Open Journal of Stomatology
Vol.05 No.11(2015), Article ID:61076,9 pages
10.4236/ojst.2015.511032
SspB Peptide Assay Reveals Saliva-Mediated Porphyromonas gingivalis Attachment
Tatsuro Ito1,2*, Hidenobu Senpuku3, Takahiro Ichinosawa1, Nana Ikematsu-Ito1, Nao Kimura1, Takehiko Shimizu1,2
1Department of Pediatric Dentistry, Nihon University School of Dentistry at Matsudo, Chiba, Japan
2Nihon University Research Institute of Oral Science, Chiba, Japan
3Department of Bacteriology, National Institute of Infectious Diseases, Tokyo, Japan

Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).


Received 17 October 2015; accepted 10 November 2015; published 13 November 2015

ABSTRACT
Background: Porphyromonas gingivalis is a major periodontal pathogen that binds efficiently to Streptococcus gordonii, which in turn binds to salivary agglutinin (gp340). The SspB of S. gordonii appears to mediate this association. We previously reported that the streptococcal SspB peptide analog, designated SspB (390-T400K-402), showed high binding activity with saliva. To understand the three-way interaction among S. gordonii, P. gingivalis and salivary gp340 as a unit, we established a peptide binding assay using SspB (390-T400K-402). Methods: The binding activity of the SspB (390-T400K-402) to P. gingivalis was detected by ELISA. Ninety-six well plates were coated with whole bacterial cell (P. gingivalis strains ATCC 33277, and W83; S. gordonii DL1) in Na2CO3 coating buffer. After blocking, bacterial cells were incubated with saliva or salivary agglutinin peptide (SRCRP2). Biotinylated SspB (390-T400K-402) was applied and incubated with 1:1000 streptoavidin-conjugated alkaline phosphatase. After development, A405 was recorded. Results: P. gingivalis 33277 showed the highest binding activity of the tested bacteria, whereas P. gingivalis W83, which was deficient in Mfa1 fimbriae, exhibited poor binding activity, as did S. gordonii. The binding of SspB (390-T400K-402) peptide in saliva- or SRCRP2-treated P. gingivalis was significantly higher than that in non-treated cells. Conclusion: The SspB (390-T400K-402) peptide binding assay revealed that initial attachment of P. gingivalis to the substrata of S. gordonii may require gp340-mediated SspB-Mfa1 interactions. The assay is available to assess the relationships among SspB, Mfa1 and salivary gp340 as a unit.
Keywords:
SspB, Biofilm, Porphyromonas gingivalis, Saliva, gp340

1. Introduction
Saliva-coated teeth and epithelium (cheeks, gums, and tongue) provide a variety of surfaces for bacterial attachment and colonization, followed by dental plaque maturation [1] . Dental plaque is a complex biofilm composed of early and late colonizing bacteria [2] [3] . Attachment of early colonizing bacteria as typified by Streptococcus gordonii to salivary components, specifically targeting innate immunity scavenger receptor glycoprotein-340 (gp340) [4] , is a key event in oral biofilm formation [5] [6] . The periodontitis-associated pathogen Porphyromonas gingivalis, one of the later biofilm inhabitants, binds and forms biofilms on the antecedent organisms, such as S. gordonii [7] .
SspB protein, a streptococcal adhesin [8] , mediates attachment of S. gordonii to saliva-coated enamel surfaces and has been shown to bind to Mfa1 fimbriae of P. gingivalis [9] . The binding domain of SspB to P. gingivalis has been mapped [10] . In addition, a discrete structural region in SspB that confers the adherence phenotype has been identified [11] [12] . However, few assays are available to assess the relationships among SspB, Mfa1 and salivary gp340 as a unit.
We previously demonstrated that analogous SspB (390-T400K-402) peptide [a substitution of threonine for lysine at 400, SspB (390-402) peptide] had the highest binding activity to the salivary components among several analogous SspB peptides [13] . The positively charged amino acid residue, i.e. lysine, is essential for binding to the negatively charged salivary components [14] and gp340 peptide (designated SRCRP2) [15] .
Salivary gp340 has a high bacteria-binding capacity, and recognizes different bacterial receptors based on whether gp340 is in the fluidphase or is bound to the hydroxyapatite surface [16] . Furthermore, the fimbriae of P. gingivalis also bind to salivary proteins, such as proline-rich salivary protein 1 and statherine [17] - [19] . Therefore, we hypothesized that P. gingivalis was able to bind to the salivary gp340 and to the SRCRP2 peptide simultaneously with Mfa1-SspB interaction.
