American Journal of Plant Sciences
Vol.4 No.12(2013), Article ID:41289,7 pages DOI:10.4236/ajps.2013.412300
Genetic Variability of Brachiaria ruziziensis Clones to Collaria oleosa (Hemiptera: Miridae) Based on Leaf Injuries
![]()
1Department of Entomology, Universidade Federal de Lavras, Lavras, Brazil; 2Entomology Laboratory, Embrapa Dairy Cattle Research Station, Juiz de Fora, Brazil.
Email: *amauad@cnpgl.embrapa.br
Copyright © 2013 Daniela Maria da Silva et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received October 5th, 2013; revised November 6th, 2013; accepted November 17th, 2013
Keywords: Signal Grass; Bug; Damage Score; Chlorophyll Content
ABSTRACT
Collaria oleosa causes leaf injuries to the main forage grasses used for cattle feeding in Brazil. The aim of this work was to determine resistance of B. ruziziensis clones against C. oleosa. Eighty B. ruziziensis clones were maintained in greenhouse, in which C. oleosa natural infestations have been occurring in previous years. After 40 days, damage score and chlorophyll content reduction were assessed for all clones. By using these parameters, genetic gain was estimated based on REML/BLUP mixed models. We observed significant differences for damage scores and chlorophyll content reduction among B. ruziziensis clones, evidencing genetic variability in this forage specie in regard to resistance against C. oleosa. Gain derived from selection of the 10 best clones was 18.2% and 5.80% when considering the damage score and chlorophyll content reduction, respectively. The clones CNPGL BR 10, CNPGL BR 64, CNPGL BR 97 and CNPGL BR 40 presented the highest genetic gain for both damage score and chlorophyll content reduction, and then they will be selected to continue the B. ruziziensis breeding program with the possibility of maximizing genetic gain for the next generations.
1. Introduction
Grasses of Brachiaria genus are agronomically welladapted to several types of soils [1], being characterized by their higher flexibility in use, management and tolerance to several limitations and/or adverse conditions when compared to other species of forage grasses [2].
Among Brachiaria species cultivated in Brazil, B. ruziziensis is diploid and it reproduces sexually, allowing selection and recombination of superior genotypes [3]. Additionally, this species is of high palatability and quality. On the other hand, this species is susceptible to insect pests, such as the grass bug Collaria oleosa (Distant, 1863) (Hemiptera: Miridae), which is widely distributed within South America including several Brazilian States [4].
This grass bug has been causing serious damage to the shoot of B. ruziziensis. Grass bug adults and nymphs cause partial or total leaf lamina desiccation, compromising the yield and plant nutritional value [5]. Under favorable conditions, grass bug populations can reach high levels within a short period of time [6]. Yet C. oleosa injuries often occur in Brachiaria and elephant grass pastures, producers and technicians are not able to associate the injury to the causing agent which leads to gradual increase in C. oleosa population every year, due to the use of wrong strategies to solve the problem.
Recently, Auad et al. [7] reported that injuries related to grass bugs were caused by the stylet insertion in leaf epidermis through stomata which leads to pigmentation loss. Plant defensive response against insect attack is an important tool to select resistant genotypes that will be less damaged by insect pests. The selection is possible given the high genetic variability in B. ruziziensis available by the breeding program from Embrapa Gado de Leite, Brazil [8]. Furthermore, we believe that the use of resistant varieties is an important component of integrated management of the grass bug by helping to keep its populations below economic threshold levels.
Therefore, the aim of this work was to assess the genetic variability and the resistance of B. ruziziensis clones to C. oleosa. The selection of resistant parents will contribute to the advance of breeding program of this grass.
2. Material and Methods
It was assessed 80 clones originated from B. ruziziensis breeding program of Embrapa Gado de Leite (Brazil), along with the controls B. brizantha cv. Marandu (resistant), B. decumbens cv. Basilisk and B. ruziziensis cv. Kennedy (susceptibles). These grasses were selected as controls in the experiment because they are resistant and susceptible to spittlebug [9,10]; and there is no previous knowledge about the resistance against C. oleosa damage.
To obtain seedlings, plants were reproduced by cloning (cuttings) and cultivated in greenhouse. After 30 days, the plants were transplanted to pots of 1 kg containing a mix of soil, sand and manure (3:1:1) and kept in the greenhouse (4.0 m high × 4.5 m wide × 6.5 m length) of Embrapa Gado de Leite/Brazil, which had presented issues in keeping C. oleosa natural infestations in previous years. Before starting the experiment, the plant height was standardized by trimming clones to reduce to 10 cm high.
