American Journal of Molecular Biology
Vol.2 No.1(2012), Article ID:16547,10 pages DOI:10.4236/ajmb.2012.21002
SPT5 affects the rate of mRNA degradation and physically interacts with CCR4 but does not control mRNA deadenylation*
![]()
Department of Molecular, Cellular, and Biomedical Sciences, University of New Hampshire, Durham, NH, USA
Email: cldenis@unh.edu
Received 12 August 2011; revised 16 September 2011; accepted 25 September 2011
Keywords: mRNA Degradation; CCR4-NOT Complex; SPT5/SPT4; Deadenylation
ABSTRACT
The CCR4-NOT complex has been shown to have multiple roles in mRNA metabolism, including that of transcriptional elongation, mRNA transport, and nuclear exosome function, but the primary function of CCR4 and CAF1 is in the deadenylation and degradation of cytoplasmic mRNA. As previous genetic analysis supported an interaction between SPT5, known to be involved in transcriptional elongation, and that of CCR4, the physical association of SPT5 with CCR4 was examined. A two-hybrid screen utilizing the deadenylase domain of CCR4 as a bait identified SPT5 as a potential interacting protein. SPT5 at its physiological concentration was shown to immunoprecipitate CCR4 and CAF1, and in vitro purified SPT5 specifically could bind to CAF1 and the deadenylase domain of CCR4. We additionally demonstrated that mutations in SPT5 or an spt4 deletion slowed the rate of mRNA degradation, a phenotype associated with defects in the CCR4 mRNA deadenylase complex. Yet, unlike ccr4 and caf1 deletions, spt5 and spt4 defects displayed little effect on the rate of deadenylation. They also did not affect decapping or 5' - 3' degradation of mRNA. These results suggest that the interactions between SPT5/SPT4 and the CCR4-NOT complex are probably the consequences of effects involving nuclear events and do not involve the primary role of CCR4 in mRNA deadenylation and turnover.
1. INTRODUCTION
The levels of eukaryotic mRNA can be regulated at many steps, from its initial synthesis to its eventual degradation in the cytoplasm. Regulation of the rate of mRNA decay is therefore an important control point in determining the abundance of cellular mRNA. A principal mRNA decay pathway in eukaryotic cells is initiated by shortening of the mRNA poly(A) tail at the 3′ end (deadenylation), followed by removal of the 5′ cap structure (decapping) and subsequent 5′ to 3′ exonuclease digestion [1-3]. Deadenylation is a key rate-limiting step in the decay of mRNA in that it is the initial step for mRNA degradation. The rate of mRNA deadenylation also plays the most important role in affecting the overall mRNA turnover rate [4], and the poly(A) tail can stimulate the translation of mRNA [5]. Regulation of poly(A) tail length can therefore play multiple roles in controlling protein expression.
In yeast, the evolutionarily conserved CCR4 protein, as part of the CCR4-NOT complex, has been identified as the major cytoplasmic deadenylase [6-8]. Deletion of both CCR4 and PAN2, a poly(A) nuclease that processes the poly(A) tails of mRNA [9], completely blocks deadenylation in vivo [8], indicating that these two deadenylases comprise the vast majority of deadenylation function in yeast. Yet, the CCR4-NOT complex acts at several junctures in the formation of mRNA. Not only can the CCR4-NOT proteins, as the cytoplasmic deadenylase complex, act to control the degradation of mRNA [6-8,10,11], these factors also function in repressing the initiation of transcription through TFIID contacts [12,13], in transcriptional elongation [14,15], in aspects of transcriptioncoupled DNA repair [16], in regulating nuclear exosome function [17], in controlling mRNA export [18], and in the activation of transcription [19-22]. Consistent with their multiple functions, the CCR4-NOT proteins have been localized to both nuclear and cytoplasmic compartments [7,8].
The CCR4-NOT complex consists of two principal forms, 1.9 × 106 daltons (1.9 MDa) and 1.0 MDa in size [21,23]. The core 1.0 MDa complex has been purified and shown to consist of CCR4, CAF1, the five NOT proteins (NOT1-5), CAF40, CAF130, and BTT1 [24,25]. The 1.9 MDa complex consists of this core and possibly several other proteins [21,26]. Within the 1.0 MDa complex the arrangement of factors has been delineated. NOT1 is the central protein in the complex [23,27]. CAF1 binds to the middle region of NOT1 and is absolutely required for linking CCR4 to the NOT1-5 proteins [21,23]. The NOT2, -4, and -5 proteins, in contrast, bind to the C-terminal region of NOT1. CAF130, BTT1, and CAF40 are located off of NOT1 in regions separate from CAF1/CCR4 and from NOT2/NOT4/NOT5 [24,25]. The multiple sizes of the CCR4-NOT complex, the numerous proteins present in it, and the distinct architecture of components within the 1.0 MDa complex are consistent with the complex, through its various interactions, playing multiple roles within the cell.
Biochemical and bioinformatic studies have shown that CCR4 protein contains three major functional domains: an N-terminal activation domain that may interact with the transcriptional machinery, a central leucine-rich repeat (LRR) domain that binds CAF1 and several other putative components of the 1.9 MDa CCR4-NOT complex and is absolutely required for CCR4 enzyme function, and a C-terminal exonuclease III like domain that comprises the CCR4 deadenylase function [6,7,26,28- 30].
Previously, genetic analysis had indicated a functional connection between the CCR4-NOT complex and SPT5 [14]. For example, a ccr4 deletion or overexpression of NOT1 suppressed the cold-sensitive phenotype of the spt5-242 allele [14]. SPT5 forms a complex with the SPT4 protein [31], called DSIF in mammalian cells [32]. SPT5/SPT4, like CCR4-NOT complex, is conserved throughout eukaryotes and has been found to be involved in multiple steps of mRNA processing. SPT5/SPT4 works in conjunction with both the positive transcription elongation factor b (P-TEFb) and RNA pol II to control transcriptional elongation [31,33,34]. SPT5/SPT4 has also been found to be involved in pre-mRNA processing and with the exosome [35,36]. These various functions indicate a role for SPT5/SPT4 in pre-mRNA processing We report here that SPT5/SPT4 can co-immunoprecipitate CCR4 and CAF1. Correspondingly, defects in SPT5 or SPT4 affected the rate of mRNA degradation. However, because these same defects did not display large effects on mRNA deadenylation, the SPT5/SPT4 effects on mRNA degradation and interactions with the CCR4-NOT complex may not be directly related to the primary function of CCR4 in mRNA deadenylation. Instead, these observations suggest that links between SPT5 and CCR4 may result from other interactions that regulate mRNA expression, processing, or transport in the nucleus.