In the present study, to establish an assay that enables us to understand a three-way interaction among S. gordonii, P. gingivalis and salivary gp340 as a unit, and to examine whether P. gingivalis is able to bind to salivary gp340 and to SRCRP2, we evaluated the binding activity of the streptococcal peptide analog SspB (390-T400K-402) to P. gingivalis in the presence of saliva or gp340 peptide SRCRP2. An understanding the association between these molecules and periodontal bacteria is essential for elucidating the mechanisms of supra- and sub-gingival oral biofilm formation.
2. Materials and Methods
2.1. Bacterial Culture
Porphyromonas gingivalis strains 33277 and W83 (afimbriated) were grown in brain heart infusion (BHI) broth supplemented with 1 µg/ml hemin (final concentration), and 5 µg/ml menadione (final concentration) (BHI-HM) under anaerobic conditions (10% CO2, 10% H2, and 80% N2) at 37˚C for 48 h. Streptococcus gordonii DL1, Streptococcus mutans MT8148, and Actinomyces naeslundii X600were cultured in BHI broth at 37˚C for 24 h.
2.2. Peptide Synthesis
The SspB peptide analog, in which the T (threonine) at position 400 has been replaced with K (lysine) in SspB (390-402), resulting in the SspB (390-T400K-402) peptide, DYQAKLAAYQKEL, and scavenger receptor cysteine-rich domain peptide 2, designated SRCRP2, QGRVEVLYRGSWGTVC, on salivary gp340 [20] were synthesized at 95% purity by Scrum, Inc. (Tokyo, Japan). Peptides were suspended in sterile distilled water (DW) at the desired concentration immediately before use.
2.3. Human Saliva Collection
As described previously [21] , saliva samples were collected from volunteers in good oral health, after stimulation with chewing paraffin gum. Volunteers had refrained from eating, drinking and brushing for at least 2 h prior to collection. Saliva was placed into ice-chilled sterile bottles for 5 min, followed by centrifugation at 10,000 × g for 10 min at 4˚C in order to remove cellular debris. Supernatant was filter sterilized through a 0.22-µm Acrodisc filter (Pall Corporation, Ann Arbor, MI) for peptide binding assay. After filtration, samples were pooled and stored at −20˚C until use.
2.4. ELISA
2.4.1. Peptide Binding Assay
Binding activity of SspB (390-T400K-402) peptide to saliva and to P. gingivalis were detected by enzyme- linked immunosorbent assay (ELISA). A previously reported method [21] was used, with some modifications. Ninety-six well H-plates (Sumitomo Bakelite, Tokyo, Japan) were coated for 1 h at 37˚C with 100 µl of whole bacterial cells (P. gingivalis strains 33277 and W83, S. gordonii DL1, S. mutans MT8148, and A. naeslundii X600) in Na2CO3 coating buffer at an optical density of 0.40 at 600 nm. After washing three times with PBS containing 0.1% Tween 20 (PBST), wells were blocked with 200 ml of 3% bovine serum albumin (BSA) in PBST at 4˚C overnight. Biotinylated SspB (390-T400K-402) peptide (12.5, 25, 50 and 100 µg/ml) in 100 µl of sterile DW was applied to the wells, followed by incubation at 37˚C for 1 h. Wells were then washed three times with PBST, and were further incubated for 1 h at 37˚C with 1:1000 streptoavidin-conjugated alkaline phosphatase (Invitrogen Corp., Carlsbad, CA). After development, absorbance at 405 nm was measured. For sandwich ELISA, we sandwiched the bacteria between biotinylated and non-biotinylated SspB (390-T400K-402) peptides. Briefly, the non-biotinylated SspB (390-T400K-402) peptide (12.5, 25, 50 and 100 µg/ml) was coated to the ELISA plate. After blocking, 100 µl of P. gingivalis 33277 cell suspension was applied to each well, followed by incubation at 37˚C for 1 h. After washing three times with PBST, 100 µl of biotinylated SspB (390-T400K-402) peptide (25 µg/ml) was applied, followed by further incubation at 37˚C for 1 h. Subsequent steps were as described above.
2.4.2. Responses of Salivary Immunoglobulins with P. gingivalis
In order to determine whether human saliva contains P. gingivalis-specific secretory IgA (sIgA) and/or IgG, we compared the ability of anti-IgA and IgG labeled antibodies to bind to P. gingivalis coated with saliva. Bacterial cells were deposited into the wells of ELISA plates. After blocking with 1% skim-milk and washing, 1:10 saliva diluted with sterile DW was added to the wells. Alkaline phosphatase-conjugated goat anti-human immunoglobulins (SIGMA) (1:1000 IgA or 1:4000 IgG) were reacted after the wells had been incubated with 1:10 saliva. Reactions were detected as mentioned above. All experiments were performed independently at least in triplicate.
