Advances in Biological Chemistry
Vol.2 No.2(2012), Article ID:18969,8 pages DOI:10.4236/abc.2012.22011
Digitonin and sodium dodecylsulfate-solubilized frog rhodopsin: Behavior under native and denaturing polyacrylamide gel electrophoresis
![]()
Sechenov Institute of Evolutionary Physiology and Biochemistry, Russian Academy of Sciences, St. Petersburg, Russia
Email: tinycapers@mail.ru
Received 9 January 2012; revised 28 February 2012; accepted 5 March 2012
Keywords: BN-PAGE; frog rhodopsin; oligomerization-dissociation of rhodopsin; SDS-PAGE
ABSTRACT
Rhodopsin oligomerization and dissociation in vivo and under experimental conditions is an important topic both for a basic understanding of photoreceptor structure-function but also as a potential eye disease mechanism. In this study, to estimate a state rhodopsin after solubilization with mild and harsh detergents, we applied the native (blue native-PAGE, BNPAGE) and denaturing electrophoresis (blue-ureaPAGE, BU-PAGE; blue-SDS-PAGE, BSDS-PAGE and SDS-PAGE). After blue BN-PAGE and BSDS-PAGE, rhodopsin and opsin, respectively, were presented in gels a major band of dimer with slight contents of higher oligomers without any traces of monomer, thus testifying in favor dimer-heteromeric state of frog rhodopsin in the photoreceptor membrane. Despite all oligomer bands gave positive staining with the rhodopsin-specific monoclonal antibodies (mAb), subsequent SDS-PAGE in combination with electroelution in denaturing conditions showed that stained bands are not homogenous and besides of opsin oligomers contain a small admixture of proteins with unknown function. Unfolding of opsin oligomers by solubilization in SDS, as compared with folded opsin in digitonin, induces their transition to a more compact conformation. It was manifested in a more rapid migration of opsin oligomers toward to anode. Cooling of digitonin/SDS mixed extracts at 4˚C for 24 hours led to a partial reverse transition of unfolded opsin dimer to initial folded conformation, thus demonstrating the entropic nature of this transition. Opsin monomer can be observed in the gels only after harsh dissociation of oligomers under BU-PAGE or SDS-PAGE. The electro elution of the individual opsin oligomers with denaturing buffer followed by SDS-PAGE resulted in dissociation of dimer to monomers. However, unexpectedly, the trimer was dissociated to a prevailing dimer and a small portion of monomer. The products dissociation of both opsin tetramer and pentamer are difficult to determine precisely, but they are neither monomer nor dimer. Dissociation data show that the degree of opsin oligomerization by unknown reasons affects the pattern of dissociation of its aggregates. Obtained in this paper data indicate a need for further detailed study the obscure mechanisms of aggregation-dissociation of rhodopsin.
1. INTRODUCTION
Rhodopsin is a G protein-coupled receptor (GPCR) that absorbs light quantum thus initiating of enzymatic cascade of phototransduction in rod photoreceptors. It consists of apoprotein opsin and bound by Shiff-base linkage chromophor, 11-cis-retinal. High expression levels of rhodopsin in the disc membranes of rod outer segments (ROSs), the lack of other highly abundant membrane proteins allows rhodopsin to be extracted from membranes with micro molar concentrations using various mild or harsh detergents (for example, digitonin or SDS, respectively). It is well known, that digitonin preserves some native properties of rhodopsin (characteristic absorbance spectrum, ability to regenerate after bleaching and to bind G-protein, etc.). In opposite to digitonin, SDS induces the deep denaturing changes in rhodopsin molecules that are accompanied by the changes in its conformation and releasing of 11-cis-retinal from chromophore-binding site, pigment bleaching and its conversion to opsin.
The question whether a native mammal rhodopsin in the photoreceptor membrane exists in the form of a monomer [1-4] or it is presented by oligomers [5-11] as many known GPCRs [12-14], are not solved finally so far. However, various approaches continue to demonstrate in mouse and bovine disk membranes of ROSs, as well as after solubilization in mild detergents, a dimeroligomer pattern of rhodopsin [7-11,15].
Rhodopsin of cool-blooded animals and, in particular, amphibians, in this respect only has started to be studied [16,17]. These data on frog visual pigment testified to a greater extend in favor of heteromeric state of rhodopsin in the membrane of discs and supported an assumption that functional unit of rhodopsin is not a monomer but a dimer [18].
However, the conditions that lead to oligomerization of rhodopsin in vivo, as well as molecular mechanisms that induce such state after solubilization by detergents in vitro are still remain to be obscure and demand deciphering.
