American Journal of Molecular Biology
Vol. 2 No. 4 (2012) , Article ID: 23910 , 14 pages DOI:10.4236/ajmb.2012.24032
SPOC domain of mint protein induces hematopoietic differentiation via Bmp4/Smad5 pathway*
![]()
1The Department of Pathology, Yale University School of Medicine, New Haven, USA
2The Department of Laboratory Medicine, Yale University School of Medicine, New Haven, USA
3Vaxinnate Corporation, New Haven, USA
4The Department of Internal Medicine, Yale University School of Medicine, New Haven, USA
Email: #xian-yong.ma@yale.edu
Received 14 August 2012; revised 13 September 2012; accepted 25 September 2012
Keywords: SPOC Domain; Hematopoiesis, Bmp4/Smad5 Pathway
ABSTRACT
Mint is a newly identified molecule that mediates signal transduction and modulates chromatin repression. Mint family members contain a highly conserved C-terminus SPOC domain (SpenParalog and OrthologsC-terminal domain) commonly associated with proliferation and related diseases (for example: cancer) due to its role in cell differentiation and apoptosis. In this study, we addressed the SPOC function using a tetracycline-inducible system to express the target domain in Ain V15 embryonic ES cells and bone marrow stem cells from SPOC transgenic mice. In vitro differentiation of Ain V15 ES cells as a model of early hematopoietic development, we found expression of SPOC domain induces hematopoietic differentiation via up-regulation of transcription factors Bmp4 and Smad5, which induce the expression of hematopoietic factors Eklf1 and hematopoietic proliferation associated factor Gata2, the SPOC domain also plays the regulation function in the differentiation of hematopoitic progenitor by colony forming Unit (CFU) assays. Further, we determined SPOC expression enhances erythrocyte and granulocyte maturation using bone marrow cells derived from tiSPOC chimeric mice. Finally, we identified that overexpression of full length Mint in ES cells drives Smad5 and Bmp4 up-regulation under culture conditions, and up-regulates endogenous Mint when induces hematopoitic differentiation of EML, M1 and WT18 cells. In summary, our study reveals the conserved SPOC domain of Mint protein induces differentiation both in the stages of embryonic stem cells and hematopoietic progenitor cells.
1. INTRODUCTION
Mint (Msx2 interacting nuclear target protein) (Newberry, et al. 1999) is homologous to Spen, a recessive-lethal gene originally identified in Drosophila [1-4]. Spen has been shown to inhibit cell division and affect cell-fate specification, survival and axonal guidance through perturbation of the Notch and Ras/Raf signaling pathways and alters expression levels of Hox, E2F, epidermal growth factor receptor (EGFR), and the expression level of transcriptional repressors Hairless and Yan [1-3].
Loss-of-function mutations for Spen displays similar thoracic and abdominal features as null-zygotic mutations of Aop/Yan [5,6], while head skeletal structures are missing. Recent research has shown that Spen participates in Wingless signaling in the eye, wing and leg imaginal discs [4]. Mint regulates skeletal and neuronal development by mediating repression of homeodomain transcriptional repressor Msx2. Human homologue to mouse Mint is SHARP (SMRT/HDAC1-Associated Repressor Protein) [7], both in vivo and in vitro experiments have shown Mint is a key component of the transcripttional repression complex SMRT/HDAC1 and is recruited by Atra nuclear receptor and the Notch-signaling mediator RBP-Jk [7-11]. Mice genetically null for Mint (deletion of 1500aa - 1650aa) are embryonic lethal, and their splenic B cells were observed to differentiate three times more efficiently into marginal zone B cells with a concomitant reduction of follicular B cells [9]. It is also known that Mint deficiency impairs differentiation of early T-cell progenitors (ETP) into double-negative (DN2) cells [10].
Mint family genes encode relatively large transcripts. Drosophila homologue Spen is approximately 5560aa; human SHARP is 3664aa, mouse Mint is 3644aa (Ref. Figure 1). It is not clear the reason that full-length protein of Mint has never been detected from in vivo expression assays, for example, the predicted size of mouse
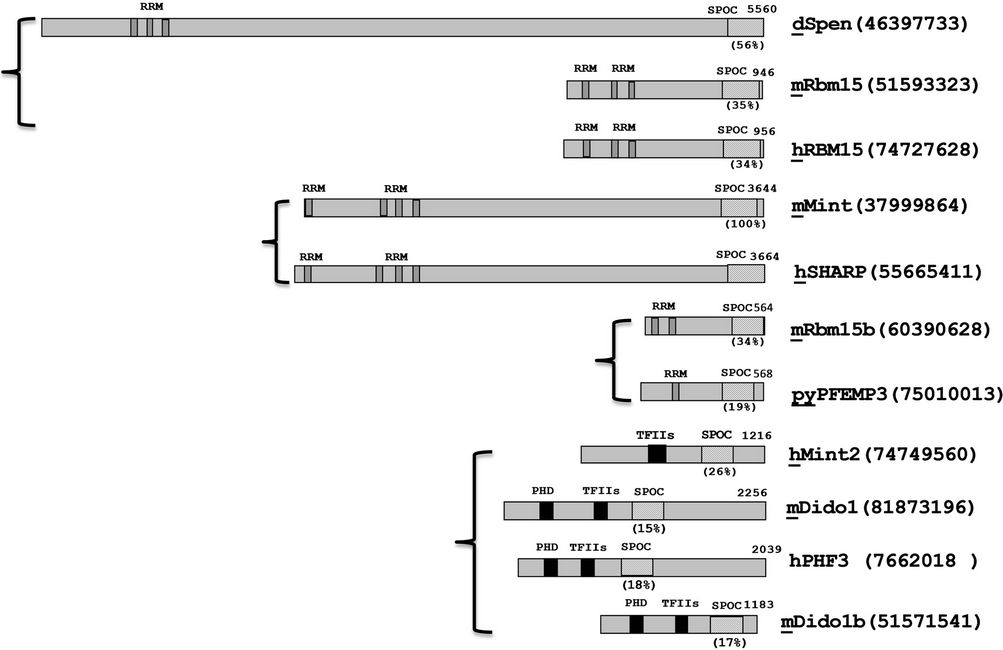
Figure 1. Domain organization and similarity analysis of Mint protein family. dSpen: DrosophilaSpen; mRbm15: mouse Rbm15 protein; hRBM15: human RBM15 protein; mMint: Mouse Mint protein; hSharp: human SHARP protein; mRbm15b: mouse Rbm15b; pyPFEMP3: plasmodium yoelii PEFMP3 protein; hMINT: human MINT protein; mDidol: mouse Didol protein; mDidolb: mouse Didol protein b. The approximate locations of RRMs and the SPOC domain of each protein are indicated in diagonal line and black boxes respectively, the TFIIs and PHD binding domain are also showed with black boxes. The percentages of identical residues in each SPOC domain homologs with human SHARP are indicated. m: mouse; h:human; d: drosophila; py: plasmodiumyoelii.
full length Mint is around 392.6 kd, but there are only two major products of 110 kd (N-terminal) and 250 kd (C-terminal) were detected using N and C terminal antibodies of Mint, based on this result the author predicated the Mint protein is proteolytically processed in vivo into these two fragments [12]. Some members with smaller size for this family have been identified, such as Spenlike protein pyPFEMP3 (568aa, plasmodium yoeli), hRBM15 (956aa, Human), Cessp (793aa, C. elegans.) and mRbm15 (946aa, mouse).All of these Mint family members contain conserved C-terminal sequence, which is called SPOC domain, and also most of these homologues contain RNA recognition motifs (RRMs) on the N-terminus and little conserved homology can be found elsewhere. RRMs were identified originally in RNA binding proteins involved in mRNA splicing, stability and translation processes, and more recently found the RRMs act as DNA binding motifs involved in the DNA binding activities of several transcription factors, these transcription factors include Mint, which binds via the RRMs to GT-rich DNA sequences [12], the RRM-containing N-terminal domain of Mint can play roles in both repression (suppresses FGF activation of the rat osteocalcin promoter) and activation (activates the HSV thymidine kinase promoter), depending on the interaction with promoters, therefore, the RRM-containing fragment can act as a repressor or an activator of transcription. Intriguingly, the C-terminal fragment of Mint binds to Msx2 homeobox protein in vitro, though not via the SPOC domain, and may coregulate the expression of genes required for craniofacial development. Thus, the C-terminal domain of spen is likely to be important for interactions with transcription factors and key signaling pathways regulating cell fate [3].