2.4.3. Statistical Analyses
Data are expressed as means standard deviation. GraphPad Prism version 5.0 d for Mac OS X (GraphPad Software, San Diego, CA) was used to assess significance. The statistical significance of differences between two groups was determined by unpaired t-test. For comparisons between multiple groups, one-way analysis of variance (ANOVA) and Tukey’s test were used. P-values less than 0.01 or 0.05 were considered to be statistically significant using two-tailed comparisons. All experiments were repeated and analyzed independently.
3. Results
3.1. Binding Properties of SspB (390-T400K-402) Peptide
In order to establish a peptide binding assay using ELISA, we examined the binding properties of SspB (390-T400K-402) peptide at various concentrations (12.5, 25, 50 and 100 µg/ml) to P. gingivalis (Figure 1(a)) or to saliva (Figure 1(b)). The binding of SspB (390-T400K-402) peptide to P. gingivalis cells at 25 µg/ml was significantly higher than that at 12.5 µg/ml. At peptide concentrations of 50 or 100 µg/ml, the peptide binding properties to P. gingivalis were comparable to those at 25 µg/ml (Figure 1(a)). Thus, SspB at 25 µg/ml was used for further studies. A binding reaction to saliva was observed at 25 µg/ml, whereas no reaction was detected when the peptide was not applied (no peptide), thus suggesting that SspB (390-T400K-402) peptide has a binding capacity for saliva (Figure 1(b)).
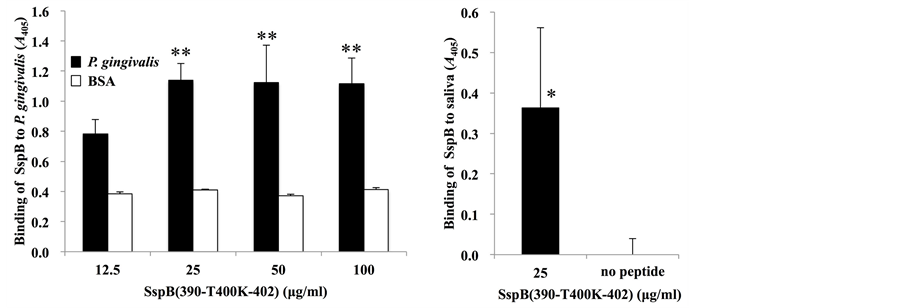
Figure 1. Binding of SspB (390-T400K-402) to P. gingivalis. (a) Binding response of biotinylated SspB (390-T400K-402) to P. gingivalis strain 33277 at various concentrations (12.5, 25, 50 and 100 µg/ml); (b) Binding response of biotinylated SspB (390-T400K-402) to saliva at a peptide concentration of 25 µg/ml. Binding is expressed as A405 values obtained from three independent experiments. Values are expressed as means ± standard deviation (SD) of triplicate assays. Asterisks denote significant differences (vs. 12.5 µg/ml, **P < 0.01; vs. no peptide, *P < 0.05).
3.2. Sandwich ELISA
As mentioned above, we utilized biotinylated SspB (390-T400K-402) peptide in the present methods. Therefore, to investigate whether biotinylation affects the binding activity of this peptide, sandwich ELISA with biotinylated and non-biotinylated SspB (390-T400K-402) peptide was performed (Figure 2). When the wells were pre-coated with non-biotinylated SspB peptide, binding activities of biotinylated SspB peptide to P. gingivalis cells increased in a dose-dependent manner, but decreased with a peptide concentration of 100 µg/ml (Figure 2(a)). On the other hand, when wells were pre-coated with biotin, binding activity of biotinylated SspB peptide to P. gingivalis cells remained unchanged (Figure 2(b)). When samples were sandwiched between pre-coated solution [DW, biotin or non-biotinylated SspB (390-T400K-402)] and biotinylated SspB (390-T400K-402), the strongest reactions were observed in P. gingivalis 33277 sandwiched between non-biotinylated and biotinylated SspB peptides (Figure 2(c)). In contrast, low reaction levels were observed in S. gordonii DL1 and BSA (Figure 2(c)).
3.3. SspB (390-T400K-402) Peptide Specificity to Mfa1 of P. gingivalis
We examined the specificity of the SspB (390-T400K-402) peptide for P. gingivalis Mfa1 fimbriae (Figure 3). P. gingivalis strains 33277 and W83, and oral commensals S. gordonii DL1, S. mutans MT8148 and A. naeslundii X600 were tested. P. gingivalis 33277 showed the highest peptide binding activity of the tested bacteria (Figure 3(a)), whereas P. gingivalis W83, which is deficient in Mfa1 fimbriae, exhibited poor binding activity, as did S. gordonii DL1 and BSA (Figure 3(b)).