In this paper, we applied a combination of native electrophoresis (BN-PAGE), denaturing electrophoresis (BSDSPAGE, BU-PAGE and SDS-PAGE) and electro elution under denaturing conditions as well as rhodopsin-specific staining by monoclonal antibodies to identify rhodopsin state after solubilization with mild and harsh detergents (digitonin and SDS, respectively). We showed that rhodopsin in digitonin extracts, as well as opsin after unfolding in denaturing SDS, are situated in gels in dimer-oligomeric state. We showed also that in mixed digitonin/SDS extracts the transition unfolded-folded opsin is reversible. In addition, we found out that rhodopsin (opsin) in monomeric state is absent in both initial detergent extracts from ROSs. Monomer was occurred in the gel only after BU-PAGE and SDS-PAGE exclusively because of denaturing dissociation of opsin oligomers. In these conditions, the dissociation of oligomers to monomer is often accompanied by a secondary self-association of monomers to oligomers and by a release of unknown concomitant proteins. Surprisingly, the final products of dissociation of oligomers, for reasons unknown, may be not only monomers, but a mixture of monomer with dimer or of trimer with higher oligomers.
2. MATERIALS AND METHODS
2.1. Chemicals and Animals
All chemicals used in the work were purchased from Sigma-Aldrich (USA) and from Fluka, Merck, Serva, and Ferak (Germany). Treatment and care of animals were conducted according to the Sechenov Institute’s Rules and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. In the experiments, the Grass frogs (Rana temporaria) were used. The procedures of isolation of ROSs as well as solubilization of rhodopsin with digitonin and SDS were described early [17]. Detergent/protein ratio in the sample extracts was 4:8 (in g/g).
2.2. BN-PAGE, BSDS-PAGE, BU-PAGE and SDS-PAGE
The original protocols of BN-PAGE [19] and SDS-PAGE [20] were taken as a basis. In order to have other denaturing electrophoresis systems in addition to SDS-PAGE, the original protocol of BN-PAGE [19] was modified by using Coomassie Brilliant Blue G-250 (CBB) in conjuction with SDS (BSDS-PAGE) instead of original mild detergents (see details in [19]) or by adding of urea (2, 4, 6, 7 M) to solubilization buffer (BU-PAGE) instead of 6-aminohexanoic acid. Sometimes urea was added also to cathode buffer for final concentration 2 М. In some SDS-PAGE or BSDS-PAGE experiments the dry SDS was added to digitonin extracts for final concentration 0.5%, thus preparing mixed digitonin/SDS micelles of opsin.
2.3. Formation of Retinyl-Opsin in ROSs
The protocol of formation stable secondary amine, retinylopsin (RO), in which retinal is still bound to denatured opsin at the chromophore-binding site, was described in details early [17]. Briefly, all ROS obtained from 1 - 2 frogs were suspended in 500 μl of mixture of 0.25 M acetate buffer with 0.7 M NaBH3CN, pH 5.0 [19], and incubated 1 h in the light at room temperature. After finishing of reaction, the suspension was sedimented at 20,000 g for 15 min and washed five times with 0.05 M imidazole buffer, pH 7.0, using the same conditions of centrifugation. The final pellet was immediately solubilized, the extract was loaded onto a gel, and after electrophoresis the fluorescence of RO on the gel was viewed and documented.
2.4. Electrophoresis Technique and Treatment of Gels
For all kinds of electrophoresis a vertical electrophoresis chamber MiniProtean 3 Cell (BioRad, USA) with 10 × 8 cm glasses, 1 mm thickness spacer, and 5.6 ml volume of gel were used. The power supply was a Power Pac HC (250 V/3 A/300 W; BioRad). A J5 Jule gradient former (Jule, USA) was used to prepare a linear gradient of polyacrylamide gel (8% - 26%) for BN-, BUor BSDSPAGEs. The aliquots (2 - 15 μl, 10 - 40 μg protein) were loaded under the cathode buffer onto the sample gel using a Guide ReadyGel Sample loading device (Bio-Rad, USA) and a syringe from Hamilton (USA). After sample loading onto the gel (5 - 10 μl, 10 - 35 μg of protein), the run parameters were kept according to original protocols [19,20]. After three kinds of electrophoresis the sample gel was cut off, and the rest of the gel, depending on the task, was subjected to: 1) Standard fixation followed by protein staining with CBB or AgNO3 [21]; 2) Electro blotting of proteins onto PVDF membrane (Sigma) and subsequent staining of bands with reaction product formed by interaction rhodopsin-specific mAb (titer 1: 1000, mouse, clone 1D4; Sigma) with anti-mouse IgG (titer 1:30,000; Sigma), containing conjugated alkaline phosphatase [17]; 3) electro elution of protein bands from the gel in denaturing buffer.