2. MATERIALS AND METHODS
2.1. Yeast Strains and Growth Conditions
Yeast strains are listed in Table 1. Growth conditions were on YEP medium (2% yeast extract/1% bactopeptone) supplemented with the appropriate carbon source as indicated below. Two-hybrid assays were conducted in diploid strain EGY188/EGY191 containing the p34 plasmid (8 LexA operators upstream of lacZ) following growth on minimal medium [21] lacking uracil, tryptophan, and histidine that was supplemented with 2% galactose/2% raffinose. Standard errors of the means for the assays were less than 20%. The plasmid pCAF1 is YEp13- CAF1-U whose LEU2 gene is now URA3.
2.2. Immunoprecipitations, in Vitro Binding Asays, and Western Analysis
Immunopreciptations with anti-Flag antibody were conducted as previously described [21,37]. Western analysis utilized standard procedures [28]. GST fusion proteins were isolated from E. coli as described previously [38] and used to bind Flag purified SPT5-Flag isolated as described [6].
2.3. RNA Analysis
Quantitative S1 nuclease protection assays were conducted as described for determining mRNA degradation rates [39]. Control experiments in each case indicated that at the concentration of S1 nuclease used, no radioactively labeled oligonucleotide remained if no RNA was present and that the S1 nuclease assay was linear over the concentration of RNAs used. Total RNA were purified as described previously [40,41]. Oligonucleotides were radiolabelled at their 5′ end with T4 polynucleotide kinase as described [6].
The deadenylation rates and deadenylation end points for GAL1 and MFA2pG mRNA were determined by using a transcriptional pulse-chase and RNase H assay as described previously [1,11,42-44]. For S1 analysis, the ADH2 probe contained the sequence +1126 to 1167 of ADH2 and the ACT1 probe contained the sequence +1039 to +1098 of ACT1. For GAL1, a 3′ GAL1 probe was used for S1 analysis [42]. A phosphorimager was used to quantitate the relative densities of the ADH2 or GAL1 mRNA levels as compared to the control ACT1.
3. RESULTS
3.1. SPT5 Can Interact Physically with CCR4
Previously, we had conducted a two-hybrid screen for proteins interacting with the C-terminal deadenylase domain of CCR4 (residues 495-837) and had identified a small piece of SPT5 (residues 137-217) that could interact with

Table 1. Yeast strains.
LexA-CCR4 (495-837) (31 U/mg b-galactosidase activity for LexA-CCR4 with B42-SPT5 (SPT5 fused to the E. coli transactivity domain B42) [21] as compared to 3.4 U/mg of activity for LexA-CCR4 with B42 alone). To verify thatthe above described two-hybrid interaction was the result of an in vivo physical association, immunoprecipitation analysis was conducted using full-length SPT5. The full length version of SPT5 when it was fused to B42 was capable of immunoprecipitating LexA-CCR4 (495-837) (Figure 1, lane 6). These data indicate that the CCR4 exonuclease domain can contact SPT5. Other comparable LexAfusion proteins, such as LexA-MOB1 or LexA-VpU were not immunoprecipitated by B42- SPT5 (Figure 1, lanes 4 and 5) and previous data has shown that CCR4 does not interact with the B42 domain alone [28].
Because these above experiments were conducted with overexpressed versions of SPT5 and CCR4, we subsequently used the SPT5-Flag allele present at a single copy at its natural location in the genome to reconduct these immunoprecipitations [31]. Immunoprecipitating SPT5-Flag with anti-Flag antibody resulted in CCR4 being co-immunoprecipitated (Figure 2, lane 7). The protein that was immunoprecipitated was identified as CCR4 as no protein co-immunoprecipitated from a strain deleted for CCR4 (lane 9). Also, a caf1 deletion eliminated the ability of CCR4 to co-immunoprecipitate with SPT5-Flag (lane 8), indicating that the CCR4 contact to at least the CAF1 component of the CCR4-NOT complex was required for CCR4 to be brought down with SPT5-
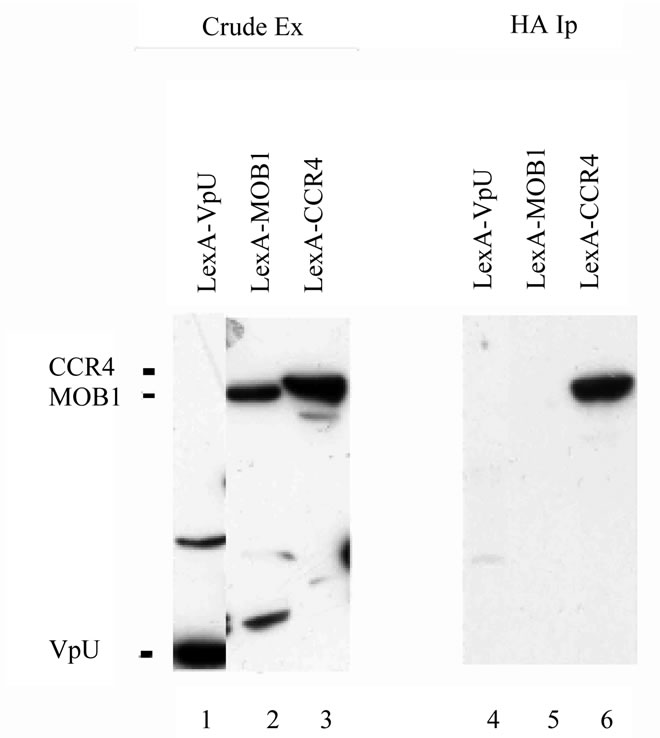
Figure 1. SPT5 co-immunoprecipitates with CCR4- NOT components. Immunoprecipitations were conducted in diploid strain EGY188/EGY191 containing the B42 and LexA fusion proteins as indicated with the HA1 antibody that recognized the HA1 epitope fused to the B42 transactivation domain. Western analysis utilized the anti-LexA antibody. Crude extracts-lanes 1 - 3; HA1 immunoprecipitateslanes 4 - 6. B42-SPT5 contained full length SPT5; LexA fusions contained full length LexA (1 - 202) fused to complete versions of the indicated protein except for CCR4 (residues 495 - 837 were present).