3.4. Salivary Components Affect the Binding of SspB (390-T400K-402) Peptide to P. gingivalis
To determine whether the saliva samples used in this study contain P. gingivalis-specific sIgA and/or IgG, the binding of anti-IgA and anti-IgG antibodies to saliva-coated bacteria was compared (Figure 4(a)). The results showed that salivary immunoglobulins bound significantly better to S. gordonii when compared with P. gingivalis strains 33277 and W83. The binding of salivary immunoglobulins to the P. gingivalis strains was comparable to BSA, which was used to demonstrate the background level of non-specific antibodies.
Hamada et al. [13] previously demonstrated that SspB (390-T400K-402) peptide had the highest binding activity to salivary components and to the salivary gp340 peptide SRCRP2. Thus, we hypothesized that when P. gingivalis was incubated with whole saliva or with SRCRP2, the binding activity of the SspB (390-T400K-402) peptide would increase. To assess this hypothesis, P. gingivalis cells incubated with saliva were studied (Figure 4(b)). When P. gingivalis was incubated with 100 µl of whole saliva, the binding activity of SspB (390-T400K-402)
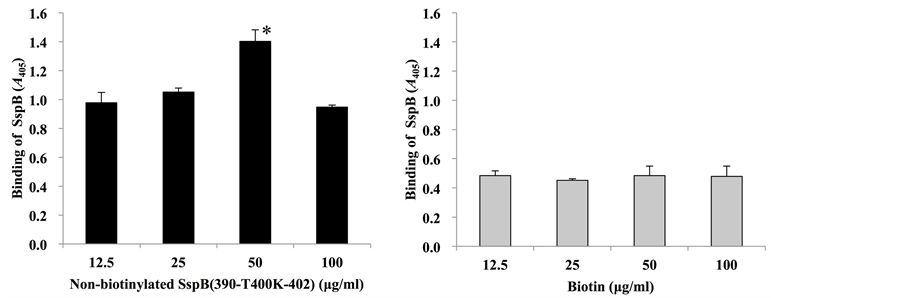
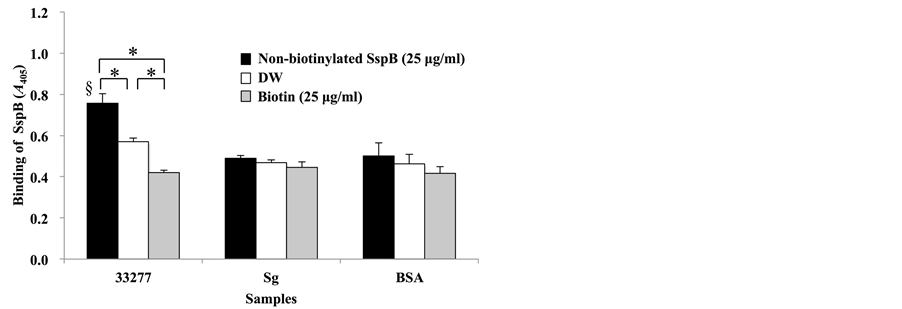
Figure 2. Sandwich assay with biotinylated and non-biotinylated SspB (390-T400K-402). (a) Microtiterplates were coated with non-biotinylated SspB (390-T400K-402) (12.5, 25, 50 and 100 µg/ml); (b) biotin (12.5, 25, 50 and 100 µg/ml), (c) DW, 25 µg/ml biotin, or 25 µg/ml of non-biotinylated SspB (390-T400K-402). P. gingivalis strain 33277 (33277), S. gordonii (Sg) or BSA were added to the wells of coated plates, and then 25 µg/ml biotinylated SspB (390-T400K-402) was added. Data are expressed as A405 values obtained from three independent experiments. Values are expressed as means ± SD of triplicate assays (*P < 0.01, § represents significant differences vs. Sg, vs. BSA; P < 0.01).
