Open Journal of Genetics
Vol.3 No.2(2013), Article ID:33610,4 pages DOI:10.4236/ojgen.2013.32011
Isolation and characterization of polymorphic microsatellite loci for the valuable medicinal plant Astragalus mongholicus
![]()
1College of Life and Environmental Sciences, Minzu University of China, Beijing, China
2Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
Email: *long@mail.kib.ac.cn
Copyright © 2013 Ao Wang et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 16 April 2013; revised 17 May 2013; accepted 2 June 2013
Keywords: Astragalus mongholicus; Medicinal Plant; Microsatellite; Molecular Marker; Population Genetics
ABSTRACT
Astragalus mongholicus (Fabaceae) is a perennial herb and a widely used medicinal plant in traditional Chinese medicine (TCM) known as Radix Astragali (Huangqi). It was reported to have hepatoprotective, cardioprotective, antidiabetic, antiaging, sedative and immunopotentiating effects, and could also be used as an adjuvant medicine during cancer therapy. Until now, there is only a little research on its population genetics and no report on development of microsatellite loci for this plant. In this study, a microsatellite-enriched genomic DNA library of A. mongholicus was developed and screened to identify marker loci. Ten polymorphic loci were isolated and analyzed by screening 30 individuals. The number of alleles per locus ranged from 4 to 19, with an average of 12.3 alleles per locus. The observed heterozygosity (HO) and the expected heterozygosity (HE) ranged from 0.367 to 1.000 and from 0.395 to 0.912, respectively. The polymorphism information content (PIC) varied from 0.361 to 0.888, with an average of 0.762. This is the first report on characterization of microsatellite loci for A. mongholicus, and these markers will be useful for population genetics and molecular ecology studies of this plant.
1. INTRODUCTION
Astragalus mongholicus Bunge (synonym, Astragalus membranaceus Bunge var. mongholicus (Bunge) P. G. Xiao), a perennial herb, is a member of Fabaceae and mainly distributed in the northeast, north, and northwest of China as well as in Mongolia [1]. It is a widely used medicinal plant in traditional Chinese medicine (TCM) known as Radix Astragali, whose roots have been used for centuries in China as a therapeutic agent for Yang weakness and Qi deficiency, and was reported to have hepatoprotective, antidiabetic, diuretic, and sedative effects [2-5]. Additionally, various Radix Astragali preparations are commercially available in the United States as a dietary supplement [4]. Recent study also demonstrated that Radix Astragali has a wide range of immunopotentiating effects and could be used as an adjuvant medicine during cancer therapy [4]. It has been reported that the pharmacological activity of A. mongholicus was due to several active components, such as saponins, isoflavonoids and polysaccharides, which have various biological activities [5]. However, human activities and increased utilization in TCM have caused a dramatic decline in the natural populations in the last few decades. Although the morphology, karyotype, molecular phylogeny, pharmacology and metabonomics of A. mongholicus have been extensively studied [3,6-9], information on population genetics is largely unknown.
In recent decades, microsatellites or SSRs (simple sequence repeats) have become the most popular genetic markers, owing to their high reproducibility, abundance across genomes, multi-allelic nature, codominant inheritance and high levels of polymorphism [10-12]. They have been successfully used for genomic mapping, population genetics, marker-assisted selection, conservation biology and other studies [10,13,14]. However, the original microsatellite locus has not been developed for A. mongholicus. In order to provide useful tools for population genetics analysis of this valuable medicinal plant, we report the isolation and characterization of polymorphic microsatellite loci from A. mongholicus for the first time, using a modified biotin-capture method [15].