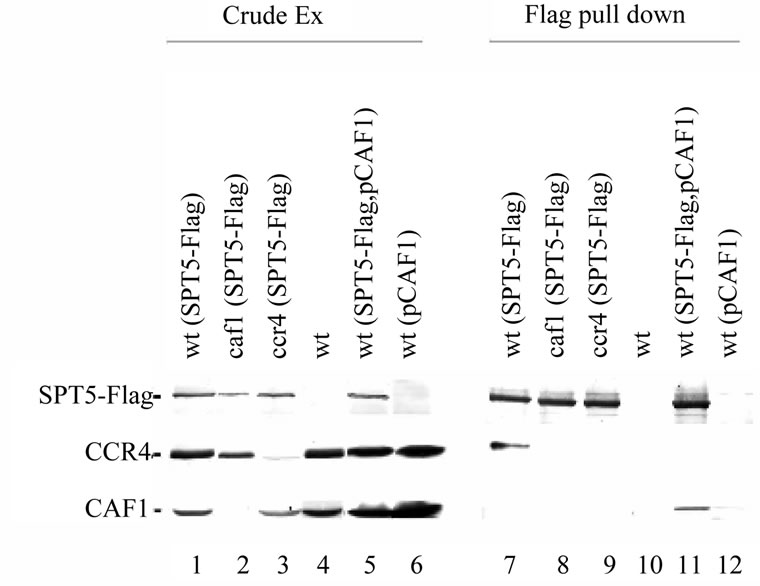
Figure 2. SPT5 immunoprecipitates CCR4 at their physiological concentrations. Immunoprecipitations were conducted with anti-Flag antibody. Crude extracts: lanes 1 - 6; Flag immunoprecipitates lanes 7 - 12. Anti-CCR4, -CAF1, and Flag antibodies were used to identify CCR4, CAF1, and SPT5-Flag, respectively. Lanes 1 and 7: strain FY1642 (wt); lanes 2 and 8: strain FY1642-c1 (caf1); lanes 3 and 9: FY1642-1a (ccr4); lanes 4 and 10: strain FY1639 (no SPT5-Flag); lanes 5 and 11: strain FY1642 with pCAF1 (YEP13-CAF1-U); lanes 6 and 12: strain FY1639 with pCAF1.
Flag. As an additional control, Flag antibody did not coimmunoprecipitate CCR4 from a strain lacking SPT5- Flag (lane 10). These results confirm that CCR4 at its physiological concentration can associate with SPT5.
Although no CAF1 protein appeared to co-immunoprecipitate with SPT5-Flag (Figure 2, lane 7), we were concerned that we might have missed observing CAF1 in the immunoprecipitate if the titer of the antiCAF1 antibody were insufficient to detect the low levels of CAF1 that might be associated with SPT5. To ensure our ability to detect CAF1 we expressed CAF1 on a high copy plasmid under the control of its own promoter. The abundance of CAF1 protein made when CAF1 is expressed from this high copy plasmid was about 3-to 5-fold more than found in a wild-type CAF1 strain (Figure 2, compare lanes 1 to 5 or 4 to 6). When SPT5-Flag was immunoprecipitated with anti-Flag antibody from a strain carrying plasmid expressing CAF1, CAF1 was coimmunoprecipitated (Figure 2, lane 11). Although we observed some CAF1 being immunoprecipitated with Flag antibody from a strain lacking SPT5-Flag, the amount was reproducibly less than observed when SPT5- Flag was present (compare lanes 11 to 12, Figure 2). While the amount of CAF1 immunoprecipitating with SPT5 is low, these results indicate that CAF1, like CCR4, can associate with SPT5. It is worth nothing, however, that overexpression of CAF1 interfered with SPT5 coimmunoprecipitating CCR4. Since nearly all of the CCR4 found in the cell is present in the CCR4-NOT complexes, the inability of SPT5 to apparently co-immunoprecipate other components of the complex along with CCR4 may imply that immunoprecipitating SPT5 helps separate CCR4 from the remainder of the complex or makes CCR4 less stably associated with other components of the complex.
We also conducted the reverse experiment. When CCR4 or CAF1 was immunoprecipitated, no SPT5-Flag protein was found to co-immunoprecipitate with the CCR4-NOT complex components (data not shown). This result is expected, however, as it has been previously established that immunoprecipitating the CCR4-NOT complex, when expressed at its physiological concentration, does not immunoprecipitate any proteins other than those which have been identified in the core 1.0 MDa CCR4-NOT complex [21,23,24].
3.2. SPT5 Does Not Associate in CCR4-NOT Complexes
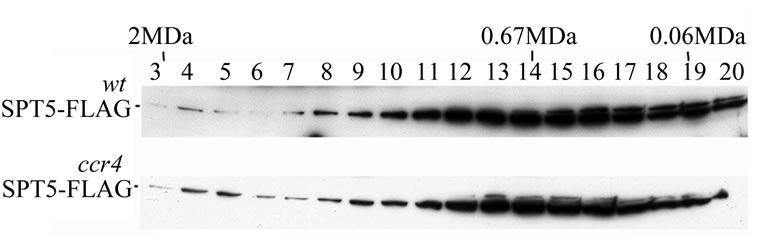
The above data indicates that SPT5 can physically interact with CCR4 and CAF1. SPT5 is unlikely, however, to be a component of the 1 MDa CCR4-NOT complex since neither SPT5 nor SPT4 were detected in the purified 1 MDa complex [24]. It remained possible that SPT5 could be a component of the 1.9 MDa CCR4-NOT complex. To address this issue we analyzed the migration of SPT5 in crude extracts following Superose 6 gel chromatography. The majority of SPT5 was found to migrate at about 0.7 MDa (Figure 3(a)), albeit a small amount was found to migrate at 1.9 MDa. We examined whether the SPT5 migrating at 1.9 MDa was affected by deleting CCR4- NOT factors. SPT5 migration at 1.9 MDa was not affected by ccr4 (Figure 3(a), bottom panel), or other deletions in CCR4-NOT components such as caf40, caf1, or not4 (data not shown), suggesting that SPT5 is not part of the 1.9 MDa CCR4-NOT complex.
3.3. SPT5 Binds CCR4 and CAF1 in Vitro
Our results indicate that SPT5 can interact with CCR4 and CAF1 in vivo but it is unlikely to be tightly associated with the CCR4-NOT complex. We conducted in vitro binding assays to examine more thoroughly the SPT5 interaction with CCR4 and CAF1. These in vitro binding assays utilized SPT5-Flag that was expressed in yeast and GST-CAF1 and GST-CCR4 (495-837) isolated from E.coli (Figure 3(b), top panel). As shown in Figure 3B, bottom panel, SPT5-Flag could be specifically retained by GST-CAF1 and GST-CCR4 (lanes 3 and 5, respectively). GST alone or other comparable GST fusions such as that of GST-CTD (C-terminal domain of RNA pol II) or GST-NOT2 were unable to retain SPT5- Flag (Figure 3(b), lanes 1, 2, and 5, respectively).