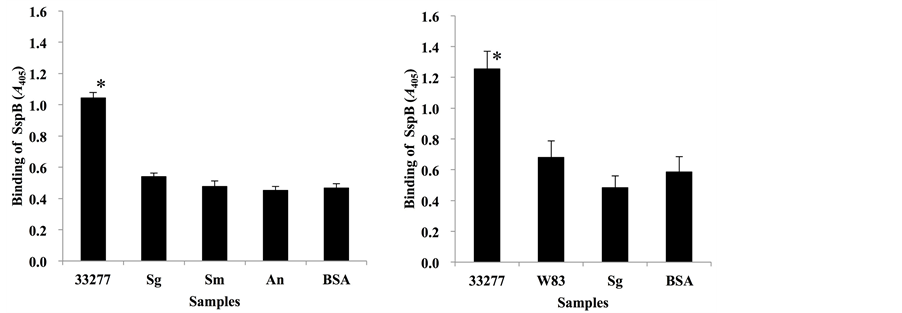
Figure 3. Binding of SspB (390-T400K-402) to P. gingivalis strain 33277. (a) Binding response of biotinylated SspB (390-T400K-402) peptide (25 µg/ml) to P. gingivalis strain 33277 (33277), S. gordonii DL1 (Sg), S. mutans MT8148 (Sm) and A. naeslundii X600 (An); (b) Comparison of SspB (390-T400K-402) peptide (25 µg/ml) binding to P. gingivalis strains between 33277 and P. gingivalis W83 (W83). Data are expressed as A405 values obtained from three independent experiments. Values are expressed as means ± SD of triplicate assays (*P < 0.01).
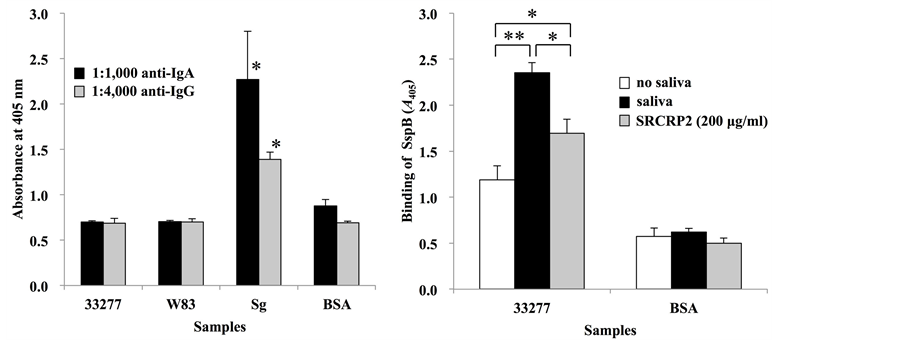
Figure 4. Analysis of salivary effects on SspB (390-T400K-402) binding to P. gingivalis. (a) Response of salivary immunoglobulins to bacterial samples with anti-human IgA or IgG (vs. 33277, vs. W83, vs. BSA; *P < 0.01); (b) Effects of SRCRP2 (200 µg/ml) on SspB (390-T400K-402) binding (*P < 0.05, **P < 0.01). Data are expressed as A405 values obtained from three independent experiments. Values are expressed as means ± SD of triplicate assays.
peptide increased markedly when compared to the cells without saliva incubation. When cells were incubated with 100 µl of SRCRP2 peptide (200 µg/ml), the binding activity of SspB peptide was significantly higher than in non-treated cells; however, it was lower than in saliva-treated cells (Figure 4(b)).
4. Discussion
At present, few assays are available to assess the relationships among streptococcal adhesin SspB, P. gingivalis minor fimbrial antigen Mfa1 and salivary agglutinin gp340. In the present study, we evaluated the binding activity of the analogous peptide SspB (390-T400K-402) to P. gingivalis in the presence of saliva or salivary gp340 peptide SRCRP2 in order to establish an assay for investigating the three-way interaction among S. gordonii, P. gingivalis and salivary gp340 as a unit, and to examine whether P. gingivalis binds to both salivary gp340 and SRCRP2.
SspB (390-T400K-402) peptide has the highest response for binding to salivary components and to SRCRP2 when compared with other SspB and streptococcal adhesin-derived peptides. Consistent with previous reports [13] [14] , our study found that SspB (390-T400K-402) showed binding activity with saliva (Figure 1(b)). Furthermore, the peptide also showed binding activity with P. gingivalis 33277 (Figure 1(a), Figure 3(a)).The present results therefore suggest that there are critical residues for binding to P. gingivalis within the synthetic peptide SspB (390-T400K-402) adhesion epitope. We believe that the SspB (390-T400K-402) is markedly superior to other SspB-derived peptides with regard to binding characteristics.
A sequence of the C-terminal region of SspB, designated BAR (amino acid residues 1167-1193), mediates binding to P. gingivalis [22] . Here we demonstrate that streptococcal peptide analog SspB (390-T400K-402) is also capable of binding to P. gingivalis 33277, although the peptide is derived from the N-terminal A region (amino acid residues 390-402). SspB (390-T400K-402) is produced by a substitution of threonine for lysine at 400 of SspB (390-402) and consequently, the adhesion epitope of the peptide to P. gingivalis may be conferred by conformation changes.