After 40 days that clones were in contact with C. oleosa, percentage of damaged leaf area was assessed by two independent evaluators using a score scale of 1 - 5 based on visual pattern, being 1 = no damage, 2 = 25% of injury in leaf area, 3 = 50% of injury in leaf area, 4 = 75% of injury in leaf area and, 5 = leaf area totally desiccated [10]. To assess chlorophyll content reduction (CCR), a portable chlorophyll meter SPAD (Soil Plant Analysis Development) was used to measure injured leaves (IL) and non-injured leaves (NIL) picked at random. The measurement was done in the mid-third of a full-expanded leaf, in a total of ten samples per plant. The percentage of chlorophyll content reduction was given by the formula CCR (%) = (NIL × IL/NIL) × 100.
The experiment consisted of three replicates in a completely randomized design. The data in chlorophyll content reduction and damage score were tested by analysis of variance and means were compared by Scott Knott test at 5% of probability using the software SISVAR 5.1 (Universidade Federal de Lavras-Minas Gerais, Brazil). Pearson’s correlation for the chlorophyll content reduction and damage score was estimated using the software BioEstat 3.0 (Universidade Federal de Belém, Brasil).
In order to estimate the genetic gain using the collected data (damage score and chlorophyll content reduction), the statistical analysis was based on REMUBLUP mixed models, carried out in the software Selegen-RemllBlup, in accordance with Resende [11]; Resende and Dias [12]. The statistical model was given by: y = Xb + Zg + e; in which y, b and e are data vectors of fixed effects (blocks), random effects (genotype accesses) and random errors, respectively; X and Z are incidence matrices for b and g, respectively.
3. Results and Discussion
We observed significant differences for damage scores (F = 1.490, P = 0.013) and chlorophyll content reduction (F = 1.288, P = 0.049) among B. ruziziensis clones (Table 1), evidencing genetic variability in this forage species for resistance against C. oleosa. This variability will be used in breeding program of B. ruziziensis grass aiming at selecting genotypes that have both favorable agronomical traits and resistance against the grass bug.
Damage inflicted by C. oleosa in the score scale ranged from 1.50 to 4.66, being clone CNPGL BR 10 the lowest rating and CNPGL BR 50 the highest rating. Thirty-six clones had the damage score significantly low (1.50 to 2.87), including the three control cultivars (B. brizantha, B. decumbens and B. ruziziensis). Forty-four clones were suitable hosts to grass bug as they had a high damage score (3.00 to 4.66) (Table 1). Surprisingly, the spittlebugs-susceptible controls were resistant to the mirid C. oleosa. This might have occurred because spittlebugs feed on the host plant xylem and phloem, while grass bugs feed on leaf parenchyma.