The most conserved SPOC domain of Mint family members is located in the C-terminal region, this domain consists of an acidic/basic domain (EK) and glutamine rich regions (Poly Q). The SPOC domain is involved in protein-protein interactions with HDAC1, HDAC2, SMRT and other corepressor components, indicating this highly conserved domain is one of the critical functional regions of Mint protein. Recent studies have also shown that SPOC is a widely distributed domain, presents in the DIO (Death inducers obliterator) family members that are related to cancer, apoptosis and gene transcription processes [13]. DIO-1 protein regulates the early stages of cell death in mice and humans. It has also been shown that PHF3 protein, another member of the DIO family, is ubiquitously expressed in normal tissues, and in glioblastoma, the most common primary tumor of human CNS [14]. Although the DIO family has been shown to be involved in apoptosis and cancer, the underlying molecular mechanisms are unclear. For these Mint family members with SPOC, the conserved domain structure suggests that they may share some common functions and molecular mechanisms. To test the role of SPOC domain in hematopoiesis from ES cells as well as from hematopoietic progenitors, we used a tetracycline inducible system to express SPOC domain in both ES cells and bone marrow cells and assayed the effect on hematopoietic differentiation. Our results demonstrated SPOC domain acts as a positive regulator for hematopoietic differentiation, and the results are supported by the expression analysis of the Mint gene in hematopoietic differentiation.
2. MATERIALS AND METHODS
2.1. Cell Culture and Differentiation Induction
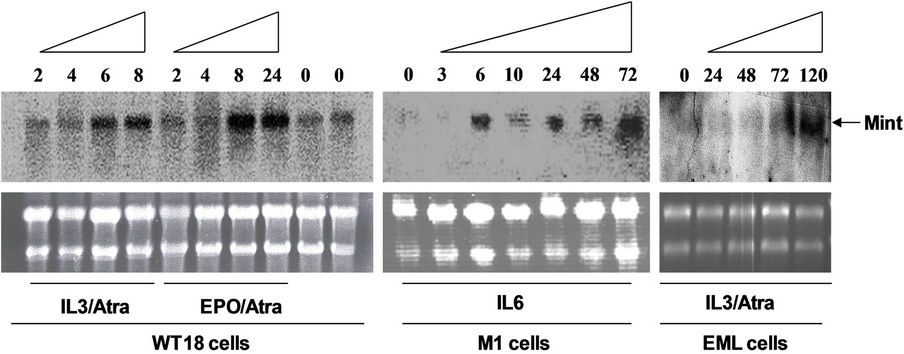
EML (Erythroid-macro-phage/lymphocyte progenitor) cells were grown in DMEM (GIBCO/BRL) supplemented with 20% horse serum (GIBCO/BRL), 15% BHK conditional medium, 0.1 mM nonessential amino acids (GIBCO/ BRL), 2 mM glutamine, 50 units/ml penicillin and 50 mg/ml streptomycin. Differentiation was induced with ATRA (10 mM) and IL6 (10 ng/ml) for indicated time points. WT18 cells (from 32D, see results) were grown in IMDM (GIBCO/BRL) supplemented with 10% IFS (Gemini Bio-Products), 10% WEHI-3B conditional medium, 2 mM glutamine, 50 units /ml penicillin and 50 mg/ml streptomycin. Differentiation was induced with IL3/Atra for indicated time points as showed in Figure 6. M1 cells (mouse monocytic leukemic cell line) were grown in RPMI 1640 supplemented with 10% IFS, 2 mM glutamine, 50 units/ml penicillin, and 50 mg/ml streptomycin, and treated with IL6 (50 ng/ml) for indicated time points. All cells were cultured at 37˚C in a 5% CO2 incubator.
2.2. RNA Preparation and Northern Blots
We prepared total RNA from cultured cells using Trizol reagent according to its manufacturer’s instruction (Life Technologies Inc.). The RNA amount was determined by using GeneQuant (Pharmacia Inc.), Northern blot and hybridization were carried out as follows: total RNA (12.0 mg) was run on a 1% agarose/0.6% formaldehyde gel, and transferred to a Hybond-N membrane (Amersham Inc.). The probe for SPOC mRNA and full length Mint detection encompasses the nucleotide sequence from 9565 to 10,995, which covers whole region of SPOC domain.
2.3. Immunoprecipitation and Immunoblots
The antibodies a-HDAC1, a-HDAC2, a-SMRT, a-Xpress (Santa Cruz Inc.) or Rabbit IgG (Sigma) were coupled to Gel 10 beads (Sigma) by mixing antibodies (or IgG) with beads in a 2 mg:1 ml ratio before incubation at 4˚C with rotation for four hours. CHO (China Hamster Ovary) cells grown in 10-cm dishes were transfected with 24 mg of appropriate plasmids using Lipofectamine 2000 (Invitrogene Inc.), at 48 hours posttransfection, the cells were rinsed once in ice-cold phosphate-buffered saline and lysed with 300 ml of IP buffer [20 mM HEPES (pH7.5), 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, 1 mM EDTA, and protease inhibitor cocktail (Sigma no. P8340)]. For Ain V15 ES cells, the lysate was prepared by using RIPA buffer, pre-cleared extracts by adding 0.25 mg of appropriate control IgG together with 20 ml of Protein L-agarose and incubating at 4˚C for 30 minutes with rotation, and cleared by centrifugation at 3000 rpm for 30 s at 4˚C. The supernatant was then incubated with antibodies of Xpress, HDAC1, HDAC2, SMRT or preserum beads at 4˚C for 2 hr. After extensive washes with IP buffer, the proteins bond to beads were eluted with sample loading buffer, analyzed by SDS-PAGE and subjected to immunoblot, then visualized using ECL system (Amersham Biosciences).
2.4. Immunofluorescence and Subcellular Localization
CHO cells were grown in 6-well plates, Lipofectamine 2000 (Invitrogen) was used to transiently transfect the CHO cells with 4 mg of pcDNA4-Xpress-SPOC or pcDNA3-HA-Mint, fixed these cells and incubated with anti-Xpress or anti-HA antibodies after 24 h transfection, followed by staining with FITC-conjugated secondary antibody. Fluorescence was detected by confocal microscopy to show the subcellular localization of target proteins, stained the nuclear DNA by DAPI as a control of subcellular localization.
2.5. Construction of Plox-SPOC Vector and Generation of Inducible ES Cells
The 1.1 kb fragment of SPOC cDNA (corresponding to amino acids 3224 to 3576 of Mint) was inserted into pCR2.1-TOPO vector, then cut with Kpn I and Not I, and inserted into pcDNA4-HisMax (Invitrogene) to generate
pcDNA4-SPOC expression vector that SPOC in-frame fused to 3’ terminus of Xpress epitope. The Kpn I/Xba I fragment of SPOC from pcDNA4-SPOC was subcloned into Plox vector (a gift from Kyba M.) to generate Plox-SPOC, and co-electroporated this plasmid together with pSalk-CRE into Ain V15 ES cells. [The targeted ES cell line Ain V15, plasmids pSalk-CRE, pLox and ploxEGFP have been described previously [15], screened co-electroporated cells with 600 mg/ml G418 (sigma) medium, the selected cells were named tiSPOC cells. All ES cells used in this study were maintained on Mitomycin C (Sigma) treated PMEF cells (mouse embryonic fibroblasts, Stem cell technologies) in DMEM supplemented with 15% inactivated fetal serum, 0.1 mM nonessential amino acids (GIBCO/BRL), 2 mM glutamine, 50 units/ml penicillin, 50 mg/ml streptomycin (GIBCO /BRL), 0.1 mM 2-mercaptoethanol (Sigma) and 1000 units/ml leukemia inhibitory factor (Chemicon International). To induce SPOC expression in ES cells, 2 mg/ml doxycycline (sigma) was added to the culture medium.