3.4. spt5-4, spt5-194, and the spt4 Deletion Can Reduce the Rate of mRNA Degradation
As the primary role for CCR4 is in the control of mRNA degradation, we subsequently examined the effect of several spt5 alleles and an spt4 deletion on the rate of mRNA decay. We observed that ADH2 mRNA was stabilized approximately by a factor of 2-fold in spt5-194, spt5-4, and spt4 deletion strain backgrounds as compared to the wild type strain (Figure 4; Table 2). In the same strain background, a ccr4 deletion also resulted in a 2-fold stabilization of ADH2 mRNA (Table 2), indicating that the magnitude of the spt5/4 effects was equivalent to deleting ccr4 the major cyloplasmic deandenylase in yeast [8]. Combining ccr4 with spt5-4 did not further slow the rate of ADH2 mRNA degradation (Table 2). In contrast, in spt5-25, spt5-8, and spt5-242 strain backgrounds, the rate of ADH2 mRNA decay was not affected (Figure 4, left panel; also Table 2). ccr4, however, in combination with spt5-242 or spt5-8 still decreased the rate of ADH2 mRNA degradation by 2-fold (Table 2). We also observed that spt5-4, like ccr4 and caf1 [8,43, 44], slowed the rate of GAL1 mRNA degradation (spt5-4: 20 min T1/2 compared to wild type: 12 min T1/2). These results indicate that certain alleles of spt5 or deletion of spt4 result in slowing the rate of mRNA decay. Since the
 (a)
(a)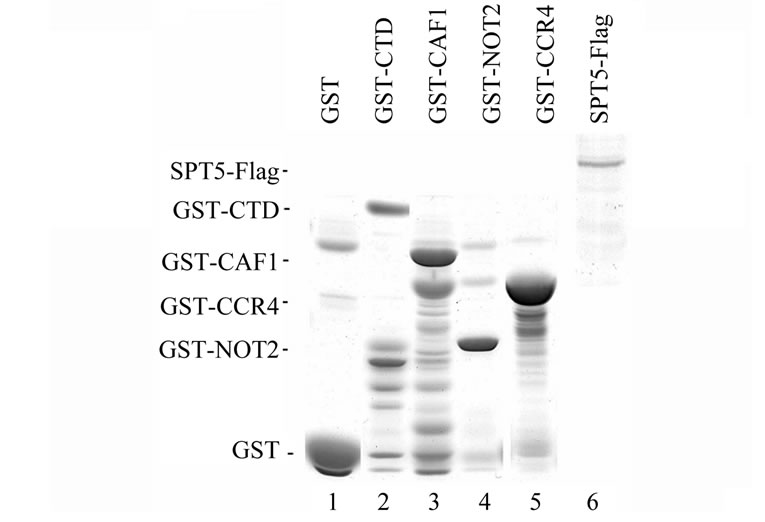 (b)
(b)
Figure 3. (a) Deletion of CCR4 does not affect SPT5 migration in a 1.9 MDa complex. Superose 6 gel chromatography was conducted on crude extracts as previously described [23]. Anti-Flag antibody was utilized to detect SPT5-Flag by Western analysis. WT-strain FY1642; ccr4-strain FY1642-1a. Molecular weight markers were Blue dextran –2 MDa; Thyroglobulin –0.67 MDa; and bovine serum albumin –0.06 MDa. (b) CCR4 and CAF1 can bind SPT5 in vitro. Purified GST-CCR4 (495-837) and GST-CAF1 (top panel) were used to bind extracts containing Flag purified SPT5-Flag (top panel). GSTpull down experiments were conducted as previously described (38). Top panel—Coomassie stained profiles of GST and Flag fusion proteins; bottom panel—Western analysis of SPT5-Flag retention on glutathine agarose columns containing GST fusion proteins as indicated. All GST fusions contained full-length proteins except for CCR4 (495-837). GST-CTD refers to the Cterminal domain of RNA pol II (52). Input SPT5-Flag represents one-twenty fifth of the amount used to bind the GST fusion proteins. About 12% of the SPT5-Flag was retained by GST-CAF1 and 2% was retained by GST-CCR4.
spt5-4 and spt5-194 alleles, in contrast to the spt5-242 allele, have been implicated in disrupting SPT5-SPT4 interactions (G. Hartzog, pers. comm.), the integrity of the SPT5-SPT4 contact may be important to mRNA degradation.
3.5. spt5 and spt4 Mutations Do Not Affect mRNA Decapping or Deadenylation
The role of SPT5/SPT4 on mRNA degradation could be occurring by effects on the deadenylation, decapping or 5′ to 3′ degradation of the RNA transcript [45]. A simple test to determine whether defects in mRNA decay occur at the decapping or later step is to analyze the degradation products of the MFA2pG RNA relative to the total
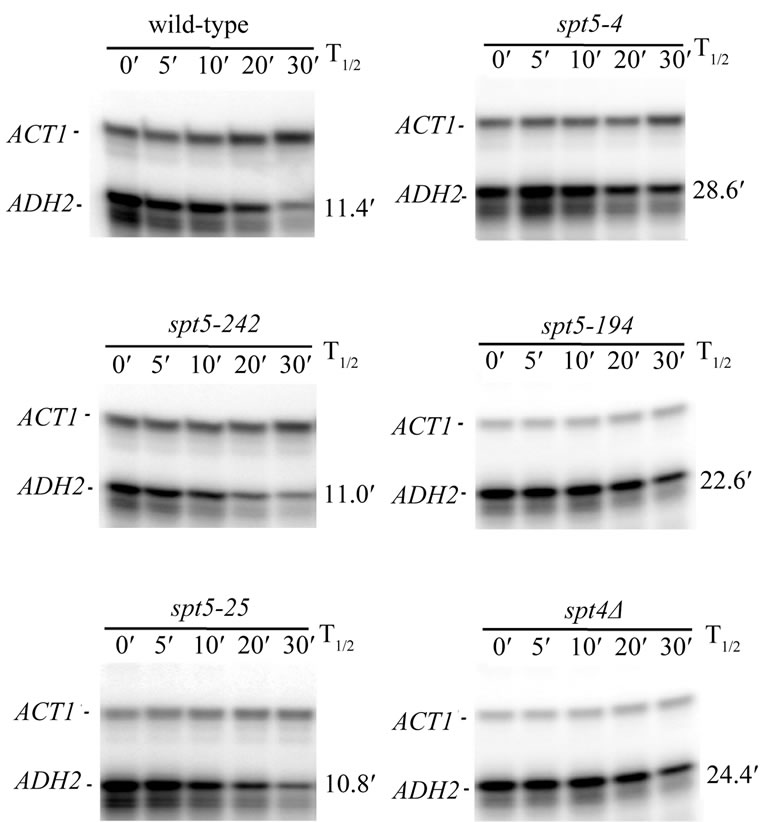
Figure 4. spt5 and spt4 alleles affect ADH2 mRNA degradation rates. Wild-type and mutant strains (see Table 1) were pregrown on YEP medium containing 2% ethanol/2% glycerol prior to shifting to medium containing 4% glucose at zero time and the taking of RNA samples at the times indicated. The ADH2 and ACT1 mRNA levels were determined by S1 nuclease protection assays. The ACT1 RNA was used to standardize loadings for RNA samples at the different time points indicated. T1/2 values are for the data shown.