From the clones studied, CNPGL BR 19; CNPGL BR 4; CNPGL BR 26; CNPGL BR 58 were reported by Souza Sobrinho et al. [3] as being resistant to spittlebugs based on damage scores. This method therefore seems to be promising to identify and select resistant clones to both pests and these clones might be used in future breeding cycles.
From all the tested genotypes, 62.5% obtained the lowest chlorophyll content reduction, with means lower than 25.21% in comparison with other tested accesses (Table 1). As these clones have not lost much chlorophyll content in injured leaves, they were less affected by herbivory and we predict that losses would be consequently lower in terms of yield and forage quality. Within this group, we can observe that the genotypes CNPGL BR 3; CNPGL BR 30; CNPGL BR 64 showed chlorophyll content reduction of 0.78%; 7.06% and 8.75%, respectively. On the other hand, in 37.5% of B. ruziziensis clones, chlorophyll content reduction was significantly high in a way that clones CNPGL BR 91; CNPGL BR 68 and CNPGL BR 74 had chlorophyll content reduction of 41.64%; 42.59% and 44.80%, respectively. There was a significant lower chlorophyll content
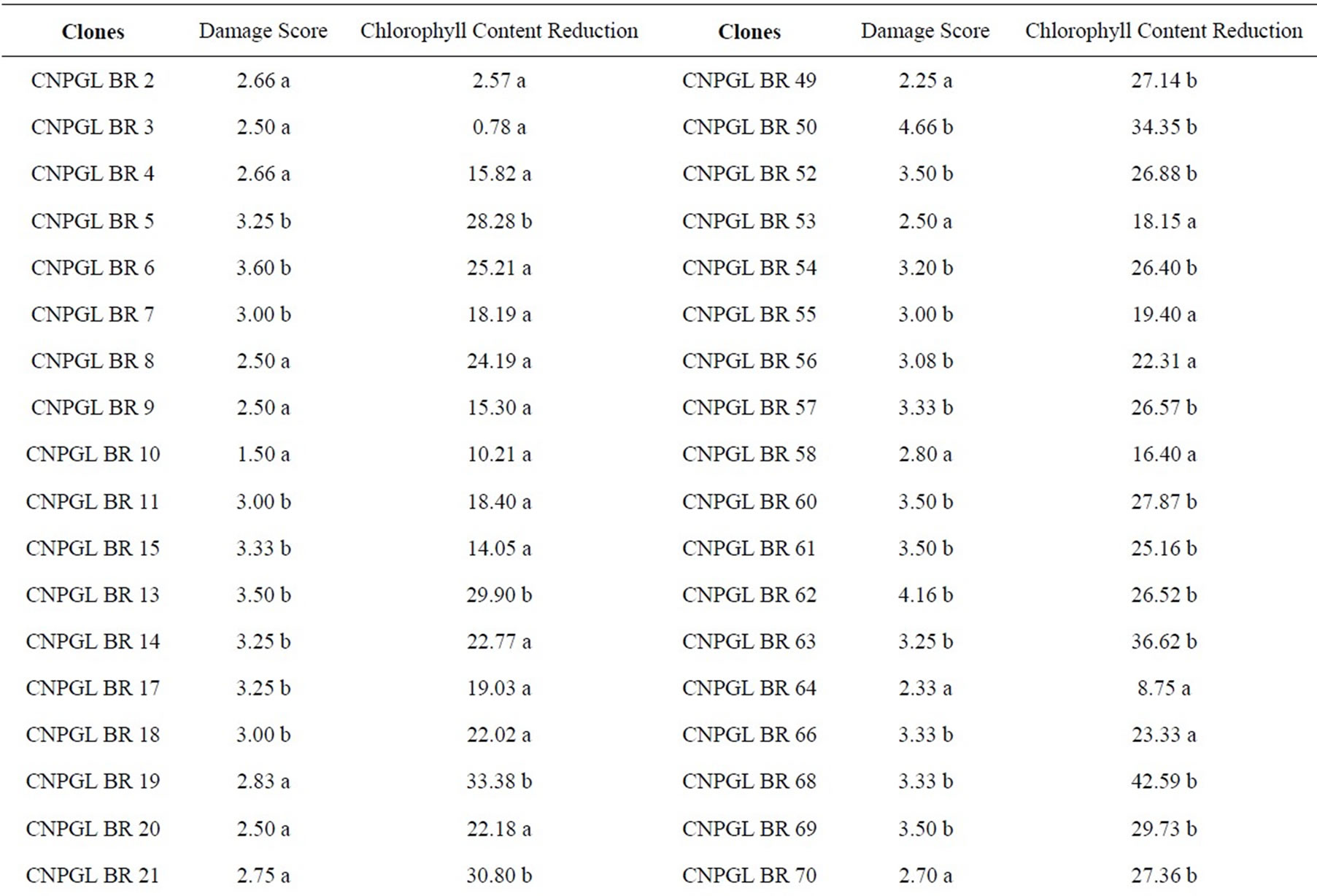
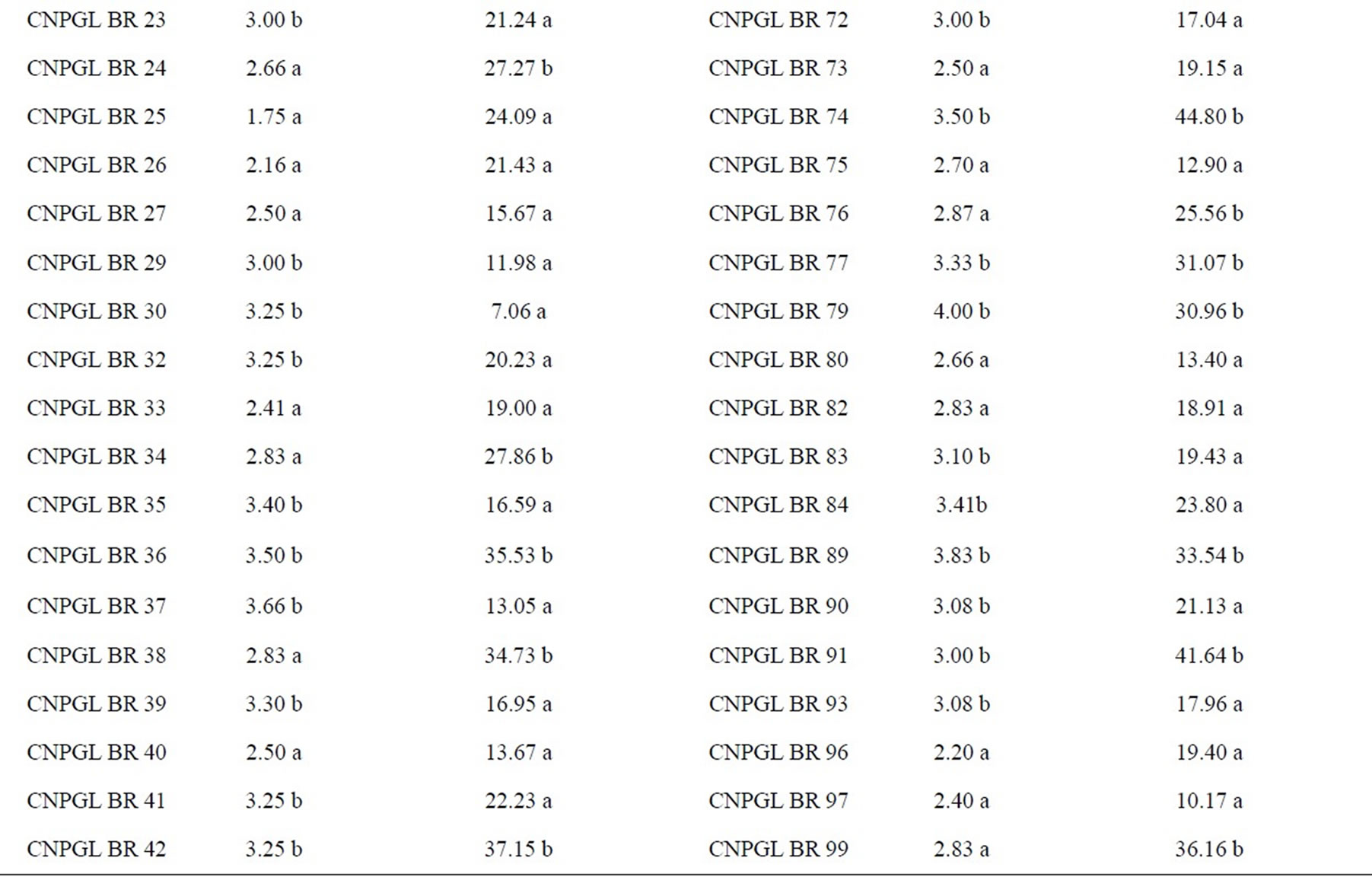
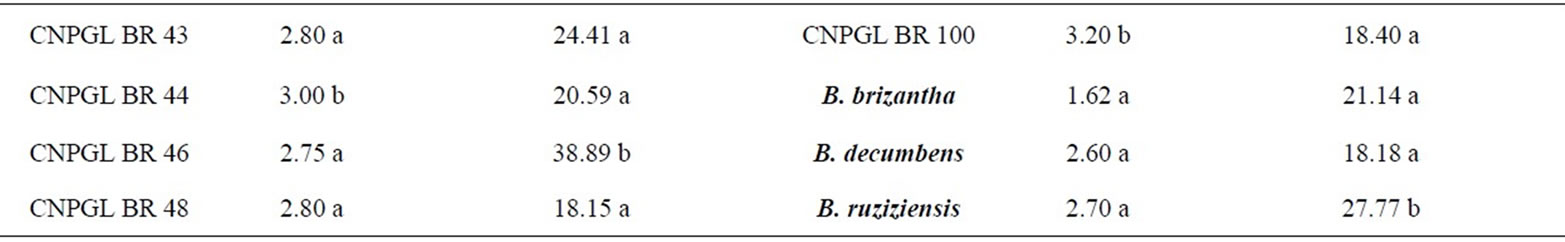
Table 1. Score damage (1 to 5) and chlorophyll content reduction (%) of B. ruziziensis clones and controls.
reduction in the controls B. decumbens and cultivar Marandu (B. brizantha), but not in B. ruziziensis (Table 1).
