Open Journal of Genetics
Vol.4 No.4(2014), Article
ID:47915,8
pages
DOI:10.4236/ojgen.2014.44027
An Autosomal STR Profile of Napoléon the First
Gérard Lucotte*, Alexandra Bouin Wilkinson
Institute of Molecular Anthropology, Paris, France
Email: *Lucotte@hotmail.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 12 May 2014; revised 10 June 2014; accepted 9 July 2014
ABSTRACT
Objective: We report the results of nuclear DNA analyses of Napoléon the First (Napoléon Bonaparte; 1769-1821). Design: His genomic DNA was extracted from dandruff adherent to his hair, coming from a lock of his hair dating from the year of 1811. Results: We obtained the complete STR (short tandem repeats) profile of Napoléon, based on fifteen autosomal loci. On this profile, ten loci (D8S1179, D21S11, D7S820, D3S1358, TH01, D16S539, D2S1338, vWa, D18S51 and FGA) are heterozygous; the most frequent alleles in Caucasians are present for only seven (allele 8 for TPOX and allele 11 for D5S818, allele 13 for D8S1179, allele 10 for D7S820, allele 9.3 for THO1, allele 12 for D16S539 and allele 24 for FGA) of the homozygous and heterozygous loci. Conclusions: So the discriminating power of this sort of genetic profile is elevated, permitting useful comparisons to other STR profiles in the future. Finally, an analysis of fifteen Y chromosomal STRs from the dandruff of this lock of hair confirms allele values of Napoléon already obtained or deduced for the corresponding loci in previous determinations.
Keywords:Napoléon the First, Hair Dandruff, Genomic DNA, Autosomal STR Profile

1. Introduction
Genetics added recently a new dimension to genealogical research; this discipline is called “Genetic genealogy”. Genetic genealogy is useful to verify maternal and paternal lineages, using haploid markers from mitochondrial DNA (mtDNA) and Y-chromosome. The usefulness of genetic genealogy was initially demonstrated by the pioneering works on the Romanov family [1] [2] and on the third US President Thomas Jefferson [3] [4] .
The enormous success of genetic genealogy today is mainly due to that it concerns personages of historical interest and, above all, royal families [5] . Concerning especially royal dynasties in France, several studies— some of which being highly controversial between them have been published successively: on Louis XVII, the second son of Louis XVI and Marie-Antoinette [6] [7] , on the King Louis XVI himself [8] , on the King Henri IV—the first reigning Bourbon in France—grand-father of the King Louis XIV [9] , and on three living males of the House of Bourbon [10] .
As part of the Napoléon I Genome (NIG) project, we have recently published some genetic genealogy data on the French Emperor Napoléon the First (Napoléon Bonaparte; 1769-1821) and his family. One of us described [11] the finding of a rare variant (16184C > T) in the sequence of the first hypervariable segment (HVS1) of mtDNA extracted from two preserved hairs conserved in the Vivant Denon reliquary; this mutation in the HVS1 sequence was also found in the mtDNA of Napoléon’s mother (Letizia). After that, our groups have described a first reconstruction of the Napoléon’s Y-chromosomal haplotype [12] , based on the genomic DNA of his indirect descendant Charles Napoléon (C.N.), the living 4th generation descendant of Jérôme Napoléon (Napoléon’s youngest brother); the Y-haplogroup of Napoléon, determined by the study of ten NRY-SNPs (non-recombinant Y-single nucleotide polymorphisms) is the E1b1b1c1* paragroup. Our third publication on the subject [13] concerns the complete reconstruction of the Y-haplotype of Napoléon, based on more than one hundred Y-STRs (Y-short tandem repeats) of C.N. and of Alexandre Colonna Walewski (A.C.W.), the living 4th generation descendant of Count Alexandre Walewski (the son born of the union between Napoléon I and Countess Maria Walewska).
Autosomal loci were rarely used to genotype of ancient famous men and Kings [14] [15] . The aim of the present study is to determine a complete autosomal STR profile of Napoléon I, based on genomic DNA extracted from his hair dandruff.