P. gingivalis 33277 has two distinct fimbriae molecules, major fimbriae FimA encoded by fimA [23] and minor fimbriae Mfa1 encoded by mfa1 [24] . The Mfa1 of P. gingivalis mediates adherence to the SspB of S. gordonii, so that the Mfa1-SspB interaction is necessary for biofilm development [7] [25] . Strain W83 expresses the type IV major fimbriae [23] , but the minor fimbriae Mfa1 has not been observed [26] . These reports, together with our observations that P. gingivalis 33277 showed the highest peptide binding activity of the bacterial samples (Figure 3(a)), whereas P. gingivalis W83 deficient in Mfa1 fimbriae exhibited poor peptide binding (Figure 3(b)), suggest that the analogous peptide SspB (390-T400K-402) binds to Mfa1. However, some limitations are worth noting. We used P. gingivalis W83 as an Mfa1-deficient strain in the present study. This assay needs to be tested further by using mfa1-knockout mutants derived from strain 33277 in order to compare binding reactions in a 33277 genetic background.
In this study, we used biotinylated SspB (390-T400K-402) peptide, which was synthesized with a single N-terminal biotinylation. Indeed, biotinylated SspB (390-T400K-402) peptide had a high binding activity against whole salivary components on Western blotting [13] . Thus, in the present study, we assessed whether biotinylation in the N-terminal of SspB (390-T400K-402) enhances binding activity to P. gingivalis (Figure 2). In Sandwich ELISA, pre-coating with biotin did not affect SspB binding to P. gingivalis (Figure 2(b)). In contrast, the SspB binding levels varied and peaked at a concentration of 50 µg/ml non-biotinylated SspB (Figure 2(a)), suggesting that the adhesion epitope of Mfa1 was occupied by the pre-coated non-biotinylated peptide. Moreover, the strongest reaction was observed in P. gingivalis 33277 sandwiched between non-biotinylated and biotinylated SspB peptides (Figure 2(c)). Overall, these results suggest that biotinylation in the N-terminal of SspB (390-T400K-402) has no promotional effect on peptide binding activity.
Salivary molecules, proline-rich protein 1 and statherin, have been reported to promote the adherence of P. gingivalis to saliva-coated oral surfaces through specific interactions [17] - [19] . In fact, saliva appears to have a significant impact on SspB binding to P. gingivalis. The increase in SspB peptide binding to saliva-incubated P. gingivalis (Figure 4(b)) suggests a saliva-P. gingivalis interaction. Moreover, the increase in SspB peptide binding to SRCRP2-incubated P. gingivalis (Figure 4(b)) indicates that P. gingivalis interacts with salivary gp340. SIgA, a salivary agglutinin, is the predominant immunoglobulin found in all mucosal secretions, including saliva. Salivary sIgA may promote colonization of certain strains of bacteria [27] . We previously demonstrated that salivary sIgA promoted initial attachment of Streptococcus mutans on the mouse tooth surface [21] . In contrast, decreased salivary sIgA negatively modulated Candida albicans populations in the oral cavity of mice [28] . In addition, IgG has been shown to be the major antibody class that mediates host immune response against P. gingialis [29] . In the present study, we confirmed that the saliva did not contain either sIgA or IgG specific to P. gingivalis (Figure 4(a)). These findings provide evidence that the SspB peptide-P. gingivalis interaction via saliva was not induced by salivary antibodies, but rather by salivary gp340.
Taken together, a peptide binding assay using SspB (390-T400K-402) presented herein may provide important insights into the mechanisms of supra- and sub-gingival oral biofilm formation.
5. Conclusion
A novel assay using the analogous SspB (390-T400K-402) peptide established in this research is available to assess the relationships among SspB, Mfa1 and salivary gp340 as a unit. The assay also reveals that P. gingivalis binds to salivary gp340 and to SRCRP2.
Acknowledgements
We would like to thank all volunteers for their cooperation in this study. This work was supported by a Grant-in-Aid for Young Scientists (B) 25862034 and a Grant-in-Aid for Exploratory Research 19659559 from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Cite this paper
TatsuroIto,HidenobuSenpuku,TakahiroIchinosawa,NanaIkematsu-Ito,NaoKimura,TakehikoShimizu, (2015) SspB Peptide Assay Reveals Saliva-Mediated Porphyromonas gingivalis Attachment. Open Journal of Stomatology,05,259-267. doi: 10.4236/ojst.2015.511032
References
- 1. Scannapieco, F.A. (1994) Saliva-Bacterium Interactions in Oral Microbial Ecology. Critical Reviews in Oral Biology & Medicine, 5, 203-248.
- 2. Kolenbrander, P.E. and London, J. (1993) Adhere Today, Here Tomorrow: Oral Bacterial Adherence. Journal of Bacteriology, 175, 3247-3252.
- 3. Marsh, P.D. (1994) Microbial Ecology of Dental Plaque and Its Significance in Health and Disease. Advances in Dental Research, 8, 263-271.