2.5. Electro Blotting and Electro Elution
The electro blotting apparatus and power supply were the Mini TransBlot Electrophoresis Transfer Cell and Power Pac HC (250 V, 3 A, 300 W), respectively (both from BioRad). The electro elution apparatus and power supply were the Mini Whole Gel Eluter equipped 14-channels chamber core and Power Pac HC (250 V, 3 A, 300 W), respectively (all from BioRad). The electro elution of digitonin or SDS solubilized protein bands from the gel slabs after BN-or BSDS-PAGEs was proceeded in denaturing buffer (43 mM Imidazole - 35 mM HEPES −1% SDS, pH = 7.4). Sometimes before electro elution a narrow strip of the gel was cut off from the slab and subjected fixation and staining to control the separation quality. Following equilibration in the denaturing buffer, the slab gel was removed from the buffer and placed on a glass plate. The template was used to trim the excess gel from the part of the gel containing the separated rhodopsin (opsin) oligomers. The Mini Whole Gel Eluter was filled with the same buffer. The gel was transferred to it and overlaid with soaked filter paper. Electro elution was performed under constant current 50 mA and voltage limited to 5 - 10 V. After 20 min elution, the reverse current was applied to Eluter for 30 sec to dislodge proteins bound to the cellophane membrane. Sometimes proteins after elution were concentrated by spin filtration on the dry Sephadex G-25 (Pharmacia) using a small column and the angle rotor N5404 of K280R centrifuge (Centurion, England).
2.6. Detection Protein Bands and Determination of Protein in Samples
Protein bands on the gels and PVDF membranes (Sigma) were viewed in scattered white light (staining by CBB or antibodies) or 365 nm UV light (RO fluorescence) emitted by the TFX-L/WL transilluminator (Vilber Lourmat, France). Because CBB completely quenched a shortlived and weak fluorescence of RO, only the gels after SDS-PAGE could be used to register fluorescence of this protein. Protein in the samples was determined by the method of Lowry et al. [22] after its precipitation by 3% CCl3COOH, washing by H2O and solution in 1 N NaOH. The standard protein was BSA.
2.7. Data Collection
After each experiment, the fresh or treated (fixed, stained and washed) gels were taken for the pictures by the PowerShot A3100 IS camera (Canon, Japan). The images as jpeg files were collected in computer hard disk and then they were transformed to tif format using Adobe Photoshop CS3 software. Each experiment was repeated 3 - 4 times and mostly typical and characteristic images were choice to illustrate the results of the experiments.
3. RESULTS AND DISCUSSION
3.1. Rhodopsin and Opsin Maintains Their Dimer-Oligomer State under BN-PAGE and BSDS-PAGE
BN-PAGE and BSDS-PAGE of digitonin and SDS extracts of rhodopsin (opsin) followed by staining with CBB or mAb have confirmed our previous data [16,17] and have shown that protein pattern of extracts in the gradient gel are presented by dimer with a slight content of higher oligomers (Figure 1, lines 1-4).
No opsin monomer and protein impurities were detected in the gels. It seems also that the content of the higher oligomers in SDS extracts is more pronounced than in digitonin one. In spite of SDS is a most harsh detergent among well-known, the dissociation of opsin oligomers to monomer in SDS extracts under BSDSPAGE does not occur. This means that the used electrophoresis system together with CBB preserve the native no covalent interactions between monomers in opsin oligomers. Furthermore, the equal presence of opsin dimers and oligomers in both detergent extracts supports conclusion that native rhodopsin forms dimeric and higher order structures in the very photoreceptor membranes [23].
3.2. SDS Induces a More Rapid Mobility toward the Anode of Opsin Oligomers, Changing Their Form
All oligomers in SDS or in digitonin/SDS mixed micelles under BSDS-PAGE, as compared to oligomers in digitonin micelles under BN-PAGE, showed a more rapid mobility toward the anode (Figures 1 and 2, lines 3, 4 compared to lines 1, 2). Analogical behavior of rhodopsin in other detergents was observed in the previous papers [16,17]. In the current work, we explain it by the graded degree of unfolding of rhodopsin in mild and harsh detergents, thus resulting in opsin structure the different changes of the compactness [16]. Indeed, references [24,25], using the biophysical methods, have recently shown that denatured states opsin in SDS micelles (0.3% - 15% SDS) are characterized a relatively greater burial of cystein and tryptophan residues and are
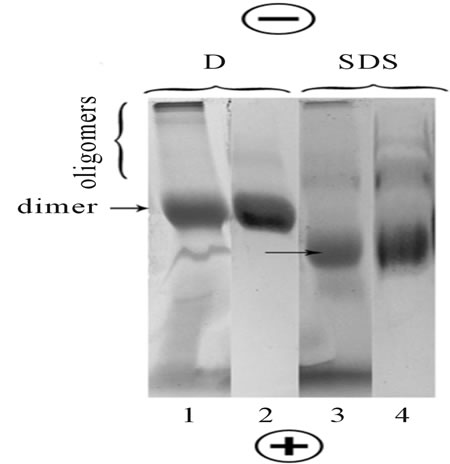
Figure 1. BN-PAGE and BSDS-PAGE separation of rhodopsin and opsin extracted with digitonin (D) and SDS, respectively. Lines 1, 2: BN-PAGE (rhodopsin); Lines 3, 4: BSDSPAGE (opsin); Lines 1, 3: The gels after CBB staining; Lines 2, 4: The blots on PVDF membrane stained by rhodopsin-specific mAb. The loaded samples contained 20 - 25 µg protein in 15 µl. Data of one typical experiment are presented.