2. The Samples
Napoléon I’s lock of hair I is kept at present in a golden frame (Figure 1), under a looking glass. The lock of hair originates from collections of the Imperial Family (last proprietor known: Princess Eugénie). Napoléon’s lock of hairdates from 1811; at this date the Emperor lived in Paris [16] : he married Marie-Louise in 1810, and their son was born during this year (on March 20, 1811).
We could observe plenty dandruff on this lock of hair (Figure 2) when the lock was taken out of the frame. So such hair, that did not seem to have required any special procedure of treatment or conservation, corresponds to a hairdresser’s lock. We have taken for analysis, with sterilized tweezers, about thirty isolated (detached from his hair) dandruffs and five bigger dandruffs coupled with hair; these dandruffs constitute the samples analyzed in this study.
Figure 3 shows an example of such dandruff, intricated between hairs, observed in optical microscopy. This same dandruff, observed in scanning electronic microscopy (SEM), is shown in Figure 4(a). The dandruff and
Figure 1. The golden frame containing Napoléon I’s hair lock.

Figure 2. Numerous dandruffs (D) are observed at the lock of hair’s periphery, when sorted out of the frame.
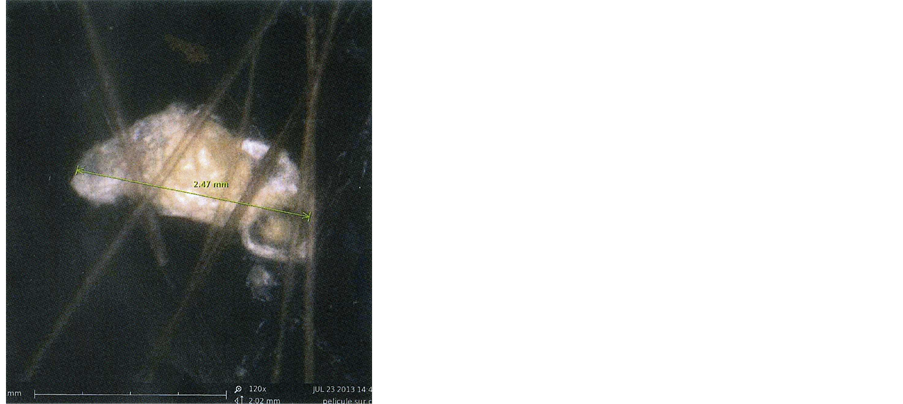
Figure 3. An example of a dandruff and its associated hairs, observed (×120) through optical microscopy (SEM Fondis Bioritech Phenom G2 Pure).
associated hair, studied by X-ray microfluorescence (X-RMF) show a similar elementary composition (Figure 4(b)): the main difference between the two concerns sulfur (S) peak, which is more elevated in value for the hair than for the dandruff.
3. Methods
All staff involved in the sampling wore protective clothing, sterile gloves and facemasks, to prevent exogenous contamination. DNA extraction and purification were performed according to our previously published protocol [11] , in a dedicated laboratory.
Fifteen polymorphic microsatellites of the nuclear genome (chosen because they represent the usual panel used in legal investigations) were amplified (Identifiler-plus and AmpF/STR Minifiler kits, Applied Biosystems), according to the instructions given by the Company. We amplified also 15 Y-chromosomal short tandem repeats (AmpF/STR Y Filer PCR amplification kit, Applied Biosystems). The Y-chromosomal haplogroup of Napoléon I was deduced from the STR allele values by using the Whit Attey’s Haplogroup Predictor [17] .
The laboratory performed DNA typing under strict precautions, following previously published criteria for ancient DNA authentication [18] .