- 4. Bikker, F.J., Ligtenberg, A.J., Nazmi, K., Veerman, E.C., van’t Hof, W., Bolscher, J.G., Poustka, A., Amerongen, A.V.N. and Mollenhauer, J. (2002) Identification of the Bacteria-Binding Peptide Domain on Salivary Agglutinin (gp-340/DMBT1), a Member of the Scavenger Receptor Cysteine-Rich Superfamily. Journal of Biological Chemistry, 277, 32109-32115.
http://dx.doi.org/10.1074/jbc.M203788200 - 5. Gibbons, R.J. (1996) Role of Adhesion in Microbial Colonization of Host Tissues: A Contribution of Oral Microbiology. Journal of Dental Research, 75, 866-870.
http://dx.doi.org/10.1177/00220345960750030201 - 6. Brady, L.J., Maddocks, S.E., Larson, M.R., Forsgren, N., Persson, K., Deivanayagam, C.C. and Jenkinson, H.F. (2010) The Changing Faces of Streptococcus Antigen I/II Polypeptide Family Adhesins. Molecular Microbiology, 77, 276- 286.
http://dx.doi.org/10.1111/j.1365-2958.2010.07212.x - 7. Park, Y., Simionato, M.R., Sekiya, K., Murakami, Y., James, D., Chen, W., Hackett, M., Yoshimura, F., Demuth, D.R. and Lamont, R.J. (2005) Short Fimbriae of Porphyromonas gingivalis and Their Role in Coadhesion with Streptococcus gordonii. Infection and Immunity, 73, 3983-3989.
http://dx.doi.org/10.1128/IAI.73.7.3983-3989.2005 - 8. Rosan, B. and Lamont, R.J. (2000) Dental Plaque Formation. Microbes and Infection, 2, 1599-1607.
http://dx.doi.org/10.1016/S1286-4579(00)01316-2 - 9. Chung, W.O., Demuth, D.R. and Lamont, R.J. (2000) Identification of a Porphyromonas gingivalis Receptor for the Streptococcus gordonii SspB Protein. Infection and Immunity, 68, 6758-6762.
http://dx.doi.org/10.1128/IAI.68.12.6758-6762.2000 - 10. Brooks, W., Demuth, D.R., Gil, S. and Lamont, R.J. (1997) Identification of a Streptococcus gordonii SspB Domain That Mediates Adhesion to Porphyromonas gingivalis. Infection and Immunnity, 65, 3753-3758.
- 11. Daep, C.A., James, D.M., Lamont, R.J. and Demuth, D.R. (2006) Structural Characterization of Peptide-Mediated Inhibition of Porphyromonas gingivalis Biofilm Formation. Infection and Immunity, 76, 5756-5762.
http://dx.doi.org/10.1128/IAI.00813-06 - 12. Daep, C.A., Novak, E.A., Lamont, R.J. and Demuth, D.R. (2011) Structural Dissection and in Vivo Effectiveness of a Peptide Inhibitor of Porphyromonas gingivalis Adherence to Streptococcus gordonii. Infection and Immunity, 79, 67- 74.
http://dx.doi.org/10.1128/IAI.00361-10 - 13. Hamada, T., Kawashima, M., Watanabe, H., Tagami, J. and Senpuku, H. (2004) Molecular Interactions of Surface Protein Peptides of Streptococcus gordonii with Human Salivary Components. Infection and Immunity, 72, 4819-4826.
http://dx.doi.org/10.1128/IAI.72.8.4819-4826.2004 - 14. Okuda, K., Hanada, N., Usui, Y., Takeuchi, H., Koba, H., Nakao, R., Watanabe, H. and Senpuku, H. (2010) Inhibition of Streptococcus mutans Ad-herence and Biofilm Formation Using Analogues of the SspB Peptide. Archives of Oral Biology, 55, 754-762.
http://dx.doi.org/10.1016/j.archoralbio.2010.06.014 - 15. Koba, H., Okuda, K., Watanabe, H., Tagami, J. and Senpuku, H. (2009) Role of Lysine in Interaction between Surface Protein Peptides of Streptococcus gordonii and Agglutinin Peptide. Oral Microbiology and Immunology, 24,162-169.
http://dx.doi.org/10.1111/j.1399-302X.2008.00490.x - 16. Loimaranta, V., Jakubovics, N.S., Hytonen, J., Finne, J., Jenkinson, H.F. and Stromberg, N. (2005) Fluid- or Surface- Phase Human Salivary Scavenger Protein gp340 Exposes Different Bacterial Recognition Properties. Infection and Immunity, 73, 2245-2252.
http://dx.doi.org/10.1128/IAI.73.4.2245-2252.2005 - 17. Amano, A., Sojar, H.T., Lee, J.Y., Sharma, A., Levine, M.J. and Genco, R.J. (1994) Salivary Receptors for Recombinant Fimbrillin of Porphyromonas gingivalis. Infection and Immunity, 62, 3372-3380.