2.6. EB Differentiation and CFU Assay of tiSPOC ES Cells
ES cells were collected and grew in EBD medium [Iscove’s modified Dulbecco’s medium (IMDM), 15% IFS, 200 mg/ml ascorbic acid (sigma), 2 mM glutamine)], then plated onto 10 cm tissue culture dish for 1 hr to allow the embryonic fibroblasts to adhere on surface of dishes, the nonadherent cells were collected and plated in 10 ml of Petri dishes, changed medium after four days by collecting the EBs in 15 ml tubes (kept at RT for 5 min), resuspended precipitated EBs with fresh EBD medium and added 2 mg/ml of doxycycline to induce the expression of SPOC. EBs were dissociated on Day 6 with trypsin/EDTA and resuspended in IMDM supplemented with 10% IFS at a concentration of 5 × 105 cells per ml. A total 100 ml of suspended cells were added to 1.5 ml of 1% methylcellulose in Iscove’s MDM supplemented with 15% IFS, 1% bovine serum albumin, 10 mg/ml recombinant human insulin, 200 mg/ml human transferring (iron saturated), 10–4 M 2-Mercaptoethanol, 2 mM L-glutamine, 50 ng/ml recombinant mouse stem cell factor, 10 ng/ml recombinant mouse IL-3, 10 ng/ml recombinant human IL-6 and 3 units/ml recombinant human erythropoietin (Stem Cell Technologies, Vancouver, Catalog no. 3434). CFU colonies were counted on day 8.
2.7. Detection of Expression of Hematopoietic Related Proteins
The following PCR primers were used in this study: BMP4 (f): 5’-TGCACTTGGAACAAC AGAACAA-3’; BMP (r): 5’-CGTGCGAGGCCTGCTT-3’; Smad5 (f): 5’-GCTGAACCCCATTTCTTCTG-3’; Smad5 (r): 5’-CGTTCCAGGTTAAGATCAATGC-3’; OCT4 (f): 5’-GGCGTTCTCTTTGGAAAGGTGTTC-3’; OCT4 (r): 5’-CTCGAACCACATCCTTCTCT-3’; GATA2 (f): 5’-TGCATGCAAGAGAAGTCACC-3’; GATA2 (r): 5’-ACCACCCTTGATGTCCATGT-3’; EKLF1 (f): 5’-CCTGTTTGCTCTTCCAGAGG-3’; EKLF1 (r): 5’-TAGCAGACCAGGCTGACC TT-3’; b-ACTIN (f): 5’-GTGGGCCGCCCTAGGCACCA-3’; b-ACTIN (r): 5’CGGTTGGCCTTAGGGTTCCAGG-3’. Total RNA was prepared as previously described. One tenth of total material was used for cDNA synthesis using oligo-dT and reverse transcriptase (Quigen) in a volume of 20 ml. Real time RT-PCR was performed using 1 ml of reverse transcription product and b-actin as an internal control.
2.8. Generation of tiSPOC Chimerical Mice and Analysis of CFU Formation of Bone Marrow
In order to generate tiSPOC chimerical mice, we microinjected the tiSPOC ES cells from a single clone into mouse blastocysts (d3.5). 12 chimerical mice were identified by Southern blot analysis (data not shown). After 2 - 3 months, the bone marrow cells from chimerical mice were prepared and seeded in complete M3434 medium, CFU colonies were counted on day 8.
2.9. FACS Analysis
Disaggregated CFU cells were washed with 1XPBS and resuspended in 0.1% trypsin/1XPBS by pipetting for 30 s, removed clumps with mesh, and collected individual cells by spin, added PE-conjugated CD45/FITC-conjugated Ter119 or PE-conjugated Mac/FITC-conjugated Gr-1 antibodies and then incubated for 20 min at RT, washed 3 times with FACS buffer, and analyzed by FACS.
2.10. Statistical Analysis
The mean value and standard deviation of combined CFUs or individual CFUs from 4 or more chimerical mice were collected. p values were calculated with a Paired Student’s T Test. The hematopoietic subpopulations of bone marrow cells that either expressed or did not express SPOC domain in SPOC transgenic mice were compared by T test (A p value of <0.05 was considered to be statistically significant). Expression level of the transcriptional factors Eklf1, Bmp4, Smad5 and Gata2 in ES cells were compared with or without SPOC expression using three independent experimental data.
3. RESULTS
3.1. The SPOC is an Evolutionary Conserved and Widely Distributed Domain
Spen family members or related proteins are existed from drosophila to human, and the sizes are varied (90 - 600 kd), most of members contain repeated RNA recognition motifs (RRM) (1 - 4 RRM) near tothe N-terminus, the hMint2 doesn’t contain RRM motif but has TFIIs (transcription elongation factor IIs) binding site in Nterminus as showed in Figure 1, similar with hMint2, mDidol has Plant Homeo domain (PHD) but no RRMs in this region, all of these proteins has conserved SPOC domain near to C-terminus, the intervening region between these conserved domains shows low homology, that reflects the strain difference from evolutionary.
3.2. Coimmunoprecipitation and Subcellular Localization of SPOC Protein
Transiently transfected CHO cells were used for co-immunopreciptation, antisera to HDAC1, HDAC2, SMRT and the pre-immune IgG were separately used to coimmunoprecipitate Xpress-SPOC from whole cell extracts, following blotted with anti-Xpress antibody (Figure 2(c)). Since previously reports [10,16] show the SPOC domain of full length of human Sharp can interact with SMRT and HDAC1/2, we also detected HDAC1, HDAC2 and SMRT proteins from precipitated lysate by a-Xpress (Figure 2(c)). To determine SPOC subcellular localization, we transfected CHO cells with expression plasmids Mint-HA and Xpress-SPOC, the expressed target proteins were localized using rabbit a-HA monoclonal antibody for full length Mint and a-Xpress monoclonal antibody for SPOC, and counterstained these
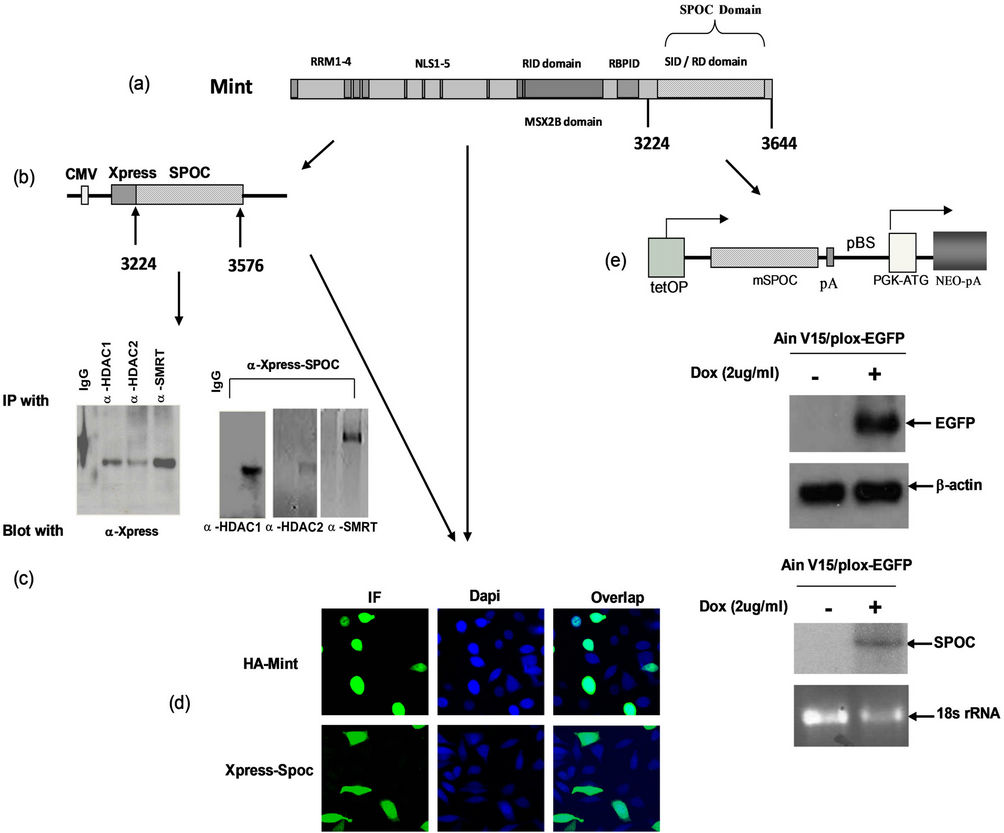