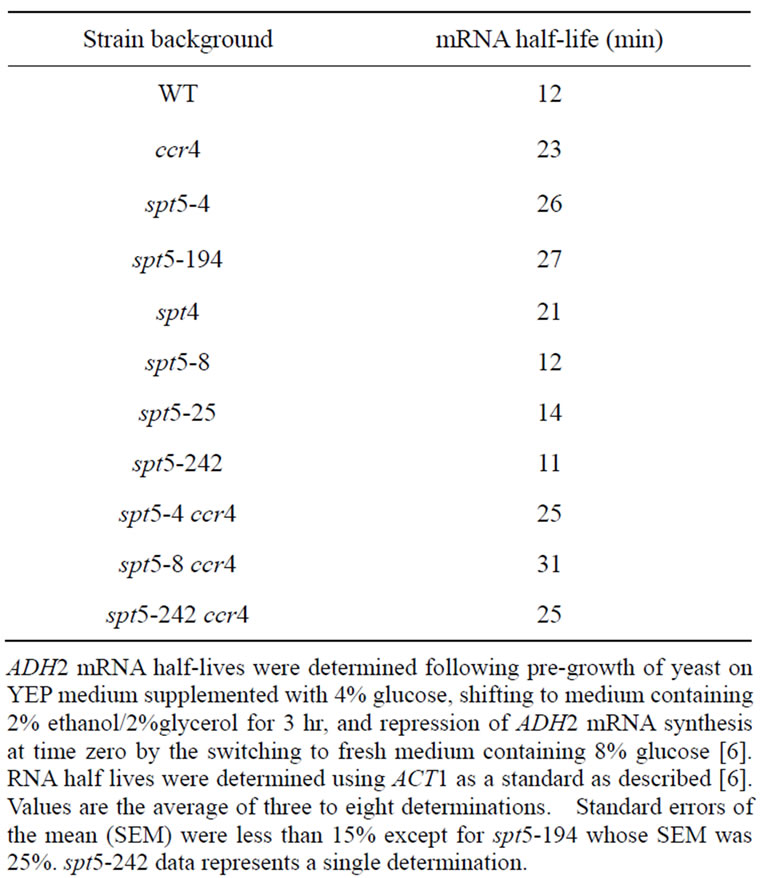
Table 2. Effect of spt5/spt4 alleles on ADH2 mRNA degradation rates.
level of MFA2pG RNA present [46]. The MFA2pG gene contains a poly(G) tract insertion in its 3′ UTR that inhibits exonuclease activity [1]. Therefore, a poly(G) to 3′ end RNA fragment will accumulate when the decapped mRNA is degraded from the 5′ to 3′ direction. However, in the absence of any decapping and 5′ to 3′ mRNA digestion, the formation of the poly(G) 3′ end RNA fragment will be blocked. As shown in Figure 5, spt5-4 had no obvious effect on the abundance of the poly(G) mRNA fragment relative to the full-length MFA2pG mRNA (Figure 5, lane 2 compared to lane 1). Careful analysis of the data indicated that spt5-4 resulted in the poly(G) fragment to be slightly longer than wild-type indicative of a very short poly(A) tail remaining on the poly(G) fragment. Over the course of several experiments, it appears that the oligo poly(A) tail remaining in an spt5-4 background is about 1-3 nucleotides longer in length as compared to that found in wild-type. Similar results were obtained with spt5-194 and spt4 (data not shown). In comparison, ccr4 also does not block decapping or 5' to 3' mRNA degradation (Figure 5, lane 3) and leaves a terminal 20 - 25 poly(A) tail on the fragment [8]. These results indicate that the spt5/spt4 alleles are not affecting decapping or subsequent 5′ RNA degradation.
We subsequently tested whether these alleles affected the rate of mRNA deadenylation, the same step that CCR4 and CAF1 control. Changes in the mRNA deadenylation rate were followed using a transcriptional pulse chase analysis [1] with the GAL1 mRNA. Expression of GAL1 was rapidly induced for 15 minutes by adding galactose to a raffinose growth culture and then repressed by adding glucose. The deadenylation of this pool of newly synthesized GAL1 mRNA was monitored at different time points by Northern analysis. GAL1 mRNA contains two polyadenylation sites [43,44,47] and the RNA corresponding to the shorter RNA is displayed in Figure 6. Similar results were obtained with the longer RNA (data not shown).
As shown in Figure 6, we observed that, in a wildtype strain, by following the shortest poly(A) tail fragments present at a particular time [1], the poly (A) tail was degraded to an oligo(A) tail length of approximately 8 - 13 A’s by around 10 minutes minutes, and by 15 minutes a significant portion of the mRNA were in the oligo(A) form. It has been shown previously that decapping and 5′ to 3′ degradation of the transcript occurs once the oligo(A) tail form (8 - 13 A’s) appears [1,2,48]. Similarly in the spt5-242 strain, which had no effect on ADH2 or GAL1 mRNA decay rate, the oligo(A) tail was formed in 10 to 15 minutes (Figure 6, left panel). In the spt5-4 and spt4 deletion strains, the oligo(A) tail form of GAL1 mRNA routinely appeared in 15 minutes and some was visible at ten minutes (Figure 6, right panel). The spt5-4 and spt4 alleles had similar effects on the deadenylation of the long GAL1 mRNA (data not shown). ccr4 effects
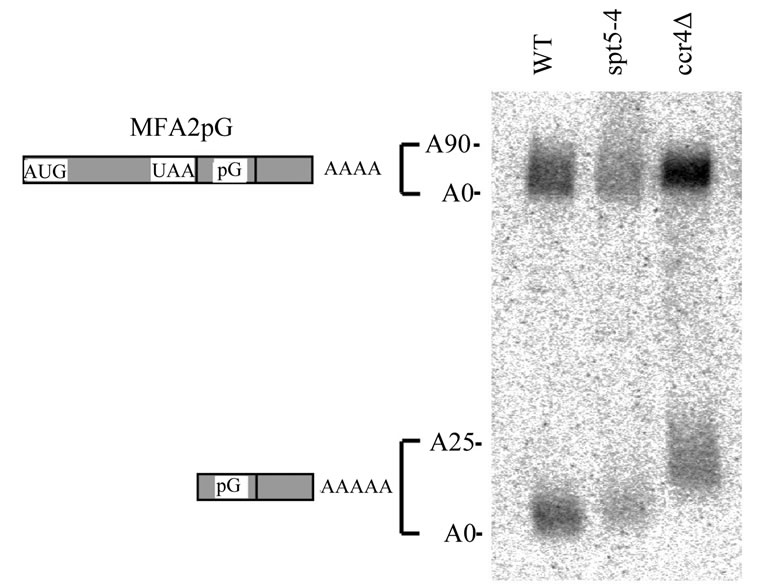
Figure 5. spt5 and spt4 alleles do not affect mRNA degradative steps after deadenylation. Wild-type (FY1642), spt5-4 (FY1668- uH), and ccr4 (FY1642-1a) strains containing plasmid RP485 (MFA2pG) [1] were grown in CAA-U- medium [21] with 4% glucose to mid-log stage. Total RNA were extracted after shifting to 2% galactose for 4 hours. MFA2pG transcripts were detected following Northern analysis [8]. The upper band represents the full-length MFA2pG transcript and the lower band corresponds to the poly(G) to 3′ end RNA fragment. The fully deadenylated MFA2pG RNAs were identified by RNase H treatment after hybridization to oligo d(T) (not shown) [8].