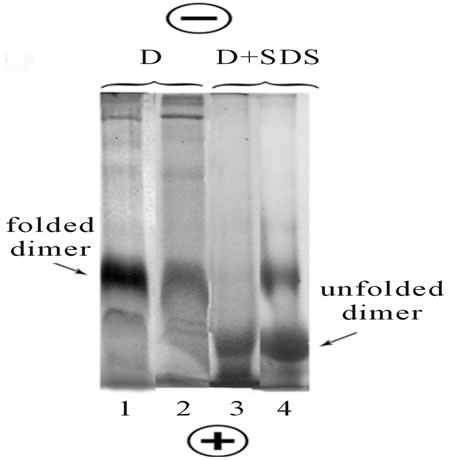
Figure 2. The stable folded state of rhodopsin in digitonin (D) micelles and temperaturedependent conformational transition of unfolded-folded opsin in mixed D/SDS micelles. Line 1: Fresh rhodopsin extract in digitonin was subjected to BN-PAGE; Line 2: Digitonin extract of rhodopsin was stayed in refrigerator for 24 h at +4˚C and then an aliquot was subjected to BN-PAGE; Line 3: Dry SDS powder was dissolved in fresh digitonin extract and an aliquot was immediately subjected to BSDSPAGE; Line 4: The same as on line 3, but the mixture was stayed in refrigerator for 24 h at +4˚C. All gels were stained with CBB. The loaded samples contained 20 µg protein in 15 µl. Data of one typical experiment are presented.
more compact as compared to the states observed in mild detergents or dodecylmaltozide/SDS mixed micelles. Thus, we concluded that opsin in SDS, as compared with rhodopsin in digitonin, has a distinctive shape. The behavior of rhodopsin (opsin) under BNand BSDSPAGEs supported the conclusion that not only the molecular mass of a protein but also its shape appears to be an important parameter determining its position in the gel [26]. From this, it follows that the use of BN-PAGE or BSDS-PAGE coupled with mild and harsh detergents for the determination of molecular mass of various proteins looks very doubtful.
3.3. SDS-Induced Conformational Transitions in Opsin Dimer Is Dependent on the Temperature
Taking in account the entropic nature of hydrophobic interactions (the gain with increasing temperature and, conversely, the decrease when the temperature is lowered), seems likely that the conformational transitions in opsin dimer under the influence of SDS should be dependable from the temperature. Indeed, our experiments have shown that in contrast to digitonin extracts, where rhodopsin is in the stable folded state (Figure 2, lines 1 and 2), opsin in mixed SDS/digitonin extracts at low temperature showed ability to conformational transition from unfolded dimer state back to the initial folded dimer conformation (Figure 2, lines 3 and 4).
3.4. Urea Induces Dissociation of Opsin Dimer to Monomer and Inverse Association of Monomer to Oligomers
Introduction of urea in solubilizing and cathode buffers under BU-PAGE, resulted in both digitonin and SDS extracts roughly dose-dependent dissociation of dimer to monomer and inverse association of monomer to oligomers. This was manifested in decrease the proportion of major band of dimer and increase in the share of monomer and higher oligomers of opsin, respectively (Figure 3, lines 2 - 5, 7 - 10).