Figure 2. Expression of SPOC domain, Binding activity assays. (a) Schematic drawing represents domain structure of mouse Mint protein,RRM: RNA recognize motif; NLS: nuclear localization signal sequence; RID: Receptor interaction domain; RBPID: RBP interaction domain; SID/RD: SMRT interaction domain/Repression domain); (b) Co-immunoprecipitation to detect the interaction between SPOC domain and interaction partners of Mint protein. Constructed pcDNA4-SPOC vector to insert the mouse Mint SPOC domain and in-frame fused with epitope tag Xpress (Figure 2(b), upper panel), co-transiently transfect SPOC domain with HDAC1, HDAC2 and SMRT expression plasmids separately in CHO cells, then precipitated and blotted with corresponding antibodies as showed in the figures, precipitated the lysates with normal rabbit serum and antibodies against HDAC1, HDAC2 and SMRT respectively and blotted with anti-Xpress antibody (Figure 2(c). Lower left), precipitated the lysates with anti-Xpress and blotted with indicated antibodies separately (Figure 2(c). Lower right); (d) Subcellular localization of Mint and SPOC, CHO cells were transfected with the plasmids expressed full length HA-Mint (upper left and right) and Xpress-SPOC (bottom left and bottom right) fragment, after 24 hour transfection, cells were fixed, premobilized and immunostained with antiserum against HA for Mint and Xpress for SPOC followed by immunofluorescence, the Dapi was used to stain the nucleus as the cellular localization control; (e) Drawing represents the Tet on inducible construct of SPOC domain (Figure 2(e). top panel), (tetOP: tetra-cycline response element, SPOC cDNA: SPOC target sequence from 3224 to 3664aa of mouse Mint, pA: poly A signal sequence; pBS: plasmid sequence; PGK-ATG: PGK promoter + initiation codon, Cre-mediated recombination of targeting vectors into the genomic homing site confers resistance to the antibiotic G418 (Neo), allowing for isolation of transgenic cells. Western blot to detect the target protein EGFP expression by adding 1mg/ml of doxycycline, b-actin used as the loading control (Figure 2(e). middle panel), Northern blot to detect the target protein SPOC expression by adding 1mg/ml of doxycycline, and stained ribosomal RNA with ethidium bromide and used as the loading control (Figure 2(e), bottom panel).
cells with Dapi to show nucleus, cofocal microscopy was used to capture the images from fluorescent stained cells. The full length Mint was localized only in the nucleus (Figure 2(d), top row), whereas SPOC was localized both in the nucleus and cytoplasm (Figure 2(d), bottom row).
3.3. Establishment of Inducible ES Cell Line and Expression of SPOC Domain
We established the Tet-on inducible ES cell line by using the Tet-on targeting cell line Ain V15 [15], which constitutively expresses reverse tetracycline transactivator from ROSA 26 locus. The tetracycline response element (TRE) was integrated into the transcriptional open chromatin 5’ to the HPRT gene on the X chromosome. A single LoxP site is present at downstream of the TRE, where we integrated the SPOC sequence by Cre-Lox recombination, the resulted ES cell line, as well as its differentiated progenies were named tiSPOC. The fulllength Mint and its domains are shown in Figure 2(a), and the tet-inducible SPOC vector was showed in Figure 2(e) (top), expression of SPOC mRNA with or without doxycycline (Dox) treatment (Figure 2(e), bottom) was compared with tiEGFP (Ain V15/plox-EGFP) protein expression which used as a control (Figure 2(e), middle).
3.4. SPOC Expression Induces the Differentiation of Embryonic Cell
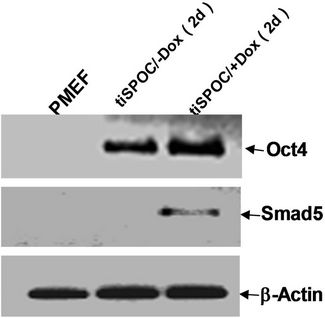
We used tiSPOC ES cells to generate EBs and applied doxycycline to the cultures for 48 hr (from day 0 of differentiation). Induction for 2 days resulted in Smad5 expression [17-19], we found there is no Smad5 expression in the control group (without Dox treatment)as shown in Figure 3(a), also no significant change was observed for Oct4 expression (octamer binding protein 4) which is a characteristic marker of ES cells [20,21] this result indicates these cells still maintenance undifferentiated state. Interestingly, Smad5 was induced by expression of SPOC domain even under this undifferentiated status. In order to identify if this induction is associated with early stage of hematopoiesis, we prepared EB and then further differentiated the EB bodies into CFU colonies using 3434 medium. The morphology of induced CFU cells was shown in Figures 3B1 to B4. Condition 1: Basic EB medium [Iscove’s modified Dulbecco’s medium IMDM, 15% IFS, 200 mg/ml ascorbic acid (sigma), 2 mM glutamine], under this conditions, 90% of cells differentiated into epithelia-like cells and are adherent to dishes (Figure 3B1); 2: Basic CFU medium plus Dox, 30% of cells were round and did not adhere to dishes (Figure 3B2); 3: Basic CFU medium plus recombinant human BMP4 protein, 40% of cells was round (Figure 3B3); 4: Basic CFU medium plus cytokine cocktails (described in methods and materials), 80% of cells were round (Figure 3B4). Further detection for these cells was performed by RT-PCR, we detected Bmp4 from Bmp4 treated group and Dox treated group, and Smd5 significantly increased (Figure 3(c)) after Dox treated. FACS sorting for induced cells were also performed to further prove the morphology observations for the hematopoietic differentiation under these conditions, stained these cells with FITC-Gr1 and FITC-Mac1 and FACS sorted to count the Gr-1 and Mac1 positive cells. Total numbers (corresponding to round cells from morphology observation, represented potential hematopoietic and hematopoietic-like cells) were compared and showed with histogram (Figure 3(d)).
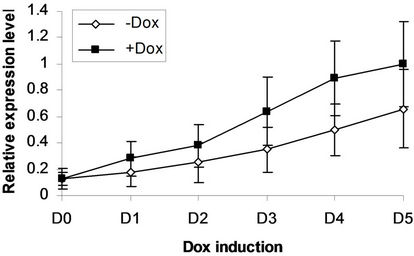
We next addressed whether the timing of SPOC expression was crucial for induction of Bmp4, Smad5, Eklf and Gata2 during hematopoiesis of embryonic stem cells. We generated EBs from tiSPOC ES cells and further cultured the CFUs with or without Dox for indicated time points, then harvested CFU cells and prepared RNA for Real time RT-PCR amplification, the expression level of target genes were normalized to an internal control (b-actin). The results indicated that Bmp4, Smad5, Gata2 and Eklf1 were significantly affected by expression of SPOC during CFU induction (Figures 4(a)-(d)). As Eklf1 and Gata2 normally come on by day 4 under CFU induction, in this inducible system the SPOC expression is earlier than Eklf1 and Gata2, then the SPOC upregulated Eklf1 and Gata2 expression around 8 and 2 fold respectively. These data indicated the expression of Smad5, Eklf1, Bmp4 and Gata2 are up regulated by SPOC inatime-dependent manner.