From the 36 genotypes classified in the group of lowest damage score, 66.7% showed the lowest chlorophyll content reduction of insect-injured leaves. Therefore, it was clear that there is a positive significant correlation (t = 3.6639; P = 0.0004) between the visual damage assessment and chlorophyll content measures of C. oleosa injured leaves. The positive correlation between insect leaf damage and chlorophyll content reduction allows a direct selection method. Thus, it is possible to estimate C. oleosa damage only by means of plant chlorophyll content readings without relying on visual and subjective damage score assessment. Furthermore, chlorophyll content readings are easy and fast and therefore it allows assessment of a high number of plant materials with low costs and no detriment in the data quality.
By analyzing the parameters damage score and chlorophyll content reduction in the current study, we found that there is a great genetic variability among B. ruziziensis clones in regard to resistance against C. oleosa. The same parameters were also used for selection of soybean [13,14] and millet [15] that were resistant to stinkbugs. Diaz-Montano et al. [16] used the same parameter and equipment to measure the chlorophyll content in order to separate resistant wheat plants against the aphids Aphis glycines Matsumura. They showed that susceptible wheat accesses had low chlorophyll content and, consequently decreased photosynthetic capacity.
Given the genetic variability of B. ruziziensis clones, it was possible to estimate genetic parameters and genetic gain from selection using damage score and chlorophyll content reduction (Tables 2 and 3). Juhász et al. [17]; Abreu et al. [18]; Faria Neto et al. [19]; Missio et al. [20]; Costa et al. [21]; Costa et al. [22] also used the genetic parameters and genetic gain with from selection to select the best genotypes of several crop plant based on plant height and diameter.
The individual heritability (h2g) was estimated between 0.16 and 0.14 for both assessed traits (Table 2), and it is considered moderate according to the classification proposed by Resende [11]. Heritability indicates the degree by which individuals pass their traits over the generations and it can also infer about the genetic control of traits. In this sense, a high heritability value indicates that there is a good chance of genetic gain from selection
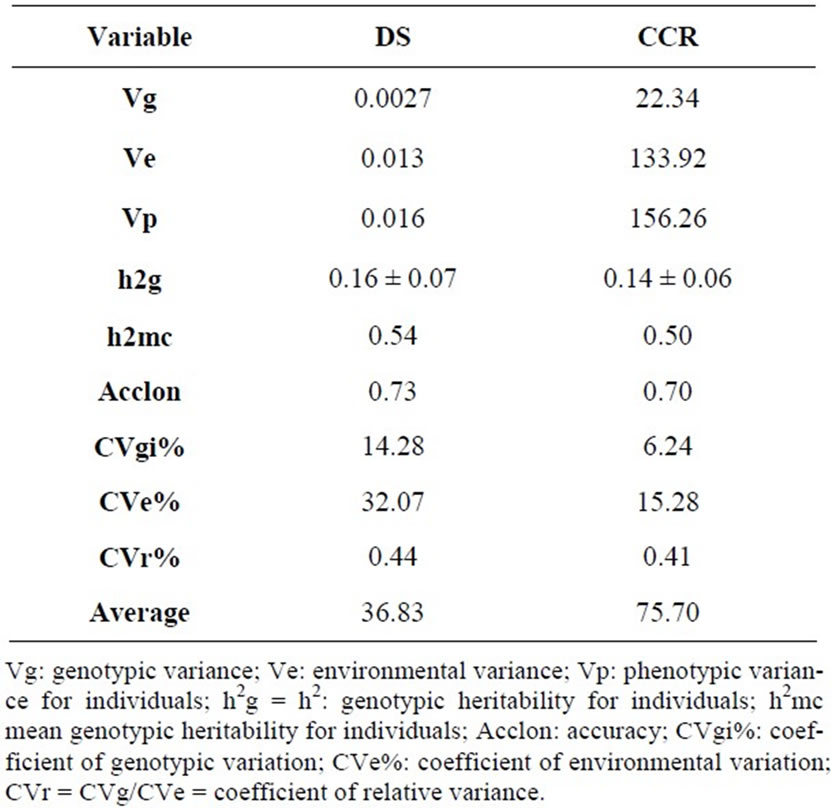
Table 2. Values of genetic parameters for chlorophyll content reduction (CCR) and damage score (DS) for B. ruziziensis clones.
[23].
The mean heritability for clones was higher than 50% for damage score and chlorophyll content reduction (Table 2) showing that the variability of plant resistance trait against C. oleosa can be transferred to next generations. Also, these results indicate the efficiency of selection within B. ruziziensis for plant resistance trait. Similarly, Sharma et al. [24] also reported a high heritability for the first (50%) and the second generations (82%) of a sorghum resistance trait against the fly Atherigona soccata (Rondani).