It is evidently, that small but distinct increase in a share of the higher oligomers in the presence of urea is caused by a secondary self-association of the formed monomer (or oligomers). It is interesting, that in the condition of BU-PAGE, opsin monomer extracted by digitonin has a tendency to be divided on two sub bands (Figure 3, lines 2 - 5). This tendency is much less pronounced in SDS extracts (Figure 3, lines 7 - 10). Note that the relative functional integrity of frog rhodopsin solubilized with digitonin in the presence 2 and 4 M urea was still demonstrated by its characteristic UV-visible absorption spectrum. However, 6 - 7 M urea caused the deep destructive changes in rhodopsin that resulted in
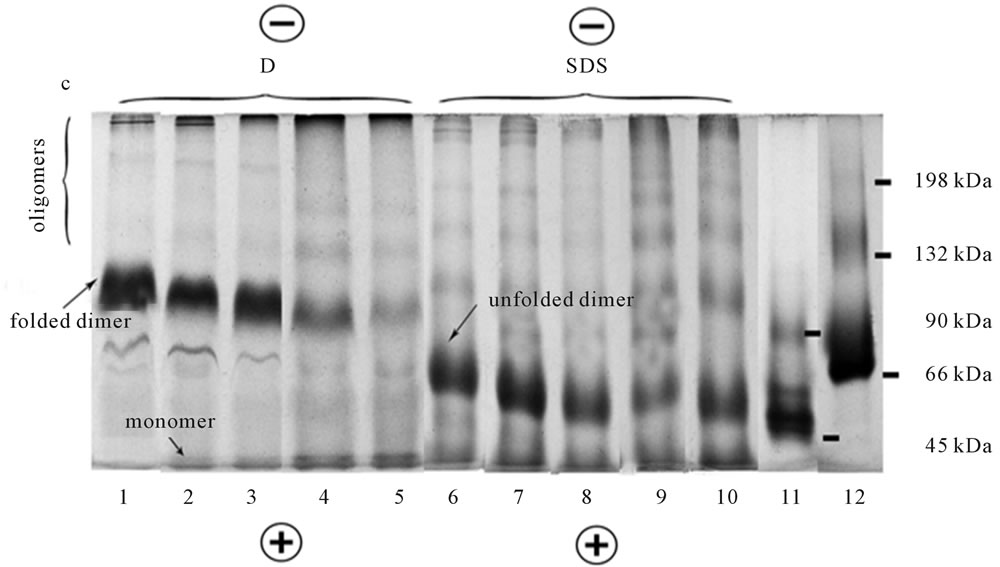
Figure 3. BUREA-PAGE of digitonin (D) and SDS extracts of rhodopsin and opsin. Lines 1 - 5, 6 - 10: Digitonin and SDS extracts, respectively; Lines 1, 6: No urea in solubilizing buffer (control); Lines 2 - 5, 7 - 10: Solubilizing buffer contains 2, 4, 6, 7 M urea, respectively; Line 11: Marker proteins: Monomer (45 kDa) and dimer (90 kDa) of ovalbumin; Line 12: Marker proteins: Monomer (66 kDa), dimer (132 kDa) and trimer (198 kDa) of BSA. The loaded marker proteins contained 30 µg in 10 µl; The samples were 20 µl in 15 µl. Data of one typical experiment are presented.
detachment of 11-cis-retinal from opsin and respectively, in bleaching of rhodopsin even in digitonin extracts (data not shown).
3.5. Retinyl-Opsin Formed in ROS Membranes Has Also the Dimer-Oligomeric State
As showed SDS-PAGE, retinyl-opsin (RO) formed directly in photoreceptor membrane, was presented in gel by fluorescent monomer, dimer and higher oligomers, regardless SDS or digitonin were initially used for solubilization (Figure 4, lines 1-7).
SDS extracts, as compared to digitonin extracts, demonstrated the larger number of protein impurities (molecular mass ~36, 50, 66 kDa), thus indicating the selectivity of digitonin for RO extraction. Similar to opsin monomer devoided of the chromophore under BU-PAGE (Figure 3, lines 2-5), RO monomer with “sewn” chromophore under SDS-PAGE (Figure 4, lines 1 - 4, 5 - 8) also demonstrated its subdivision on two sub bands. The phenomena of “splitting” (or vice versa “doubling”) which was detected under denaturing electrophoresis in a single-chain protein, opsin, cannot be interpreted now and requires further study.
The presence of opsin aggregates in SDS as well as in SDS + urea extracts, as revealed by BSDS-PAGE, BU-PAGE and SDS-PAGEs, contradicts to the conclusions of other authors obtained after use of biophysical methods [24,25]. They are as follows: 1) “SDS is able to prevent the exposure of large hydrophobic regions present in rhodopsin which otherwise leads to aggregation”, 2) “3% SDS + 8 M urea are the denaturing conditions of choice to study maximally unfolded rhodopsin without aggregation”. The reasons of such discrepancy are not clear and cannot be commented on now.
3.6. Dissociation of Oligomers Does Not Necessarily Lead to the Appearance of Only the Monomers
Can the various opsin oligomers obtained by BN-PAGE or BSDS-PAGE entirely dissociate to monomer?