4. Results
In a three-replicate assay concerning various dandruffs, all the 15 STR loci of the Identifiler and Minifiler kits
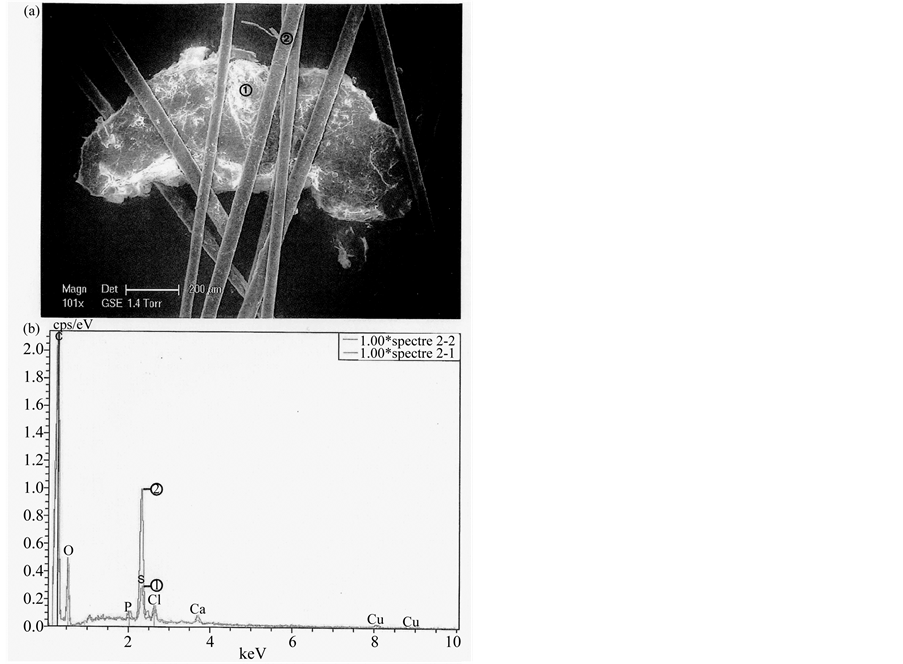
Figure 4. The same dandruff (a) observed (×101) through electronic microscopy (Philips XL30 model, environmental version). Above (b), comparisons between dandruff (in red) and hairs’ (in blue) spectras (Bruker probe AXS, energy dispersive X-ray; PGT-Spirit Model, Princeton gamma technology-system of analysis).
gave genotyping results (Table 1 and Figure 5): D8S1179-13,14 ; D21S11-27,29; D7S820-9,10; CSF1PO-11; D3S1358-14,16; THO1-8,9.3; D13S317-13; D16S539-11,12; D2S1338-17,23; D19S433-13; vWA-16,18; TPOX-8; D18S51-11,16; D5S818-11; FGA-23,24. PCR products were also obtained for the amelogenin sex marker (Figure 5), showing that the gender of the samples corresponds to a male (XY).
The 15 loci of the Y Filer amplification kit gave the following allele values (Table 2): DYS19-13; DYS385.a, b-16; DYS389.I-14 and DYS389.II-31; DYS390-24; DYS391-10; DYS392-11; DYS393-14; DYS438-10; DYS439-12; DYS448-20; DYS456-15; DYS458-16; DYS635-23; GATA-H4-11. These allele values are identical to those previously known for Napoléon, but the allele value = 23 for DYS635.
Using the Whit Athey’s Haplogroup Predictor [17] determined that, with these fifteen allele values, the corresponding Y chromosomal haplogroup is (with 100% of probability) E1b1b.
5. Discussion
Using peculiar kits permitting amplification of microsatellite markers from ancient (degraded) genomic DNA, we were able to obtain from his dandruff a complete genetic profile of Napoléon I consisting of fifteen variable STR autosomal loci.
Such a genetic profile (when obtained) is rarely complete for ancient DNA samples [14] [15] , because the shortest amplicons can only be usually obtained in this sort of degraded material. This complete STR profile of Napoléon, completing because it is based on the total of the 15 variable autosomal STRs tested, permits discrimination performances comparable to those used today for living persons in forensic medicine cases. In France for example [19] , such a battery of these fifteen autosomal STRs is routinely employed in legal investigations.
Table 1. Autosomal microsatellite data analysis.