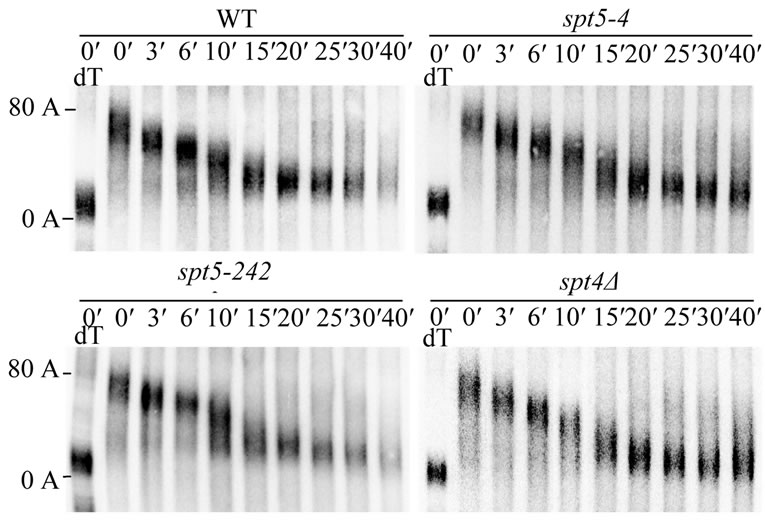
Figure 6. The effect of spt5/spt4 alleles on the rate of GAL1 mRNA deadenylation. Wild-type (FY1642), spt5-4 (FY1668- uH), spt5-242 (FY1635), and spt4 (GHY180) strains was shifted from raffinose-containing YEP medium to galactosecontaining YEP medium for 15 minutes to induce GAL1 gene expression. After transcriptional repression by the addition of glucose, total RNA were extracted at different time points as indicated. GAL1 mRNA was detected by Northern analysis following RNase H treatment of the RNA after hybridization to a 3′ GAL1 RNA probe (Materials and Methods) [42]. The GAL1 mRNA shown is the short GAL1 RNA which is 110 nt shorter at its 3′ end than the long GAL1 mRNA [47]. dT-RNase treatment of the RNA following hybridization to oligo d(T) to identify the fully deadenylated species.
on the rate of GAL1 deadenylation, however, have been shown to be much more severe in which the oligo (A) did not appear until after at least 40 minutes [8,43,44]. These observations indicate that the deadenylation rate of GAL1 mRNA is not significantly reduced by the spt5-4 and spt4 defects, although they do slow the rate of mRNA degradation to an extent comparable to that observed for ccr4. It should be noted that we also examined the ability of the spt5-4 and spt4 defect on the ability of SPT5-Flag to associate with CCR4. However, neither of these defects had any effect on the ability of CCR4 to associate with SPT5 (data not shown), suggesting that the effect of these alleles on mRNA degradation was not simply due to their inability to bind CCR4.
4. DISCUSSION
4.1. SPT5 Interacts Physically with CCR4 and CAF1
The SPT5 protein was shown to physically associate in vivo and in vitro with CCR4 and CAF1. Initially, a portion of the SPT5 protein was identified as interacting with the exonuclease domain of CCR4 using a two-hybrid screen, a result verified by co-immunoprecipitation analysis. We subsequently showed that full-length SPT5 could immunoprecipitate full-length CCR4 when both were expressed at their physiological concentrations. CAF1 could also immunoprecipitate with SPT5, although CAF1 needed to be slightly overexpressed for this effect to be observed. In addition, overexpression of CAF1 interfered with CCR4 interaction with SPT5. These results indicate a close proximity in the cell of CCR4 and CAF1 with SPT5. However, the interaction of CCR4 and CAF1 with SPT5 may not be very tight. The fact that other CCR4-NOT components at their physiological concentrations were not co-immunoprecipitated with SPT5 suggests that the CCR4-SPT5 interaction is not bringing along the whole complex.
Several pieces of evidence indicate that SPT5 is not in the most common complexes associated with CCR4 and CAF1. The purified 1.0 MDa CCR4-NOT complex does not contain SPT5 or SPT4 [24]. Also, immunoprecipitating any of the 1.0 MDa CCR4-NOT protein components does not co-immunoprecipitate any other proteins (data not shown) [49]. SPT5 was shown to migrate in complexes sized at both 1.9 and 0.7 MDa, although the bulk of the SPT5 protein is in the smaller complex. Deleting CCR4, CAF1, CAF40 or NOT4 was found to have no effect on SPT5 migration in either of these complexes. Deleting CAF1 has previously been shown to reduce CCR4 presence in the 1.9 MDa CCR4-NOT complex [21] and deleting CCR4 eliminates the association of CAF16 with the 1.9 MDa complex [26]. It is, therefore, unlikely that SPT5 is part of the 1.9 MDa CCR4-NOT complex unless it were retaining its association in this complex through other as yet undetermined protein interactions. We conclude, therefore, that the interaction between SPT5 and that of CCR4 and CAF1 is of a weaker and/or more transient nature than the interactions observed for components within the CCR4-NOT complex.
4.2. SPT5 Plays a Role in the Control of mRNA Degradation But Not That of Deadenylation
The SPT5 protein has been ascribed several roles in mRNA metabolism including that of affecting transcriptional initiation, mRNA elongation, and possibly splicing and capping [31,35,50]. Based on our previous data supporting a genetic interaction between SPT5 and CCR4 and the current data supporting their physical interaction, we have examined the role of SPT5/SPT4 in mRNA degradation. Defects in both SPT5 and SPT4 were found to reduce the rate of degradation of mRNA. The magnitude of this effect on the rate of ADH2 mRNA degradation was equivalent to the effect of deleting CCR4, the major cytoplasmic deadenylase in yeast [8]. However, SPT5 or SPT4 defects that affected the degradation rate did not appreciably affect the rate of deadenylation. Also, SPT5 defects did not result in a dramatic deadenylation end-point defect, as has been observed with either ccr4 or caf1 [8]. As observed in the deadenylation of MFA2pG, spt5-4 resulted in a deadenylation end-point with 1 - 3 A’s remaining in contrast to the 20 - 25 A’s remaining typically observed in a ccr4 strain. Similarly, in the deadenylation of GAL1 mRNA, SPT5 or SPT4 defects resulted in the complete or nearly complete deadenylation of the RNA (Figure 6) whereas ccr4 and caf1 defects result in the accumulation of only partially deadenylated species [8,11,43,44]. Moreover, defects in either SPT5 or SPT4 had no apparent effects on decapping or 5′ exonuclease activity. These data indicate that SPT5/SPT4 are involved in affecting mRNA degradation, but do so by a mechanism that is not clearly related to effects on CCR4 or on cytoplasmic mRNA degradation.