In our experiments we showed that electro elution in denaturing conditions followed by SDS-PAGE induced complete dissociation of opsin dimers visible in the Figures 5(a), 6(a) to opsin monomers with molecular mass close to 40 kDa, regardless of BN-PAGE (run native rhodopsin) or BSDS-PAGE (run denatured opsin) were initially used for their isolation (Figure 5(b), lines 4-7; Figure 6(b), lines 9-12). Because of the proportion of higher rhodopsin oligomers (trimer and >) in digitonin extracts is sufficiently low (Figure 5(a)), it is difficult to see and understand on what products they dissociate (Figure 5(b)). Surprisingly, but clearly visible after BSDS-PAGE opsin trimer (Figure 6(a)), under similar conditions dissociated to a small fraction of monomer and a predominant share of dimer (Figure 6(b), lines 7,8), thus clearly indicating that dissociation in these conditions does not necessarily leads to the formation only monomers. The products of dissociation of opsin
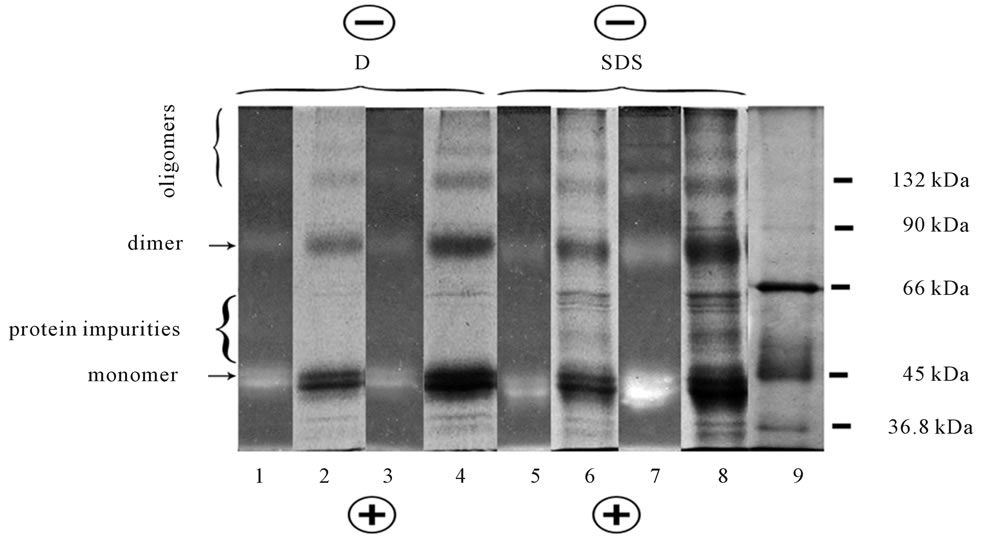
Figure 4. SDS-PAGE of digitonin (D) and SDS extracts from NaBH3CN reduced ROSs. Lines 1 - 4 were digitonin extracts: Line 1 was unfixed gel with fluorescent RO oligomers viewed in UV light, 20 µg protein of extract was loaded in 10 µl; Line 2 was the same as line 1, but after gel fixation and staining by CBB; Line 3 was the same as line 1 but 37 µg protein was loaded in 10 µl; Line 4 was the same as line 3 but after gel fixation and staining by CBB. Lines 5 - 8 were SDS extracts: Line 5 was unfixed gel with fluorescent RO oligomers viewed in UV light, 26 µg protein was loaded in 10 µl; Line 6 was the same line 5 but after fixation and staining by CBB; Line 7 was the same line 5 but 45 µg protein was loaded in 10 µl; Line 8 was the same as line 7 but after fixation and staining by CBB; Line 9 was the mixture of protein markers: Dimer of β-lactoglobulin (36.8 kDa), monomer of ovalbumin (45 kDa), monomer of BSA (66 kDa), dimer of ovalbumin (90 kDa), dimer of BSA (132 kDa). The loaded marker protein contained 15 - 25 µg in 10 µl. Data of one typical experiment are presented.

Figure 5. (a) BN-PAGE separation of digitonin extracts from frog ROSs. Following BN-PAGE separation of digitonin extracts, the narrow strip from the slab gel was cut, fixed and stained by CBB. The loaded sample contained 20 µg protein in 15 µl. The typical separation on the gel is presented. The rest gel was subjected to electro elution followed by SDS-PAGE separation (see Figure 5(b)); (b) SDS-PAGE separation of dissociated opsin oligomers initially extracted with digitonin and separated by BN-PAGE. The rest unfixed portion of the slab gel, containing 9 slots was subjected to electro elution in denaturing conditions using Mini Whole Gel Eluter unit. After electro elution fourteen fraction of 0.5 ml each were harvested from the Mini Whole Gel ELutor unit, concentrated by spin-gel filtration on Sephadex G-25 and then were analyzed by separation with SDS-PAGE followed by silver staining. Line 15 was the mixture of protein markers: monomer of β-lactoglobulin (18.4 kDa), dimer of β-lactoglobulin (36.8 kDa), monomer of ovalbumin (45 kDa), monomer of BSA (66kDa), dimer of ovalbumin (90 kDa). The loaded samples contained 20 - 25 µg protein in 15 µl. The typical separation is presented.