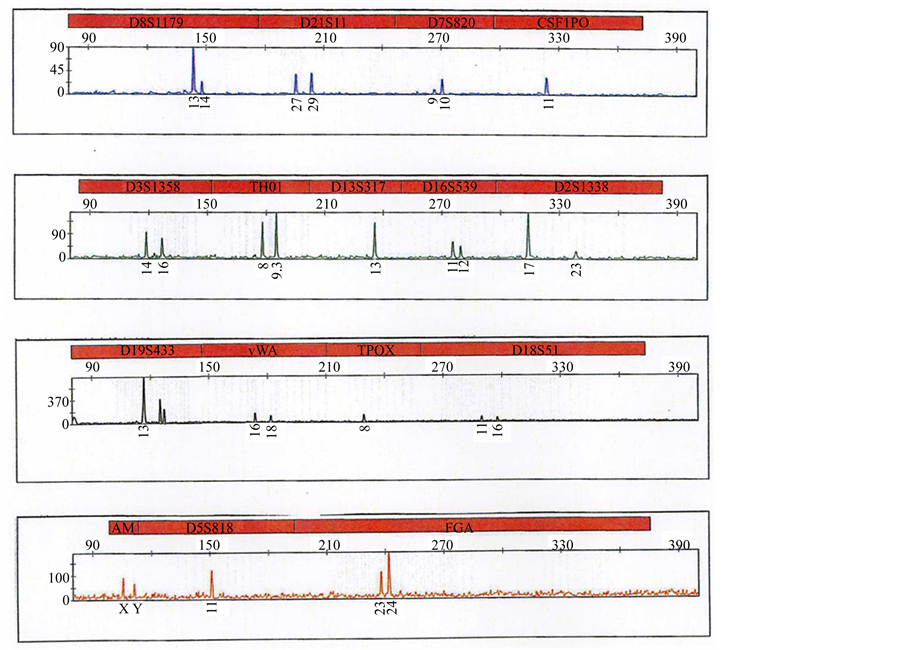
Figure 5. The 15-STRs autosomal loci profile of Napoléon I.
Table 2. Y-chromosomal microsatellite data analysis.
N.: Napoléon; C.N.: Charles Napoléon; A.C.W.: Alexandre Colonna Walewski. Asterisk indicates DYS635, with variable allele values between individuals.
The main characteristics of Napoléon’s autosomal STRs profile (Table 1) are as follows: 1) five loci only (CSF1PO, D13S317, D19S433, TPOX and D5S818) are homozygous, the other ten (D8S1179, D21S11, D7S- 820, D3S1358, TH01, D16S539, D2S1338, vWA, D18S51 and FGA) being heterozygous; 2) the most common allele in Caucasians is present for two (allele 8 for TPOX and allele 11 for D5S818) of the homozygous loci, and for five (allele 13 for D8S1179, allele 10 for D7S820, allele 9.3 for THO1, allele 12 for D16S539 and allele 24 for FGA) of the heterozygous loci; 3) the most discriminant heterozygous loci (with allele values differing for more than one step mutation compared to the most frequent one) are D21S11 (allele 27), D2S1338 (alleles 17 and 23) and D18S51 (alleles 11 and 16).
Knowledge of the autosomal STR profile of Napoléon should permit a very precise genetic identification of his remains (bones, teeth, hair bulbs, tendons, skin…). The knowledge of this profile will also allow us to compare it to those (to be established) of his four brothers: Joseph (1768-1844), Lucien (1775-1840), Louis (1778- 1846) and Jérôme (1784-1860). For the moment, we have already obtained, from Letizia’s hair [11] (1750- 1836), Napoléon’s mother, her genomic DNA; a comparison between her STR profile with that of Napoléon can allow deducing, at the corresponding autosomal heterozygous loci, alleles of the paternal line (and, consequently, some of those of Napoléon’s real father).
Results reported here on Y-STRs of Napoléon (Table 2) confirm the allele value = 13 for DYS19 previously obtained [12] by direct detection in his genome. These results establish that the allele values are 16 for DYS385, and that DYS389.I = 14, DYS389.II = 31, DYS390 = 24, DYS391 = 10, DYS392 = 11, DYS393 = 14, DYS438 = 10, DYS439 = 12, DYS448 = 20, DYS456 = 15, DYS458 = 16 and GATA-H4 = 11, as deduced from our study [13] comparing Y-STR profiles of Charles Napoléon (C.N.) and Alexandre Colonna Walewski (A.C.W.). For GATA-C4, the present study establishes that the real allele value is 23, and not = 21 as previously supposed [13] . So for this genetic marker, the allele value of Napoléon corresponds to that of the modal class in the distribution of alleles for Caucasians [20] ; we have previously shown [13] that GATA-C4 allele values = 22 for C.N. and = 21 for A.C.W.