3.5. SPOC Induced Hematopoietic Differentiation in Bone Marrow Cells
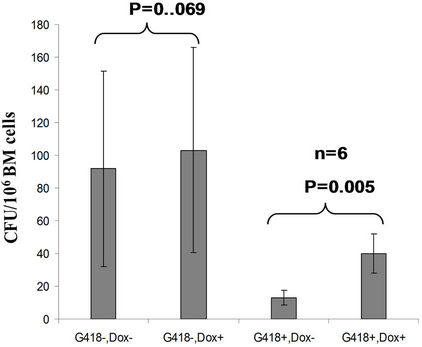
The tiSPOC chimeric transgenic mice were analyzed for the integrations of target gene by Southern blot, the result showed a single copy of the target SPOC sequence was integrated into mouse genomic DNA (data not shown). The integration percentages in founder chimeric mice were approximately 20%. The germline transmission of modified alleles was only partial function of pluripotency of the ES cells, resulted in no germ line transmission. The bone marrow cells produced from these transgenic mice and their wild-type siblings were used to induce CFU formation following standard procedure of CFU induction. Based on previous description [15] that the critical precursor of the hematopoietic and endothelial lineages, the hemangioblast, peaks at day 3.75 and is no longer present by day 5, whereas hematopoietic colony-forming cell (CFC), particularly the mixed erythroidmyeloid colonies, first become detectable between day 5 and day 6 [22], thus we applied Dox (2 mg/ml) at Day 4 of CFU induction, therefore SPOC was induced at the time point of specification of hematopoietic linage. As shown in Figure 5(a), we tested bone marrow cells (unselected with G418) from 6 of tiSPOC chimeric mice, the results showed there was no significant difference in total CFU numbers for treated with and without Dox (p > 0.05), this data was due to the fact that most populations
 (a)
(a)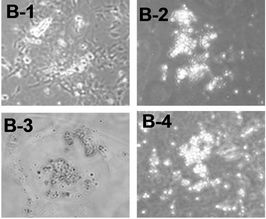 (b)
(b)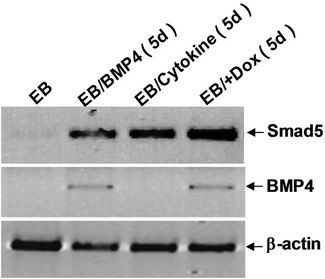 (c)
(c)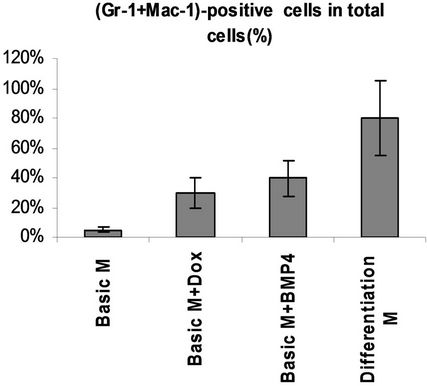 (d)
(d)
Figure 3. SPOC induces the expression of hematopoietic transcription factors during EB-CFU formation. (a) Treated tiSPOC ES cells were with Dox for 48 hr, then detected the expression of ES cell specific marker Oct4 and hematopoietic factor Smad5, (PMEF is Primary Mouse Embryo Fibroblasts, mytomycin C treated, used as the control in this experiment); (b) The effect of expression SPOC domain in cell differentiation during CFU formation. Figures B-1 to B-4 represented the morphology difference of growing cells from different CFU induction conditions, basic CFU medium (Figure B1), basic CFU medium +Dox (Figure B2), basic CFU medium + BMP4 (Figure B3) and basic CFU medium + cytokine cocktails (Figure B4) respectively, the CFU colonies were pictured from invert microscopy; (c) The tiSPOC ES cells were induced into embryonic bodies and further differentiated into CFU for 5 days with indicated culture conditions, the gene expression of Smad5 and BMP4 were detected by RT-PCR (Figure 4(c)); (d) FACS analysis of hematopoietic cells from four different mediums, the Gr-1 and Mac-1 positive cells were counted by FACS sorting and subject to compare the hematopoietic differentiation, three independent experiments data were used (mean ± SD).
in the bone marrow were derived from wild type ES cells which is no SPOC induction (not pure). After application of selection drug G418 to EB-CFU induction medium, the total CFU numbers was significant difference for treated with and without Dox (p = 0.0055), results were shown as mean ± Std (Figure 5(a)). We counted CFU numbers based on the morphology of the colonies and showed as a histogram (Figure 5(b)), the result indicated induction of SPOC expression increased all of hematopoietic lineages, including pre-B colony forming unit (CFU-preB); granulo-monocyte colony forming unit (CFUGM); granulocyte-erythrocyte-monocyte and megarkaryocyte colony forming unit (GEMM); megakaryocytic colony forming unit (CFU-M); Burst and colony forming unit erythrocyte (BFU-E/CFU-E), etc., and significant difference was determined between the groups of SPOC expression and no SPOC expression (p = 0.003). In order to confirm the increased cells, we stained the G418- positive CFU cells with PE-CD45 (a marker for all hematopoietic cells)/FITC-Ter119 (a marker for erythrocytes) (Figure 5(c)), PE-Mac (a marker for myeloid cells)/FITC-Gr1 (a marker for granulocytes) (Figure 5(d)), the Dox treated CFU cells were increased in matured erythrocytes and granulocytes from 3.48% and 2.32% to 16.5% and 13.7% respectively (Figures 5(e) and (f)) (Data represent three independent experiments, mean ±
 (a) BMP4
(a) BMP4 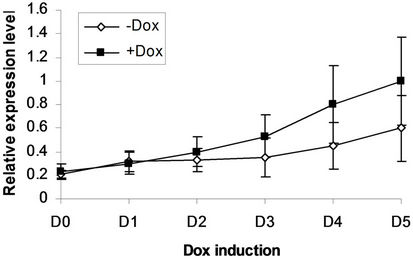 (b) Smad5
(b) Smad5
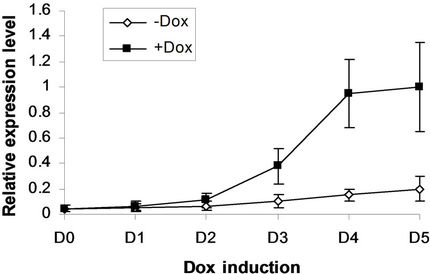 (c) EKLF
(c) EKLF 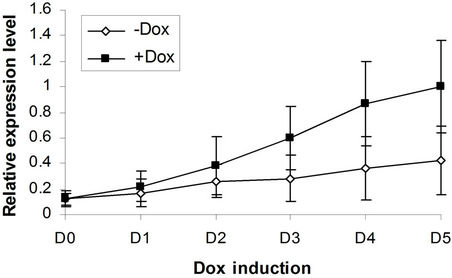 (d) Gata2
(d) Gata2
Figure 4. BMP4, Smad5, EKLF and Gata2 are all induced by SPOC expressionduring CFU formation. The EB bodies from tiSPOC ES cells were suspended and grew with standard CFU medium, applied Dox at indicated time points (from D1 to D5, and D0 is as the control of Dox treatment), harvested cells and prepared the total RNA, detected the expression level of target genes by real time RT-PCR (normalized to b-actin), the Figures (a), (b), (c) and (d) presented the relative expression level of BMP4, Smad5, EKLF1 and Gata2 respectively, three independent experiments data were used (mean ± SD).
SD). This assay confirmed that SPOC enhances hematopoietic cell maturation during ES cell induction by SPOC expression.