- 18. Amano, A., Sharma, A., Lee, J.Y., Sojar, H.T., Raj, P.A. and Genco, R.J. (1996) Structural Domains of Porphyromonas gingivalis Recombinant Fimbrillin That Mediate Binding to Salivary Proline-Rich Protein and Statherin. Infection and Immunity, 64, 1631-1637.
- 19. Lamont, R.J. and Jenkinson, H.F. (2000) Subgingival Colonization by Porphy-romonas gingivalis. Oral Microbiology and Immunology, 15, 341-349.
http://dx.doi.org/10.1034/j.1399-302x.2000.150601.x - 20. Oho, T., Bikker, F.J., Nieuw Amerongen, A.V. and Groe-nink, J. (2004) A Peptide Domain of Bovine Milk Lactoferrin Inhibits the Interaction between Streptococcal Surface Protein Antigen and a Salivary Agglutinin Peptide Domain. Infection and Immunity, 72, 6181-6184.
http://dx.doi.org/10.1128/IAI.72.10.6181-6184.2004 - 21. Ito, T., Maeda, T. and Senpuku, H. (2012) Roles of Salivary Components in Streptococcus mutans Colonization in a New Animal Model Using NOD/SCID. e2f1-/- Mice. PLoS ONE, 7, e32063.
http://dx.doi.org/10.1371/journal.pone.0032063 - 22. Demuth, D.R., Irvine, D.C., Costerton, J.W., Cook, G.S. and Lamont, R.J. (2001) Discrete Protein Determinant Directs the Species-Specific Adherence of Porphyromonas gingivalis to Oral Streptococci. Infection and Immunity, 69, 5736-5741.
http://dx.doi.org/10.1128/IAI.69.9.5736-5741.2001 - 23. Amano, A., Nakagawa, I., Okahashi, N. and Hamada, N. (2004) Variations of Porphyromonas gingivalis Fimbriae in Relation to Microbial Pathogenesis. Journal of Periodontal Research, 39, 136-142.
http://dx.doi.org/10.1111/j.1600-0765.2004.00719.x - 24. Hamada, N., Sojar, H.T., Cho, M.I. and Genco, R.J. (1996) Isolation and Characterization of a Minor Fimbria from Porphyromonas gingivalis. Infection and Immunity, 64, 4788-4794.
- 25. Lamont, R.J., El-Sabaeny, A., Park, Y., Cook, G.S. and Costerton, J.W. (2002) Role of the Streptococcus gordonii SspB Protein in the Development of Porphyromonas gingivalis Biofilms on Streptococcal Substrates. Microbiology, 148, 1627-1636.
http://dx.doi.org/10.1099/00221287-148-6-1627 - 26. Hasegawa, Y., Iwami, J., Sato, K., Park, Y., Nishikawa, K., Atsumi, T., Moriguchi, K., Murakami, Y., Lamont, R.J., Nakamura, H., Ohno, N. and Yoshimura, F. (2009) Anchoring and Length Regulation of Porphyromonas gingivalis Mfa1 Fimbriae by the Downstream Gene Product Mfa2. Microbiology, 155, 3333-3347.
http://dx.doi.org/10.1099/mic.0.028928-0 - 27. Lamont, R.J., Demuth, D.R., Davis, C.A., Malamud, D. and Rosan, B. (1991) Salivary-Agglutinin-Mediated Adherence of Streptococcus mutans to Early Plaque Bacteria. Infection and Immunity, 59, 3446-3450.
- 28. Kanaguchi, N., Narisawa, N., Ito, T., Kinoshita, Y., Kusumoto, Y., Shinozuka, O. and Senpuku, H. (2012) Effects of Salivary Protein Flow and Indigenous Microorganisms on Initial Colonization of Candida albicans in an in Vivo Model. BMC Oral Health, 12, 36.
http://dx.doi.org/10.1186/1472-6831-12-36 - 29. Donley, C.L., Badovinac, R., Sapir, S., Shapira, L., Houri, Y., Kantarci, A., Warbington, M.L., Dibart, S., Van Dyke, T.E., Needleman, H.L., Karimbux, N. and Bimstein, E. (2004) IgG Antibody Levels to Porphyromonas gingivalis and Clinical Measures in Children. Journal of Periodontology, 75, 221-228.
http://dx.doi.org/10.1902/jop.2004.75.2.221
NOTES
*Corresponding author.