The Y chromosomal haplogroup deduced from STR allele values (Table 2) using The Whit Athey’s Haplogroup Predictor is E1b1b. Initial research on this subject based on SNPs [12] established that the Y haplogroup of Napoléon is E1b1b1c1 and further studies [13] showed that he is L791 and L792+. Using M34 (the ultimate SNP of the terminal differentiation of this haplogroup), we studied frequencies of this genetic marker for various populations in Europe, in Northern Africa and in the Near East, in order to determine more precisely the ultimategeographic origins of the House of Bonaparte [21] .
It is the first time to our knowledge that hairdandruff is used for DNA extraction in humans. The reason why we chose dandruff as the study material is that there are numerous preserved hairs of Napoléon in museums and private collections; some of them, non-specially prepared, are rich in dandruff. So, as demonstrated here, hair dandruff can be taken as bones, teeth, skin and other biological materials—as a useful source to obtain genomic DNA in ancient remains.
Acknowledgements
We thank Pierre d’Harville (Belgium) for reference material; Jacques Macé for his help in taking the samples and for photography; Khalid Rbii (Fondis Bioritech, Guyancourt) for dandruff photography with portable SEM; Thierry Thomasset (UST of Compiègne) for SEM utilization and X-RMF analysis; Dr Soisic Le Guiner-Lebeau (IGNA of Nantes), who initiated analysis of the autosomal STR profile; Peter Hrechdakian for using the Whit Athey’s Haplogroup Predictor program.
Funding
The present work was realized with the continuous financial help of the “Souvenir Napoléonien” Foundation (Paris).
Conflict of Interest
The authors declare no conflict of interest.
References
- Gill, P., Ivanov, P.L., Kimpton, C., Piercy, R., Benson, N., Tully, G., et al. (1994) Identification of the Remains of the Romanov Family by DNA Analysis. Nature Genetics, 6, 130-135. http://dx.doi.org/10.1038/ng0294-130
- Rogaev, E.I., Grigorenko, A.P., Moliaka, Y.K., Faskhutdinova, G., Goltsov, A., Lahti, A., et al. (2009) Genomic Identification in the Historical Case of the Nicholas II Royal Family. Proceedings of the National Academy of Sciences of the United States of America, 106, 5258-5263. http://dx.doi.org/10.1073/pnas.0811190106
- Foster, E.A., Jobling, M.A., Taylor, P.G., Donnelly, P., de Knijff, P., Mieremet, R., et al. (1998) Jefferson Fathered Slave’s Last Child. Nature, 396, 27-28. http://dx.doi.org/10.1038/23835
- King, T.E., Bowden, G.R., Balaresque, P.L., Adams, S.M., Shanks, M.E. and Jobling, M.A. (2007) Thomas Jefferson’s Y Chromosome Belongs to a Rare European Lineage. American Journal of Physical Anthropology, 132, 584-589. http://dx.doi.org/10.1002/ajpa.20557
- (2013) List of Haplogroups of Notable People (from Wikipedia).