2. MATERIALS AND METHODS
The leaves of A. mongholicus from a natural population in Inner Mongolia Autonomous Region were collected, dried in silica gel in sealed polyethylene bags and stored at room temperature until use. Genomic DNA was extracted from dry leaf tissue using a Plant Genomic DNA Kit (TIANGEN, Beijing). About 600 ng genomic DNA was digested by Sau 3AI (TaKaRa), and the fragments ranging from 300 to 1000 bp were purified and ligated to the Sau 3AI adaptors (oligoA: 5’-GGCCAGAGACCCCAAGCTTCG-3’, oligoB: 5’PO4-GATCCGAAGCTTGGGGTCTCTGGCC-3’). The ligated DNA products were then hybridized with 3’-biotinylated (AG)15 oligonucleotide probes and captured by streptavidin-coated magnetic beads (Promega). The microsatellite-enriched fragments were amplified by PCR using oligoA as the primer, and ligated to pMD18-T vector (TaKaRa). The recombinant plasmid was then transformed into Escherichia coli DH5α competent cells and the transformants were distinguished by blue-white screening. The positive clones were identified by PCRs using oligoA and oligonucleotides (AG)15 as the primers. The clones yielding two or more bands contained microsatellite fractions. Totally, ninety-six positive clones were sequenced and sixty-five contained microsatellites. Fifty-two pairs of primers were then designed using PRIMER PREMIER 5.0 software
(http://www.premierbiosoft.com/) and synthesized. Of these, 29 pairs successfully amplified products of expected sizes, and ten of the most promising loci were selected for further testing (Table 1). We attached the universal M13 sequence tag to the 5’ end of the forward primer for each potential locus, allowing efficient fluorescent labeling of the products using the method described by Schuelke [16]. Potential markers were tested on five individuals from Inner Mongolia. PCR amplifications (15 µl) contained 6.25 µl GoTaq® Green Master Mix (Promega), 0.8 µmol of each reverse and FAM-M13 primer, 0.2 µmol of the M13-forward primer and 20 - 50 ng template DNA, and were performed in an iCycler thermocycler (Bio-Rad). The cycling parameters were: one cycle at 94˚C for 5 min; 35 cycles at 94˚C for 30 s, optimized locus-specific annealing temperature (Table 1) for 45 s and 72˚C for 45 s; followed by 8 cycles at 94˚C for 30 s, 53˚C for 45 s and 72˚C for 45 s, and a final cycle at 72˚C for 12 min. Loci that amplified in all five initial samples were then tested in 25 additional individuals. The PCR products were analyzed with an ABI 3100 automated sequencer using HiDi and size standard 400 HD ROX (Applied Biosystems). Genotypes were assessed using GeneMapper v3.2 (Applied Biosystems). The number of alleles (NA), expected heterozygosity (HE) and observed heterozygosity (HO), Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium (LD) at each locus were tested using GENEPOP on the Web [17,18]. Polymorphism information content (PIC) was caculated
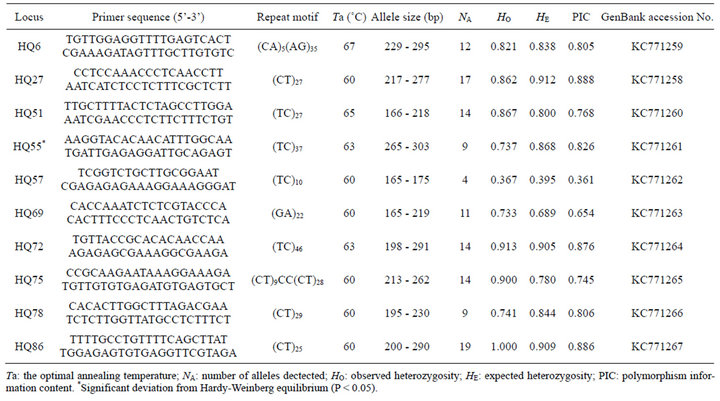
Table 1. Characteristics of ten microsatellite loci isolated from Astragalus mongholicus.
by Cervus 3.0 [19]. MICRO-CHECKER software was used to determine the presence of null alleles [20]. All results for multiple tests were corrected using Bonferroni’s correction.
3. RESULTS AND DISCUSSION
Ten polymorphic microsatellite loci were isolated and deposited in GenBank (KC771258 - KC771267; Table 1). The number of alleles per locus ranged from 4 to 19, with an average of 12.3 alleles per locus. The observed heterozygosity (HO) and the expected heterozygosity (HE) ranged from 0.367 to 1.000 and from 0.395 to 0.912, respectively. The polymorphism information content (PIC) varied from 0.361 to 0.888, with an average of 0.762. Deviations from Hardy-Weinberg expectation (HWE) were detected for each locus, nine loci conformed to HWE and one loci (HQ55) significantly deviated from HWE in the sampled population after Bonferroni’s correction (P < 0.05), which could be due to the presence of null alleles that was confirmed by the analyses using MICRO-CHECKER software. None of the loci showed significant linkage disequilibrium.
This is the first report on microsatellite marker development of A. mongholicus. These microsatellite markers showed high polymorphism and would be useful for further population genetics studies and conservation decision-making for this valuable medicinal plant.
4. ACKNOWLEDGMENTS
This work was supported by the Ministry of Education of China through its 111 and 985 projects (B08044, MUC985-9, MUC98506- 01000101).
REFERENCES
- Xu, L.R. and Podlech, D. (2010) Astragalus. In: Wu, Z.Y. and Raven, P.H., Eds., Flora of China, Science, Beijing, Missouri Botanical Garden Press, St Louis, 328-453.