According to Juhász et al. [17], accuracy (Acclon) refers to the correlation between the expected genetic values and the true genetic value of an individual. The higher the accuracy to a given individual, the higher is the reliability of the assessment and the expected genetic value, as well as the higher is the gain from selection. In the current work, we estimated in 73% for the damage score and 70% for the chlorophyll content reduction of insect-injured leaves (Table 2), reinforcing the reliability of the results.
It was found higher values of environmental variance (Ve) in comparison to the genotypic variance (Vg) for
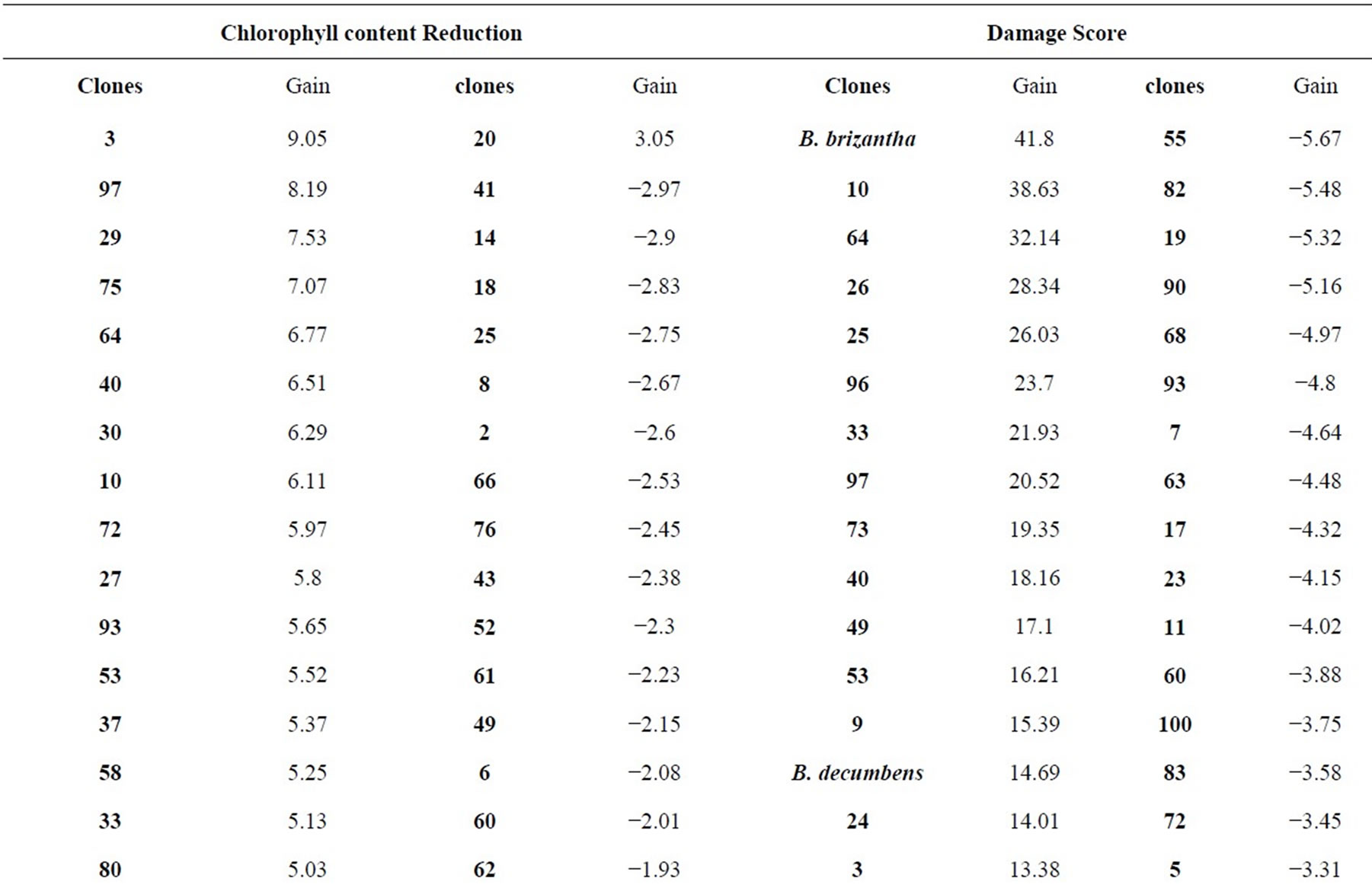

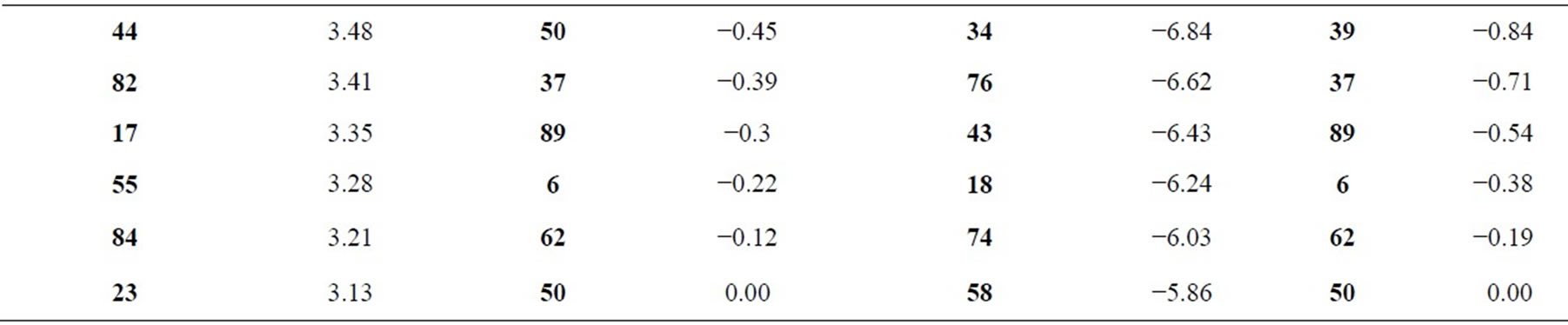
Table 3. Values of expected genetic gain for damage score (DS) and chlorophyll content reduction (CCR) of B. ruziziensis clones and controls.
both characteristics. None of the characteristics showed coefficient values of relative variation above one (Table 2). According to Vencovsky [25], for experiments with two or three replicates, values for the ratio CVgi/CVe closer to one mean higher chances of gain from progeny selection.
Numbers in Table 3 demonstrate the improvement of population average for the assessed trait. In regard to DS (damage score), gain was substantially higher compared to those assessed for CCR (chlorophyll content reduction). The material assessed in this work showed, on average, that gain varied between 41.80% and 0% for damage score and 9.05% and 0% for chlorophyll content reduction (Table 3). It is expected that desirable alleles will build up in the population over the selection cycles and then there will be a higher number of resistant plants to C. oleosa.