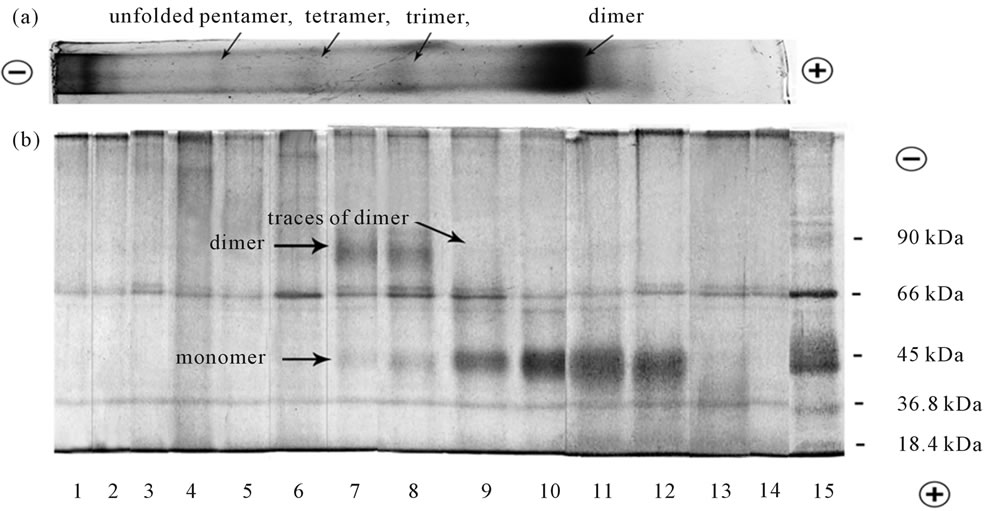
Figure 6. (a) BSDS-PAGE separation of SDS extracts from frog ROSs; (b) SDS-PAGE of dissociated opsin oligomers initially extracted by SDS and separated by BSDS-PAGE. The protocols used were the same as in Figures 5(a) and (b). Line 15 was the same as line 15 in Figure 5(b).
tetramer and pentamer are difficult to identify, but they are evidently neither the monomer nor the dimer (Figure 6(b), lines 4 - 6). The data ambiguities obtained under dissociation of Op oligomers require further experiments.
3.7. Unknown Proteins Co-Run with Rhodopsin (Opsin) Oligomers under BN-PAGE and BSDS-PAGE
As seen from the Figures 5(b), 6(b), all fractions of oligomers contain at least 2 unknown proteins with molecular mass close to 36 and 66 kDa. They first were noticed in our experiments under SDS-PAGE of RO and, as compared to digitonin extracts, were prevailing in SDS extracts (Figure 4). It means that both BN-PAGE and BSDS-PAGE do not provide a complete separation of complexes of opsin oligomers/concomitant proteins or opsin oligomers/intrinsic monomers, thus demonstrating that mAb-positive bands are non-homogenous. Complete homogeneity of Op oligomers is ensured only after use SDS-PAGE. The physiological function these concomitant proteins remain to be obscure.
4. CONCLUSION
The dimer-oligomeric forms of rhodopsin (opsin) in both mild and harsh detergents revealed by BNand BSDSPAGE are evidence that native rhodopsin in the photoreceptor membrane is located in a relatively stable state of the dimers and oligomers. In spite of all oligomer bands give the positive staining with the rhodopsin-specific mAb, they are actually no homogenous and except of opsin oligomers, contain unknown concomitant proteins. It means that in the cases of digitonin and SDS extracts both BN-PAGE and BSDS-PAGE do not ensure the intermolecular interaction breakage, thus demonstrating the inability complete separation of opsin oligomers from related proteins and opsin monomers from its oligomers. Our experiments first showed that unfolding of opsin oligomers by SDS (or mixture of digitonin/SDS) under BSDS-PAGE leads to a more rapid migration these proteins toward to anode owing to a more compact opsin conformation in SDS. The entropic nature of transition unfolded-folded opsin in cooled digitonin/SDS extracts was also first demonstrated by BSDS-PAGE. In addition we showed that opsin monomer is formed because of dissociation of oligomers exclusively in harsh denaturing condition (BU-PAGE and SDS-PAGE) and that the formation of monomers is accompanied by a secondary self-association of their into new oligomers. The degree of opsin oligomerization by unknown reasons affects the pattern of dissociation of its aggregates. The mechanisms of association-dissociation of opsin oligomers in vivo and in vitro are remain to be obscure and require further undivided attention.