- Jehaes, E., Decorte, R., Peneau, A., Petrie, J.H., Boiry, P.A., Gilissen, A., et al. (1998) Mitochondrial DNA Analysis on Remains of a Putative Son of Louis XVI, King of France and Marie-Antoinette. European Journal of Human Genetics, 6, 383-395. http://dx.doi.org/10.1038/sj.ejhg.5200227
- Jehaes E., Pfeiffer, H., Toprak, K., Decorte, R., Brinkmann, B. and Cassiman, J.J. (2001) Mitochondrial DNA Analysis of the Putative Heart of Louis XVII, Son of Louis XVI and Marie-Antoinette. European Journal of Human Genetics, 9, 185-190. http://dx.doi.org/10.1038/sj.ejhg.5200602
- Lalueza-Fox, C., Gigli, E., Bini, C., Calafell, F., Luiselli, D., Pelotti, S., et al. (2011) Genetic Analysis of the Presumptive Blood from Louis XVI, King of France. Forensic Science International: Genetics, 5, 459-463. http://dx.doi.org/10.1016/j.fsigen.2010.09.007
- Charlier, P., Olalde, I., Solé, N., Ramírez, O., Babelon, J.P., Galland, B., et al. (2013) Genetic Comparison of the Head of Henri IV and the Presumptive Blood of Louis XVI (Both Kings of France). Forensic Science International: Genetics, 226, 38-40. http://dx.doi.org/10.1016/j.forsciint.2012.11.018
- Larmuseau, M.H., Delorme, P., Germain, P., Vanderheyden, N., Gilissen, A., Van Geystelen, A., et al. (2013) Genetic Genealogy Reveals True Y-Haplogroup of the House of Bourbon Contradicting Recent Identification of the Presumed Remains of Two French Kings. European Journal of Human Genetics, 22, 681-687. http://dx.doi.org/10.1038/ejhg.2013.211
- Lucotte, G. (2010) A Rare Variant of the mtDNA HVS1 Sequence in the Hairs of Napoléon’s Family. Investigative Genetics, 1, 7-10. http://dx.doi.org/10.1186/2041-2223-1-7
- Lucotte, G., Thomasset, T. and Hrechdakian, P. (2011) Haplogroup of the Y Chromosome of Napoléon the First. Journal of Molecular Biology Research, 1, 12-19. http://dx.doi.org/10.5539/jmbr.v1n1p12
- Lucotte, G., Macé, J. and Hrechdakian, P. (2013) Reconstruction of the Lineage Y Chromosome of Napoléon the First. International Journal of Sciences, 2, 127-139.
- Bogdanowicz, W., Allen, M., Branicki, W., Lembring, M., Gajewska, M. and Kupiec, T. (2009) Genetic Identification of Putative Remains of the Famous Astronomer Nicolaus Copernicus. Proceedings of the National Academy of Sciences of the United States of America, 106, 12279-12282. http://dx.doi.org/10.1073/pnas.0901848106
- Hawass, Z., Ismail, S., Selim, A., Saleem, S.N., Fathalla, D., Wasef, S., et al. (2012) Revisiting the Harem Conspiracy and Death of Ramesses III: Anthropological, Forensic, Radiological and Genetic Study. British Medical Journal, 345, e8268. http://dx.doi.org/10.1136/bmj.e8268
- Tulard, J. and Carros, C. (1992) Itinéraire de Napoléon au jour le jour (1769-1821). Library Jules Tallandier, Paris.
- Athey, T.W. (2006) Haplogroup Prediction from Y-STR Values Using a Bayesian Allele Frequency Approach. Journal of Genetic Genealogy, 2, 34-39.
- Hofreiter, M., Serre, D., Poinar, H.N., Kuch, M. and Pääbo, S. (2001) Ancient DNA. Nature Reviews Genetics, 2, 353-359. http://dx.doi.org/10.1038/35072071
- Doutremépuich, C., Doutremépuich, F., Roca, I., Beaufils, M. and Esponda, A. (2008) Principes de Médecine Légale. Chapter 24, Arnette Editor, Paris, 295-305.
- Butler, J.M., Decker, A.E., Vallone, P.M. and Kline, M.C. (2006) Allele Frequencies for 27 Y-STR Loci with U.S. Caucasian, African, American and Hispanic Samples. Forensic Science International, 156, 250-260.http://dx.doi.org/10.1016/j.forsciint.2005.02.011
- Lucotte, G. and Diéterlen, F. Frequencies of M34, the Ultimate Genetic Marker of the Terminal Differenciation of the Napoléon the First’s Y-Chromosome Haplogroup E1b1b1c1, in Europe, Northern Africa and the Near East. International Journal of Anthropology (in Press).
NOTES

*Corresponding author.