The cultivar Marandu (B. brizantha) and the clones CNPGL BR 10, CNPGL BR 64, CNPGL BR 26, CNPGL BR 25, CNPGL BR 96, CNPGL BR 33, CNPGL BR 97, CNPGL BR 73, CNPGL BR 40 were classified as the best ten given their high genotypic values for score damage. Values of gain from selection were for some of these clones higher than 18.2% (Table 3). Regarding chlorophyll content reduction, the gain from selection of the best genotypes (CNPGL BR 3, CNPGL BR 97, CNPGL BR 29, CNPGL BR 75, CNPGL BR 64, CNPGL BR 40, CNPGL BR 30, CNPGL BR 10, CNPGL BR 72, CNPGL BR 27) were higher than 5.80% (Table 3).
For damage score, the extension between averages was 0.77 (Table 3). The cultivar Marandu (B. brizantha) presented, on average, 2.61 for damage score and the gain from selection was 41.80% based on the genotypic values. The extension between chlorophyll content reduction averages was 0.35 and the genotype CNPGL 3 showed the highest average (4.13) and gain from selection of 9.05% (Table 3), considering the genotypic values. All controls presented genotypic values higher than the overall average for damage score, what was not true only for chlorophyll content reduction of B. ruziziensis cv. Kennedy (Tables 3).
We reported considerable genetic variability in B. ruziziensis resistance against C. oleosa herbivory. The genotypes CNPGL BR 10, CNPGL BR 64, CNPGL BR 97, CNPGL BR 40 showed the highest values of genetic gain from selection for damage score and chlorophyll content reduction.
4. Conclusion
The genotypes CNPGL BR 10, CNPGL BR 64, CNPGL BR 97, CNPGL BR 40 therefore will be selected for continuing B. ruziziensis breeding program with a possible increase in gain over the next generations.
5. Acknowledgements
We thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil) and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG, Brazil) for supporting our research.
REFERENCES
- I. M. Rao, J. W. Miles, R. Garcia and J. Ricaurte, “Selección de Hibridos de Brachiaria con Resistencia a Alumínio,” Pasturas Tropicales, Vol. 26, No. 3, 2006, pp. 12-15.
