant, M. B. Soares, E. G. Ruby and M. J. McFallNgai, “Effects of Colonization, Luminescence, and Autoinducer on Host Transcription during Development of the Squid-Vibrio Association,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 105, No. 32, 2008, pp. 11323-11328. http://dx.doi.org/10.1073/pnas.0802369105
- S. Bulgheresi, I. Schabussova, T. Chen, N. P. Mullin, R. M. Maizels and J. A. Ott, “A New C-Type Lectin Similar to the Human Immunoreceptor DC-SIGN Mediates Symbiont Acquisition by a Marine Nematode,” Applied and Environmental Microbiology, Vol. 72, No. 4, pp. 2950- 2956. http://dx.doi.org/10.1128/AEM.72.4.2950-2956.2006
- J. P. Gourdine and E. J. Smith-Ravin, “Analysis of a cDNA-Derived Sequence of a Novel Mannose-Binding Lectin, Codakine, from the Tropical Clam Codakia orbicularis,” Fish & Shellfish Immunology, Vol. 57, No. 5, 2007, pp. 498-509. http://dx.doi.org/10.1016/j.fsi.2006.06.013
- S. V. Nyholm and M. J. McFall-Ngai, “Dominance of Vibrio fischeri in Secreted Mucus outside the Light Organ of Euprymna scolopes: The First Site of Symbiont Specificity,” Applied and Environmental Microbiology, Vol. 69, No. 7, 2003, pp. 3932-3937. http://dx.doi.org/10.1128/AEM.69.7.3932-3937.2003
NOTES
*Equal contributors.
#Corresponding author.