SPT5 hence may associate with CCR4 in the nucleus and affect any number of transcriptional or post-transcriptional roles assigned to these factors. It is also possible, given the variety of roles ascribed to SPT5, that its effects on mRNA degradation are indirect. A key conclusion from these studies is that analysis of mRNA degradation rates alone is insufficient to determine whether particular factors are actually involved in the cytoplasmic control of mRNA turnover.
5. ACKNOWLEDGEMENTS
We would like to thank F. Winston and G. Hartzog for providing strains used in this report. The technical assistance of G. Quigley was appreciated. This research was supported by NIH grants GM41215 and GM87684 to C.L.D. Partial funding was provided by the New Hampshire Agricultural Experiment Station. This is Scientific Contribution No. 2458.
REFERENCES
- Decker, C. J. and Parker, R. (1993) A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Development, 7, 1632-1643. doi:10.1101/gad.7.8.1632
- Muhlrad, D., Decker, C.J. and Parker, R. (1994) Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5'-->3' digestion of the transcript. Genes & Development, 8, 855-866. doi:10.1101/gad.8.7.855
- Muhlrad, D., Decker, C.J. and Parker, R. (1995) Turnover mechanisms of the stable yeast PGK1 mRNA. Molecular Cell Biology, 15, 2145-2156.
- Cao, D., and Parker, R. (2001) Computational modeling of eukaryotic mRNA turnover. RNA, 7, 1192-1212. doi:10.1017/S1355838201010330
- Sachs, A.B., Sarnow, P. and Hentze, M.W. (1997) Starting at the beginning, middle, and end: Translation initiation in eukaryotes. Cell, 89, 831-838. doi:10.1016/S0092-8674(00)80268-8
- Chen, J., Chiang, Y.C. and Denis, C.L. (2002) CCR4, a 3' - 5' poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO Journal, 21, 1414-1426. doi:10.1093/emboj/21.6.1414
- Tucker, M., Staples, R.R., Valencia-Sanchez, M.A., Muhlrad, D. and Parker, R. (2002) Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO Journal, 21, 1427- 1436. doi:10.1093/emboj/21.6.1427
- Tucker, M., Valencia-Sanchez, M.A., Staples, R., Chen, J., Denis, C.L. and Parker, R. (2001) The transcription factor associated, Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell, 104, 377-386. doi:10.1016/S0092-8674(01)00225-2
- Brown, C.E. and Sachs, A.B. (1998) Poly(A) tail length control in Saccharomyces cerevisiae occurs by messagespecific deadenylation. Molecular Cell Biology, 18, 6548- 6559.
- Daugeron, M.C., Mauxion, F. and Seraphin, B. (2001) The yeast Pop2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Research, 29, 2448- 2455. doi:10.1093/nar/29.12.2448
- Lee, D., Ohn, T., Chiang, Y.-C., Liu, Y., Quigley, G., Yao, G. and Denis, C.L. (2010) PUF3 acceleration of deadenylation in vivo can operate independently of CCR4 activity, possibly involving effects on the PAB1-mRNP structure. Journal of Molecular Biology, 399, 562-575. doi:10.1016/j.jmb.2010.04.034
- Badarinarayana, V., Chiang, Y.-C. and Denis, C.L. (2000) Functional interaction of Ccr4-not proteins with TATAA-Binding Protein (TBP) and its associated factors in yeast. Genetics, 155, 1045-1054.
- Deluen, C., James, N., Maillet, L., Molinete, M., Theiler, G., Lemaire, M., Paquet, N. and Collart, M. A. (2002) The Ccr4-not complex and yTAF1 yTaf(II)130p/yTaf(II)145p) show physical and functional interactions. Molecular Cell Biology, 22, 6735-6749. doi:10.1128/MCB.22.19.6735-6749.2002
- Denis, C.L., Chiang, Y.-C., Cui, Y. and Chen, J. (2001) Genetic evidence supports a role for the yeast Ccr4-not complex in transcriptional elongation. Genetics, 158, 627-634.
- Kruk, J.A., Dutta, A., Fu, J., Gilmour, D.S. and Reese, J.C. (2011) The multifunctional Ccr4-not complex directly promotes transcription elongation. Genes & Development, 25, 581-593. doi:10.1101/gad.2020911
- Gaillard, H., Tous, C., Botet, J., Gonzalez-Aguilera, C., Quintero, M.J., Viladevall, L., Garcia-Rubio, M.L., Rodriguez-Gil, A., Marin, A., Anno, J., revuelta, J.L., Chavez, S. and Aguilera, A. (2009) Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-not in transcription-coupled repair. PLoS Genetics, 5, e1000364. doi:10.1371/journal.pgen.1000364
- Azzouz, N., Panasenko, O.O., Calau, G. and Collart, M.A. (2009) The Ccr4-not complex physically and functionally interacts with TRAMP and the nuclear exosome. PLoS One, 4, e6760. doi:10.1371/journal.pone.0006760
- Derr, S.C., Azzouz, N., Fuchs, S.M., Collart, M.A., Strahl, B.D., Corbett, A.H. and Laribee, R.N. (2011) The Ccr4- not complex interacts with the mRNA export machinery. PLoS One, 6, e18302. doi:10.1371/journal.pone.0018302
- Govindan, M., Meng, X., Denis, C.L., Webb, P., Baxter, J. and Walfish, P. (2009) Identification of Ccr4 and other essential thyroid hormone receptor coactivators by modified yeast synthetic genetic array analysis. Proceedings of the National Academy of Sciences USA, 106, 19854- 19859.
- Denis, C.L. (1984) Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics, 108, 833-844.
- Liu, H.-Y., Badarinarayana, V., Audino, D., Rappsilber, J., Mann, M. and Denis, C.L. (1998) The not proteins are part of the Ccr4 transcriptional complex and affect gene expression both positively and negatively. EMBO Journal, 17, 1096-1106. doi:10.1093/emboj/17.4.1096
- Sakai, A., Chibazakura, T., Shimizu, Y. and Hishinuma, F. (1992) Molecular analysis of Pop2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Research, 20, 6227 -6233. doi:10.1093/nar/20.23.6227
- Bai, Y., Salvadore, C., Chiang, Y.-C., Collart, M., Liu, H.Y. and Denis, C.L. (1999) The Ccr4 and Caf1 proteins of the Ccr4-not complex are physically and functionally separated from not2, not4, and not5. Molecular Cell Biology, 19, 6642-6651.