4. DISCUSSION
4.1. The SPOC Domain Interacts with Co-Repressor Complex to Modulate Chromatin Remodeling and Associates with Hematopoietic Differentiation
Chromatin modification by histone acetylates (HATs) or histone deacetylases (HDACs) represents one of the fundamental mechanisms for transcriptional regulation. It has been proposed that histone acetylation weakens interactions of histones with DNA, induces alterations in nucleosome structure, and enhances accessibility of targeted promoters to components of the transcription machinery, thus increasing transcription activity [23,24]. Conversely, decreased histone acetylation due to the action of HDACs is thought to lead to a less accessible chromatin conformation, resulting in repression of transcription [25,26]. Even the biochemical function of full length Mint has been described, and crystal structure analysis also indicates the surface of SPOC domain has a strikingly nonuniform charge distribution, the positive charge distribution on the surface is responsible for protein-protein interaction with the LSD motif of SMRT and NcoR, but not for protein-DNA interaction [13], therefore our study firstly showsthe SPOC domain directly interacts with corepressors SMRT, HDAC1 and HDAC2, it means SPOC sequence possess the critical function of full length Mint protein.
The retinoid signaling acts through heterodimers consisting of retinoic acid receptor and retinoid X receptor (RAR/RXR), plays a crucial role during hematopoietc cell differentiation, the RAR/RXR heterodimers bind to naturally occurring response elements in the promoter regions of target genes, determining whether the gene is transcribed, in the absence of RAR-specific ligand all trans retinoic acid, RAR/RXR heterodimers are associated with the nuclear receptor corepressor N-CoR and SMRT and acts as repressor, both natural RAR/RXR and fusion protein PML-RAR or PLZFRAR repress transcription by recruiting HDAC complex via interaction with the nuclear repressors NcoR and SMRT, which directly contact HDACs [27], thus we speculate SPOC domain plays a role for hematopotic gene expression by the interaction with SMRT and HDAC1/2. SMRT and
 (a)
(a)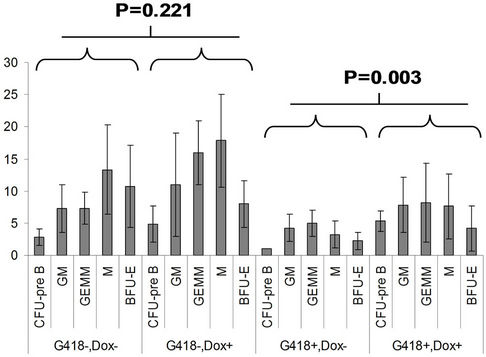 (b)
(b)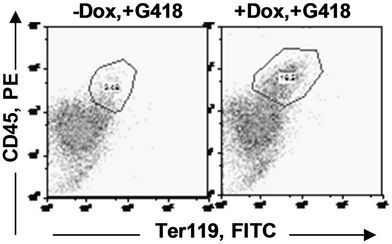 (c)
(c)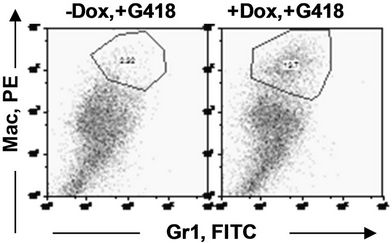 (d)
(d)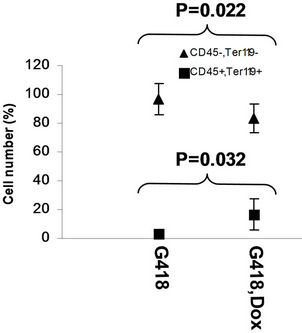 (e)
(e)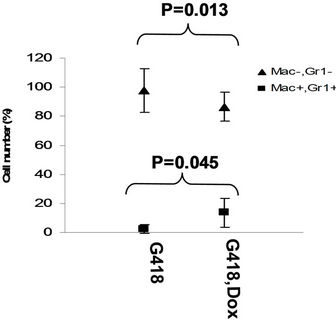 (f)
(f)
Figure 5. SPOC expression enhances hematopoietic differentiation of mouse bone marrow cells. The bone marrow cells of tiSPOC chimeric transgenic mice were used in this assay. (a) The total CFU numbers of four groups (G418−/Dox−, G418−/Dox+, G418+/Dox− and G418+/Dox+) were showed respectively, standard errors for six-independent experiments were showed for combined CFU, (N = 6 for each bar; p > 0.05 for unselected group and p < 0.05 for G418 selected group); (b) Comparison of hematopoietic CFU colony numbers counted from four different experimental groups, three independent experimental data were used (mean ± SD), the BFU-E, CFU-G, Pre-B, CFU-GM, CFU-GEMM, CFU-M were counted in this study, there is no significant difference between G418−/Dox− and G418-/Dox+ (p = 0.22 > 0.05), but G418+/Dox− is statistically different from G418+/Dox+ (p = 0.003 < 0.05); (c) FACS Characterization of hematopoietic differentiation promoted by SPOC expression, the bone marrow cells (G418 positive cells) of tiSPOC mice were treated with or without Doxycycline for 8 days, stained the cells with PE conjugated CD45 and FITC conjugated Ter119; (d) Stained the cells (same procedure as above description) with PE conjugated Mac1 and FITC conjugated Gr1, and then performed FACS analysis; (e) Analyzed CD45+/ Ter119+ cell differentiation caused by expression of SPOC domain in bone marrow cells, hematopoietic populations from Dox treated or untreated are significantly difference (p = 0.022); (f) Analyzed Mac+/Gr1+ cells induced by expression of SPOC fragment domain, hematopoietic populations from Dox treated or untreated are significantly difference (p = 0.013).
N-CoR function is not merely as repressors of basal transcription, but as modulators of both basal and ligandactivated transcription of genes controlled by RAR /RXR heterodimers in a dose-dependent manner [28], the binding between SPOC and SMRT may alter the binding activities of SMRT with the RAR/RXR heterodimer in vivo, resulted in enhancing hematopoietic differentiation. Five different fusion partners of RARa have been identified, including promyelocytic leukemia protein (PML) [29], promyelocytic leukemia zinc finger protein (PLZF) [30], nucleophosminNPM [31], nuclear mitotic apparatus (Numa), and the Stat5b gene [32]. These fusion proteins regulate hematopoiesis by similar mechanisms, in which SPOC may play a role. On the other hand, there is a specific RAR binding site in Mint protein that indicates Mint also directly binds to RAR/RXR complex. It is not clear the relationship between these two interactions and how they synergistically control hematopoietic differentiation. More work is necessary to elucidate the molecular mechanisms controlled by SPOC domain and RAR/RXR domain of Mint protein. In summary, the SPOC domain associates with several critical cellular processes, and plays an important role in cell proliferation and differentiation.
4.2. SPOC Domain is Involved in Bmp4/Smad5 Pathway and Regulates ES Cell Differentiation and Early Hematopoiesis
Embryonic stem (ES) cells derived from inner cell mass of mammalian blastocysts grow rapidly and infinitely while maintaining pluripotency, several transcription factors have been shown to be essential for pluripotency, including Nanog, Oct 3/4, and POU family members specifically express in ES cells, preimplantation embryos, epiblast, and germ cells [21,33]. We used Oct 4 as an ES cell marker to determine the status of differentiation in our experiments. Previous studies [18,19,34] indicated upregulation of Smad5 can serve as a marker for hematopoiesis, Smad5 is involved in the intracellular signaling pathways that mediates the regulation of bifunctional regulator TGF-b in the proliferation of hematopoietic progenitor. Our study showed expression of Smad5 is induced in tiSPOC ES cells after 2 days of treatment with Dox (Figure 3(c)), but there are no similar phenomena for −Dox group, this result indicates expression of SPOC can initiate intracellular Bmp4/Smad5 signaling pathway to promote hematopoietic differentiation. To further detect if early Smad5 activation resultes in the expression of hematopoietic genes, we treated tiSPOC ES cells to induce EB-CFU differentiation, and treated these cells from CFU formation using Dox (Figure 4), the results showed SPOC induces the Bmp4, Smad5, Eklf and Gata2 expression during CFU formations, and Bmp4 is induced in earliest time points compared to other three proteins, the Eklf is the latest protein induced by SPOC expression, Smad5 and Gata2 are induced in similar stage, these observations strongly support the SPOC domain play a crucial role in hematopoiesis, as Gata2 plays a critical role in the early stages of hematopoiesis by regulating the genes that control growth factor responsiveness and capacity for proliferate [35,36], therefore, the increase in Gata2 level was likely responsible for the enhanced differentiation of pluripotent hematopoietic stem cells into hematopoietic cells. Furthermore, the upregulation of Eklf1 and Gata2 is directly controlled by activation of Bmp4/Smad5 pathway in hematopoietic differentiation [37,38], and our data showed expression of SPOC induces Gata2 and Eklf1, so we conclude that SPOC regulates hematopoietic differentiation via the Bmp4/Smad5 pathway. Interestingly, the cytokine SCF/ IL3/IL6/ did not induce Bmp4 expression in our experiment, thereafter the Bmp4/Smad5 pathway that regulates hematopoietic differentiationis independence from SCF/ IL3/IL6.