- The State Pharmacopoeia Commission of PR China. (2010) Pharmacopoeia of People’s Republic of China. Chemical Industry Press, Beijing.
- Sinclair, S. (1998) Chinese herbs: A clinical review of Astragalus, Ligusticum, and Schizandra. Alternative Medicine Review, 3, 338-344.
- Xiao, W.L., Motley, T.J., Unachukwu, U.J., San Lau, C.B., Jiang, B., Hong, F., Leung, P.C., Wang, Q.F., Livingston, P.O., Cassileth, B.R. and Kennelly, E.J. (2011) Chemical and genetic assessment of variability in commercial Radix astragali (Astragalus spp.) by ion trap LCMS and nuclear ribosomal DNA barcoding sequence analyses. Journal of Agricultural and Food Chemistry, 59, 1548-1556. doi:10.1021/jf1028174
- Chu, C., Qi, L.W., Liu, E.H., Li, B., Gao, W. and Li, P. (2010) Radix Astragali (Astragalus): Latest advancements and trends in chemistry, analysis, pharmacology and pharmacokinetics. Current Organic Chemistry, 14, 1792-1807. doi:10.2174/138527210792927663
- Dong, T.T.X., Ma, X.Q., Clarke, C., Song, Z.H., Ji, Z.N., Lo, C.K. and Tsim, K.W.K. (2003) Phylogeny of Astragalus in China: Molecular evidence from the DNA sequences of 5S rRNA spacer, ITS, and 18S rRNA. Journal of Agricultural and Food Chemistry, 51, 6709-6714. doi:10.1021/jf034278x
- Yan, L., Wan, T., Zhang, Z., Wang, X. and Sun, W. (2001) Analysis on botanical characters of Astragalus membranacens Bunge. and A. membranacens var. mongolicus (Bunge) Hsiao. Journal of Inner Mongolia Agricultural University (Natural Science Edition), 22, 71-77.
- Wu, S., Wang, L., Sun, L., Bu, Y. and Wu, J. (2006) Karyotype analysis of Astragalus membranaceus. Hubei Agricultural Sciences, 45, 631-633.
- Duan, L.X., Chen, T.L., Li, M., Chen, M., Zhou, Y.Q., Cui, G.H., Zhao, A.H., Jia, W., Huang, L.Q. and Qi, X.Q. (2012) Use of the metabolomics approach to characterize Chinese medicinal material Huangqi. Molecular Plant, 5, 376-386. doi:10.1093/mp/ssr093
- Li, Y.C., Korol, A.B., Fahima, T., Beiles, A. and Nevo, E. (2002) Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Molecular Ecology, 11, 2453-2465. doi:10.1046/j.1365-294X.2002.01643.x
- Sharma, P.C., Grover, A. and Kahl, G. (2007) Mining microsatellites in eukaryotic genomes. Trends in Biotechnology, 25, 490-498. doi:10.1016/j.tibtech.2007.07.013
- Wang, H., Chen, N.F., Zheng, J.Y., Wang, W.C., Pei, Y.Y. and Zhu, G.P. (2012) Isolation and characterization of eleven polymorphic microsatellite loci for the valuable medicinal plant Dendrobium huoshanense and crossspecies amplification. International Journal of Molecular Sciences, 13, 16779-16784. doi:10.3390/ijms131216779
- Yu, K., Park, S., Poysa, V. and Gepts, P. (2000) Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). Journal of Heredity, 91, 429-434. doi:10.1093/jhered/91.6.429
- Ellegren, H. (2004) Microsatellites: Simple sequences with complex evolution. Nature Reviews Genetics, 5, 435- 445. doi:10.1038/nrg1348
- Bloor, P., Barker, F., Watts, P., Noyes, H. and Kemp, S. (2001) Microsatellite libraries by enrichment. http://www.genomics.liv.ac.uk/animal/Protocol1.html
- Schuelke, M. (2000) An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnology, 18, 233-234. doi:10.1038/72708
- Raymond, M. and Rousset, F. (1995) GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. Journal of Heredity, 86, 248-249.
- Rousset, F. (2008) Genepop’007: A complete reimplementation of the genepop software for Windows and Linux. Molecular Ecology Resources, 8, 103-106. doi:10.1111/j.1471-8286.2007.01931.x
- Kalinowski, S.T., Taper, M.L. and Marshall, T.C. (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology, 16, 1099-1106. doi:10.1111/j.1365-294X.2007.03089.x
- Van Oosterhout, C., Hutchinson, W.F., Wills, D.P.M. and Shipley, P. (2004) Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4, 535-538. doi:10.1111/j.1471-8286.2004.00684.x
NOTES
*Corresponding author.