- Chen, J., Rappsilber, J., Chiang, Y.-C., Russell, P., Mann, M. and Denis, C.L. (2001) Purification and characterization of the 1.0 MDa Ccr4-not complex identifies two novel components of the complex. Journal of Molecular Biology, 314, 683-694. doi:10.1006/jmbi.2001.5162
- Cui, Y., Ramnarain, D.B., Chiang, Y.-C., Ding, L.-H., McMahon, J.S. and Denis, C.L. (2008) Genome wide expression analysis of the Ccr4-not complex indicates that it consists of three modules with the not module controlling SAGA-responsive genes. Molecular Genetics and Genomics, 279, 323-337. doi:10.1007/s00438-007-0314-1
- Liu, H.-Y., Chiang, Y.-C., Pan, J., Chen, J., Salvadore, C., Audino, D.C., Badarinarayana, V., Palaniswamy, V., Anderson, B. and Denis, C.L. (2001) Characterization of Caf4 and Caf16 reveal a functional connection between the Ccr4-not complex and a subset of SRB proteins of the RNA polymerize II holoenzyme. Journal of Biological Chemistry, 276, 7541-7548. doi:10.1074/jbc.M009112200
- Maillet, L., Tu, C., Hong, Y.-K., Shuster, E.O. and Collart, M.A. (2000) The essential function of Not1 lies within the Ccr4-not complex. Journal of Molecular Biology, 303, 131-143. doi:10.1006/jmbi.2000.4131
- Draper, M.P., Salvadore, C. and Denis, C.L. (1995) Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the Ccr4 transcriptional regulatory complex. Molecular Cell Biology, 15, 3487-3495.
- Liu, H.-Y., Toyn, J.H., Chiang, Y.-C., Draper, M.P., Johnston, L.H. and Denis, C.L. (1997) DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the Ccr4 transcriptional regulatory complex. EMBO Journal, 16, 5289-5298. doi:10.1093/emboj/16.17.5289
- Clark, L.B., Viswanathan, P., Quigley, G., Chiang, Y.-C., McMahon, J.S., Yao, G., Chen, J., Nelsbach, A. and Denis, C.L. (2004) Systematic mutagenesis of the leucine-rich repeat (LRR) domain of Ccr4 reveals specific sites for binding to Caf1 and a separate critical role for the LRR in Ccr4 deadenylase activity. Journal of Biological Chemistry, 279, 13616-13623. doi:10.1074/jbc.M313202200
- Hartzog, G.A., Wada, T., Handa, H. and Winston, F. (1998) Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes & Development, 12, 357-369. doi:10.1101/gad.12.3.357
- Wada, T., Takagi, T., Yamaguguhi, Y., Ferdous, A., Imai, T., Hirose, S., Sugimoto, S., Yanu, K., Hartzog, G.A., Winston, F. and Handa H. (1998) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Development, 12, 343-356. doi:10.1101/gad.12.3.343
- Kim, T.-K., Ebright, R.H. and Reinberg, D. (2000) Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science, 288, 1418-1421. doi:10.1126/science.288.5470.1418
- Yamaguchi, Y., Takagi, T., Wada, T., Yano, K., Fuquay, A., Sugimoto, S., Hasegawa, J. and Handa, H. (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell, 97, 41-51. doi:10.1016/S0092-8674(00)80713-8
- Lindstrom, D.L., Squazzo, S.L., Muster, N., Burckin, T.A., Wachter, K.C., Emigh, C.A., McCleery, J.A., Yates 3rd, J.R. and Hartzog, G.A. (2003) Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Molecular Cell Biology, 23, 1368-1378. doi:10.1128/MCB.23.4.1368-1378.2003
- Andrulis, E.D., Werner, J., Nazarian, A., ErdjumentBromage, H., Tempst, P. and Lis, J.T. (2002) The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature, 420, 837-841. doi:10.1038/nature01181
- Draper, M.P., Liu, H.Y., Nelsbach, A.H., Mosley, S.P. and Denis, C.L. (1994) CCR4 is a glucose-regulated transcription factor whose leucine-rich repeat binds several proteins important for placing Ccr4 in its proper promoter context. Molecular Cell Biology, 14. 4522-4531.
- Chiang, Y. C., Komarnitsky, P., Chase, D. and Denis, C.L. (1996) ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. Journal of Biological Chemistry, 271, 32359- 32365. doi:10.1074/jbc.271.50.32359
- Collart, M.A. and Struhl, K. (1993) CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO Journal, 12, 177-186.
- Cook, W.J. and Denis, C.L. (1993) Identification of three genes required for the glucose-dependent transcription of the yeast transcriptional activator ADR1. Current Genetics, 23, 192-200. doi:10.1007/BF00351495
- Denis, C.L., Ferguson, J. and Young, E.T. (1983) mRNA levels for the fermentative alcohol dehydrogenase of Saccharomyces cerevisiae decrease upon growth on a nonfermentable carbon source. Journal of Biological Chemistry, 258, 1165-1171.
- Cui, Y. and Denis, C.L. (2003) In vivo evidence that defects in the transcriptional elongation factors RPB2, TFIIS, and SPT5 enhance upstream poly(A) site utilization. Molecular Cell Biology, 23, 7887-7901. doi:10.1128/MCB.23.21.7887-7901.2003
- Yao, G., Chiang, Y.-C., Zhang, C., Lee, D.J, Laue, T.M. and Denis, C.L. (2007) PAB1 self-association precludes its binding to poly(A), thereby accelerating Ccr4 deadenylation in vivo. Molecular Cell Biology, 27, 6243-6253. doi:10.1128/MCB.00734-07
- Ohn, T., Chiang, Y.-C., Lee, D.J., Yao, G., Zhang, C. and Denis, C.L. (2007) CAF1 plays an important role in mRNA deadenylation separate from its contact to Ccr4. Nucleic Acids Research, 35, 3002-3015. doi:10.1093/nar/gkm196
- Beelman, C.A. and Parker, R. (1995) Degradation of mRNA in eukaryotes. Cell, 81, 179-183. doi:10.1016/0092-8674(95)90326-7
- Tharun, S. and Parker, R. (1999) Analysis of mutations in the yeast mRNA decapping enzyme. Genetics, 151, 1273- 1285.
- Miyajima, A., Nakayama, N., Miyajima, I., Arai, N., Okayama, H. and Arai, K. (1984) Analysis of full-length cDNA clones carrying GAL1 of Saccharomyces cerevisiae: A model system for cDNA expression. Nucleic Acids Research, 12, 6397-6414. doi:10.1093/nar/12.16.6397
- Couttet, P., Fromont-Racine, M., Steel, D., Pictet, R. and Grange, T. (1997) Messenger RNA deadenylylation precedes decapping in mammalian cells. Proceedings of the National Academy of Sciences USA, 94, 5628-5633. doi:10.1073/pnas.94.11.5628
- Denis, C.L. and Chen, J. (2003) The Ccr4-not complex plays diverse roles in mRNA metabolism. Progress in Nucleic Acid Research & Molecular Biology, 73, 221- 250. doi:10.1016/S0079-6603(03)01007-9
- Pei, Y. and Shuman, S. (2002) Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. Journal of Biological Chemistry, 277, 19639-19648. doi:10.1074/jbc.M200015200
NOTES
*This research was supported by NIH and the USDA.
1These authors contributed equally to this work.
2To whom correspondence should be addressed.