4.3. Overexpressed Full Length Mint Induces Smad5 and Bmp4 Expression in ES Cells
Since SPOC can induce the Bmp4/Smad5 expression, we are interested if full length Mint can also induce these proteins expression, we overexpressed full length Mint protein by transiently transfecting HA-Mint construct into ES cells (see Figure 7(b)). Smad5 is significantly upregulated after 48 hours Mint transfection, and Bmp4 was induced after 24 hours, indicating Bmp4 protein level can be immediately up regulated by full length Mint expression. These results were consistent with the finding that SPOC domain induces Bmp4 and Smad5 in bone marrow cells of SPOC transgenic mice. This study also supported that SPOC regulates hematopoiesis via the Bmp4/Smad5 pathway, therefore we conclude SPOC domain has a similar function with full length Mint protein for hematopoiesis.
4.4. SPOC Enhanced CFU Formation and Hematopoietic Maturation in Vitro
Our studies in cultured ES cells proved that SPOC drives hematopoietic differentiation via the Bmp4/Smad5 pathway, however, it is important to show if the primary bone marrow stem cells derived from tiSPOC chimeric mice can also be differentiated into hematopoietic cells with a similar pattern. In contrast to cultured ES cells, bone marrow cells can be easily induced to produce all hematopoietic lineages and the ex vivo experiment will give a direct evidence for the SPOC function, using transgenic model of induced cell, we found expression of SPOC enhances CFU formation and hematopoietic maturation, therefore this study offered us with a new view for
 (a)
(a)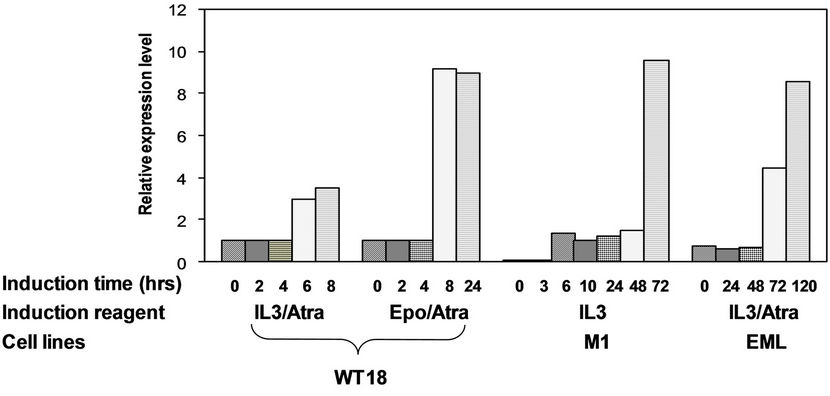 (b)
(b)
Figure 6. Effect of differentiation inducers of hematopoietic cells on Mint expression level. (a) Left panel: WT18 cells were treated with IL3/Atra or Epo/ATRA for indicated time points (hours), Middle panel: M1 cells were treated with IL3 for indicated time points (hours), Right panel: EML cells were treated with IL3/Atra for indicated time points (hours). Total RNA was prepared and analyzed by Northern blot, ethidium bromide-stained ribosomal RNA were used to indicate the loading amount of each wells; (b) Semi-quantitative analysis of relative expression level for Mint gene (normalized to 28S ribosomal RNA, for further details, see materials and methods).
 (a)
(a)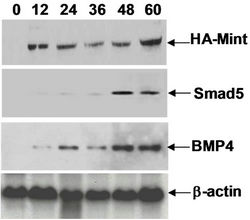 (b)
(b)
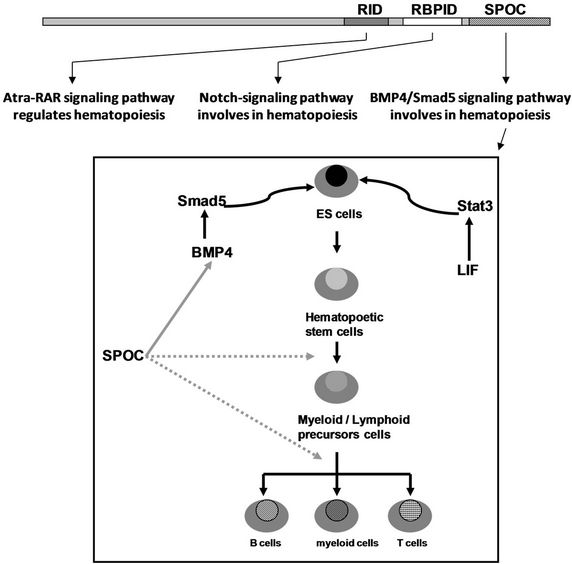
Figure 7. (a) Working model for activation of hematopoiesis by SPOC domain. RID, RBPID and SPOC domain of Mint protein involve in different signaling pathways that regulate hematopoiesis and stem cell differentiation. The RID domain (retino acid receptor interaction domain) interacts with retinoic acid receptor and thus regulates the hematopoietic differentiation of ES or bone marrow cells, the RBPID domain (RBPJk interaction domain) plays role by regulation of Notch signaling that is a critical pathway for cell fate decision during hematopoietic differentiation, the SPOC domain interacts with SMRT, HDAC and other corepressors, and these interactions regulate ES cell and bone marrow cell differentiation and hematopoiesis through epigenetic remodeling to activate the target genes (for example: Bmp4, Smad5); (b) Detected Smad5 and Bmp4 induction in HA-Mint transfected Ain V15 ES cells by Western blot, b-actin were used as loading control.
the function of SPOC domain and full length Mint protein. Our studies have provided the evidence that SPOC activates hematopoietic factors Gata2 and Eklf1, and associated factors Bmp4 and Smad5, finally, SPOC domain is apparently functional in a variety of molecular niches, involves in controlling hematopoiesis and ES cell differentiation (See Figure 7(a)). It is interesting that the Mint protein has three domains (SPOC, RBPID and RID) involve in three different pathways that have the potential ability to regulate the same cellular procedure (hematopoietic differentiation). The SPOC domain enhances hematopoietic differentiation via interaction with corepressor components (HDAC1, 2; Sin3A and SMRT) [7,12]. The RBPID domain in the middle of Mint protein regulates hematopoietic differentiation through interaction with RBPJk, which is a critical regulator for the Notch signaling pathway, since it is well known that Notch is involved in hematopoiesis [39,40], Mint also inhibit Notch signaling by binding to RBPJk [8,9,11], activation of Gata2 expression by Notch1/RBPJk pathway is a crucial event for the onset of definitive hematopoiesis in embryonic development [41]. The RID domain plays role in hematopoietic cell differentiation via the ATRA/RA pathway, the retinoid signaling acts through heterodimers consisting of the retinoic acid receptor and the retinoid X receptor (RAR/RXR), also playing a crucial role in hematopoiesis. Future investigation will be necessary to elucidate how these pathways synergistically regulate hematopoiesis in bone marrow cells.
5. ACKNOWLEDGEMENTS
We thank Dr. Diane Krause for her generous and excellent help. We also thank Dr. Kyba M for kindly providing the Ain V15 ES cell line, Professor Bernard G Forget, Nancy Berliner, and Khanna-Gupta, A. for critical discussions during this research.
REFERENCES
- Wiellette, E.L., Harding, K.W., et al. (1999) Spen encodes an RNP motif protein that interacts with Hox pathways to repress the development of head-like sclerites in the drosophila trunk. Development, 126, 5373- 5385.