5. ACKNOWLEDGEMENTS
This work at 2011 was in part supported by the Russian Foundation for Basic Research (grant No. 11-04-01319a) and by the Program of the Russian Academy of Sciences (Division of Biological Sciences, grants No. 2B-05)
![]()
![]()
REFERENCES
- Blasie, J.K. and Worthington, C.R. (1969) Molecular localization of frog retinal receptor photopigment by electron microscopy and low-angle X-ray diffraction. Journal of Molecular Biology, 39, 407-416. doi:10.1016/0022-2836(69)90135-1
- Chabre, M. (1975) X-ray diffraction studies of retinal rods. I. Structure of the disc membrane, effect of illumination. Biochimica et Biophysica Acta, 382, 322-335. doi:10.1016/0005-2736(75)90274-6
- Edrington, T.C., Bennett, M. and Albert, A.D. (2008) Calorimetric studies of bovine rod outer segment disk membranes supports a monomeric unit for both rhodopsin and opsin. Biophysical Journal, 95, 2859-2866. doi:10.1529/biophysj.108.128868
- Chabre, M., Deterre, P. and Antonny, B. (2009) The apparent cooperativity of some GPCRs does not necessarily imply dimerization. Trends in Pharmacological Sciences, 30, 188-197. doi:10.1016/j.tips.2009.01.003
- Fotiadis, D., Liang Y., Filipek S., et al. (2003) Atomic force microscopy: Rhodopsin dimers in native disc membranes. Nature, 421, 127-128. doi:10.1038/421127a
- Fotiadis, D., Liang, Y., Filipek S., et al. (2003) The G protein-coupled receptor rhodopsin in the native membrane. FEBS Letters, 564, 281-288. doi:10.1016/S0014-5793(04)00194-2
- Liang, Y., Fotiadis D., Liang Y., et al. (2003) Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. The Journal of Biological Chemistry, 278, 21655-21662. doi:10.1074/jbc.M302536200
- Filipek, S., Krzysko, K.A., Fotiadis, D., et al. (2004) A concept for G protein activation by G protein-coupled receptor dimers: The transducin/rhodopsin interface. Photochemical & Photobiological Sciences, 3, 628-638. doi:10.1039/b315661c
- Jastrzebska, B., Fotiadis, D., Jang, G.F., et al. (2006) Functional and structural characterization of rhodopsin oligomers. The Journal of Biological Chemistry, 281, 11917-11922. doi:10.1074/jbc.M600422200
- Morris, M.B., Dastmalchi, S. and Church, W.B. (2009) Rhodopsin: Structure, signal transduction and oligomerisation. The International Journal of Biochemistry & Cell Biology, 41, 721-724. doi: 10.1016/j.biocel.2008.04.025
- Gurevich, V.V. and Gurevich, E.V. (2008) How and why do GPCRs dimerize? Trends in Pharmacological Sciences, 29, 234-240. doi:10.1016/j.tips.2008.02.004
- Milligan, G. (2009) G protein-coupled receptor heterodimerization: Contribution to pharmacology and function. British Journal of Pharmacology, 158, 5-14. doi:10.1111/j.1476-5381.2009.00169.x
- Chamber, M., Deterred, P. and Antonny, B. (2009) The apparent cooperativity of some GPCRs does not necessarily imply dimerization. Trends in Pharmacological Sciences, 30, 188-197.
- Liu, Z., Zhang, J. and Zhang, A. (2009) Design of multivalent ligand targeting G-protein-coupled receptors. Current Pharmaceutical Design, 15, 682-718. doi:10.2174/138161209787315639
- Medina, R., Promo, D. and Bubis, J. (2004) The hydrodynamic properties of darkand light-activated states of n-dodecyl-beta-D-maltoside solubilized bovine rhodopsin support the dimeric structure of both confomations. The Journal of Biological Chemistry, 279, 39565-39573. doi:10.1074/jbc.M402446200
- Shukolyukov, S.A. (2009) Aggregation of frog rhodopsin to oligomers and their dissociation to monomer: Application of BNand SDS-PAGE. Biochemistry (Moscow), 74, 599-604. doi:10.1134/S0006297909060029
- Shukolyukov, S.A. (2010) Proof of oligomeric state of frog rhodopsin: Visualization of dimer and oligomers on gels after BNand HRCN-PAGE using Antibodies to rhodopsin and by retinylopsin fluorescence. Biochemistry (Moscow), 75, 1045-1051. doi:10.1134/S0006297910080146
- Neri, M., Vanni, S., Tavernelli, I. and Rothlisberger, U. (2010) Role of aggregation in rhodopsin signal transduction. Biochemistry, 49, 4827-4832. doi:10.1021/bi100478j
- Wittig, I., Braun, H.P. and Schägger, H. (2006) Blue native PAGE. Nature Protocols, 1, 418-428. doi:10.1038/nprot.2006.62
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680-685. doi:10.1038/227680a0
- Schägger, H. (2006) Tricine-SDS-PAGE. Nature Protocols, 1, 16-22. doi:10.1038/nprot.2006.4
- Lowry, O.H., Rosenbrough, N.I., Farr, A.L. and Randall, R.J. (1951) Protein measurement with Folin phenol reagent. The Journal of Biological Chemistry, 193, 265-290.
- Jastrzebska, B., Maeda, T., Zhu, L., et al. (2004) Functional characterization of rhodopsin monomers and dimers in detergents. The Journal of Biological Chemistry, 279, 54663-54675. doi:10.1074/jbc.M408691200
- Dutta, A., Tirupula, K.C., Alexiev, U., et al. (2010) Characterization of membrane protein non-native states. 1. Extent of unfolding and aggregation of rhodopsin in the presence of chemical denaturants. Biochemistry, 49, 6317- 6328. doi:10.1021/bi100338e
- Dutta, A., Kim, T.Y., Moeller, M., Wu, J., et al. (2010) Characterization of membrane protein non-native states. 2. The SDS-unfolded states of rhodopsin. Biochemistry, 49, 6329-6340. doi:10.1021/bi100339x
- Niepmann, M. and Zheng, J. (2006) Discontinuous native protein gel electrophoresis. Electrophoresis, 27, 3949-3951. doi:10.1002/elps.200600172