- Chen, F. and Rebay, I. (2000) Split ends, a new component of the drosophila EGF receptor pathway, regulates development of midline glial cells. Current Biology, 10, 943-946. doi:10.1016/S0960-9822(00)00625-4
- Kuang, B., Wu, S.C., et al. (2000) Split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension in the Drosophila embryo. Development, 127, 1517-1529.
- Lin, H.V., Doroquez, D.B., et al. (2003) Splits ends is a tissue/promoter specific regulator of Wingless signaling. Development, 130, 3125-3135. doi:10.1242/dev.00527
- Kolodziej, P.A., Jan, L.Y., et al. (1995) Mutations that affect the length, fasciculation, or ventral orientation of specific sensory axons in the Drosophila embryo. Neuron, 15, 273-286. doi:10.1016/0896-6273(95)90033-0
- Icht, J.D., Chomienne, C., et al. (1995) Clinical and molecular characterization of a rare syndrome of acute promyelocytic leukemia associated with translocation (11; 17). Blood, 85, 1083-1094.
- Shi, Y., Downes, M., et al. (2001) Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes & Development, 15, 1140-1151. doi:10.1101/gad.871201
- Oswald, F., Kostezka, U., et al. (2002) SHARP is a novel component of the Notch/RBP-Jkappasignalling pathway. The EMBO Journal, 21, 5417-5426. doi:10.1093/emboj/cdf549
- Kuroda, K., Han, H., et al. (2003) Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity, 18, 301-312. doi:10.1016/S1074-7613(03)00029-3
- Tsuji, M., Shinkura, R., et al. (2007) “Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/ RBP-J signaling. Proceedings of the National Academy of Sciences USA, 104, 1610-1615. doi:10.1073/pnas.0610520104
- Ma, X., Renda, M.J., et al. (2007) Rbm15 modulates notch-induced transcriptional activation and affects myeloid differentiation. Molecular and Cellular Biology, 27, 3056-3064. doi:10.1128/MCB.01339-06
- Newberry, E.P., Latifi, T., et al. (1999) The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry, 38, 10678-10690. doi:10.1021/bi990967j
- Sanchez-Pulido, L., Rojas, A.M., et al. (2004) SPOC: A widely distributed domain associated with cancer, apoptosis and transcription. BMC Bioinformatics, 5, 91. doi:10.1186/1471-2105-5-91
- Fischer, U., Struss, A.K., et al. (2001) PHF3 expression is frequently reduced in glioma. Cytogenetics and Cell Genetics, 94, 131-136. doi:10.1159/000048804
- Kyba, M., Perlingeiro, R.C., et al. (2002) HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell, 109, 29-37. doi:10.1016/S0092-8674(02)00680-3
- Ariyoshi, M. and Schwabe, J. W. R (2003) A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes & Development, 17, 1909- 1920. doi:10.1101/gad.266203
- Bruno, E., Horrigan, S.K., et al. (1998) The Smad5 gene is involved in the intracellular signaling pathways that mediate the inhibitory effects of transforming growth factor-beta on human hematopoiesis. Blood, 91, 1917- 1923.
- Fuchs, O., Simakova, O., et al. (2002) Inhibition of Smad5 in human hematopoietic progenitors blocks erythroid differentiation induced by BMP4. Blood Cells, Molecules and Diseases, 28, 221-233. doi:10.1006/bcmd.2002.0487
- Liu, B., Sun, Y., et al. (2003) Disruption of Smad5 gene leads to enhanced proliferation of high-proliferative potential precursors during embryonic hematopoiesis. Blood, 101, 124-133.
- Scholer, H.R., Balling, R., et al. (1989) Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. The EMBO Journal, 8, 2551-2557.
- Okamoto, K., Okazawa, H., et al. (1990) A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell, 60, 461-472. doi:10.1016/0092-8674(90)90597-8
- Choi, K., Kennedy, M., et al. (1998) A common precursor for hematopoietic and endothelial cells. Development, 125, 725-732.
- Roth, S.Y. and Allis C.D. (1996) Histone acetylation and chromatin assembly: A single escort, multiple dances? Cell, 87, 5-8. doi:10.1016/S0092-8674(00)81316-1
- Uteley, R.T., Ikeda, K., et al. (1998) Transcriptional factors direct histone acetyltransferase complexes to nucleosomes. Nature, 394, 498-502. doi:10.1038/28886
- Grunstein, M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349-352. doi:10.1038/38664
- Kadonaga, J.T. (1998) Eukaryotic transcription: An interlaced network of transcription factors and chromatinmodifying machines. Cell, 92, 307-313. doi:10.1016/S0092-8674(00)80924-1
- Minucci, S., Nervi, C., et al. (2001) Histone deacetylases: A common molecular target for differentiationtreatment of acute myeloid leukemias? Oncogene, 20, 3110-3115. doi:10.1038/sj.onc.1204336
- Loinder, K. and Soderstrom, M. (2003) The nuclear receptor corepressor (N-CoR) modulates basal and activated transcription of genes controlled by retinoic acid. The Joural of Steroid Biochemistry & Molecular Biology, 84, 15-21. doi:10.1016/S0960-0760(03)00007-4
- Lin, R.J. and Evans, R.M. (2000) Acquisition of oncogenic potential by RAR chimeras in acute promyelocytic leukemia through formation of homodimers. Molecular Cell, 5, 821-830. doi:10.1016/S1097-2765(00)80322-6
- Petti, M C., Fazi, F., Gentile, M., et al. (2002) Complete remission through blast cell differentiation in PLZF/ RARα-positive acute promyelocytic leukemia: In vitro and in vivo studies. Blood, 100, 1065-1067. doi:10.1182/blood-2001-12-0368
- Redner, R.L., Rush, E.A., et al. (1996) The t(5;17) variant of acute promyelocytic leukemia expresses a nucleophosmin-retinoic acid receptor fusion. Blood, 87, 882- 886.
- Arnould, C., Philippe, C., et al. (1999) The signal transducer and activator of transcription STAT5b gene is a new partner of retinoic acid receptor alpha in acute promyelocytic-like leukaemia. Human Molecular Genetics, 8, 1741-1749. doi:10.1093/hmg/8.9.1741
- Schöler, H.R., Ruppert, S., Suzuki, N., Chowdhury, K. and Gruss, P. (1990) New type of POU domain in germ line-specific protein Oct-4. Nature, 344, 435-439. doi:10.1038/344435a0
- Singbrant, S., Moody, J.L., et al. (2006) Smad5 is dispensable for adult murine hematopoiesis. Blood, 108, 3707-3712. doi:10.1182/blood-2006-02-003384
- Tsai, F.Y., Keller, G., et al. (1994) An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature, 371, 221-226. doi:10.1038/371221a0
- Lepage, A., Uzan, G., et al. (1999) Functional characterization of the human platelet glycoprotein V gene promoter: A specific marker of late megakaryocytic differentiation. Blood, 94, 3366-3380.
- Crossley, M., Tsang, A.P., Bieker, J.J. and Orkin, S.H. (1994) Regulation of the erythroid Kruppel-like factor (EKLF) gene promoter by the erythroid transcription factor GATA-1. The Journal of Biological Chemistry, 269, 15440-15444.
- Adelman, C.A., Chattopadhyay, S., et al. (2002) The BMP/BMPR/Smad pathway directs expression of the erythroid-specific EKLF and GATA1 transcription factors during embryoid body differentiation in serum-free media. Development, 129, 539-549.
- Bigas, A., Martin, D.I., et al. (1998) Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Molecular and Cellular Biology, 18, 2324- 2333.
- Carlesso, N., Aster, J.C., et al. (1999) Notch1-induced delay of human hematopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. Blood, 93, 838-848.
- Robert-Moreno, A., Espinosa, L., et al. (2005) “RBP jkappa-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development, 132, 1117-1126. doi:10.1242/dev.01660
NOTES
*This work was support by grant from the NIH (to D.S.K).
#Corresponding author.