- J. R. Valério, “Considerações Sobre a morte de Pastagens de Brachiaria brizantha Cultivar Marandu em Alguns Estados do Centro e Norte do Brasil: Enfoque Entomológico,” In: R. A. Barbosa, Ed., Morte de Pasto de Braquiárias. Embrapa Gado de Corte, Campo Grande, 2006, 206 Pages.
- F. Souza Sobrinho, A. M. Auad and F. J. S. Lédo, “Genetic Variability in Brachiaria ruziziensis for Resistance to Spittlebugs,” Crop Breeding Applied Biotechnology, Vol. 10, No. 1, 2010, pp. 83-88.
- J. C. M. Carvalho and A. V. Fontes, “Mirideos Neotropicais, CCXXV: Revisão do Gênero Collaria Provancher no Continente Americano (Hemiptera),” Experientiae, Vol. 27, No. 2, 1981, pp. 1-46.
- M. Menezes, “Collaria oleosa (Distant, 1883) (Hemiptera: Miridae), nova Praga de Gramíneas Forrageiras no Sudeste da Bahia, Brasil,” Agrotrópica, Vol. 2, No. 2, 1990, pp. 113-118.
- R. R. Vergara, “Collaria spp. Insecto Dañino del Kikuyo: métodos de Control,” Seminario Internacional Competitividad en Carne y Leche, 5, 2006, Colanta, Medellín, 2006, pp. 197-231.
- A. M. Auad, D. S. Pimenta, D. M. Silva, P. H. Monteiro and T. T. Resende, “Collaria oleosa (Hemiptera: Miridae) on Brachiaria ruziziensis and Penissetum Purpureum (Poaceae): Characterization of Injury and Biological Aspects,” Revista Colombiana de Entomologia, Vol. 37, No. 2, 2011, pp. 80-81.
- F. Souza Sobrinho, “Melhoramento de Forrageiras No Brasil,” In: A. R. Evangelista, P. N. C. Amaral, R. F. Padovani, V. B. Tavares, F. M. Salvador and A. J. Perón, Eds., Forragicultura e Pastagens: Temas em Evidência. UFLA, Lavras, 2005, pp. 65-120.
- J. R. Valério, H. Jeller and J. Peixer, “Seleção de Introduções do Gênero Brachiaria (Griseb) Resistentes à Cigarrinha Zulia entreriana (Berg) (Homoptera: Cercopidae),” Anais da Sociedade Entomológica do Brasil, Vol. 26, No. 2, 1997, pp. 383-387. http://dx.doi.org/10.1590/S0301-80591997000200023
- C. Cardona, P. Fory, G. Sotelo, A. Pabon, G. Diaz and J. W. Miles, “Antibiosis and Tolerance to Five Species of Spittlebug (Homoptera: Cercopidae) in Brachiaria spp.: Implications for Breeding for Resistance,” Journal Economic Entomology, Vol. 97, No. 2, 2004, pp. 635-645. http://dx.doi.org/10.1603/0022-0493-97.2.635
- M. D. V. Resende, “Software Selegen-Reml/Blup,” Embrapa Floresta, Colombo, No. 77, 2002, p. 67.
- M. D. V. Resende and L. A. S. Dias, “Aplicação da Metodologia de Modelos Mistos (REML/BLUP) na Estima- ção de Parâmetros Genéticos e Predição de Valores Genéticos em Espécies Frutíferas,” Revista Brasileira de Fruticultura, Vol. 22, No. 1, 2000, pp. 44-52.
- A. L. Lourenção and M. A. C. Miranda, “Resistência de Soja a Insetos: VII. Avaliação de Danos de Percevejos em Cultivares e Linhagens,” Bragantia, Vol. 46, No. 1, 1987, pp. 65-72. http://dx.doi.org/10.1590/S0006-87051987000100008
- A. L. Lourenção, N. R. Braga, M. A. C. Miranda, P. C. Reco, C. G. Q. Fugi and J. C. V. N. A. Pereira, “Avalia- ção de Danos de Insetos e de Severidade de Oídio Em Genótipos de soja,” Bragantia, Vol. 64, No. 3, 2005, pp. 423-433. http://dx.doi.org/10.1590/S0006-87052005000300012
- X. Ni, J. P. Wilson and D. G. Buntini, “Differential Responses of Forage Pearl Millet Genotypes to Chinch Bug (Heteroptera: Blissidae) Feeding,” Journal Economic Entomology, Vol. 102, No. 5, 2009, pp. 1960-1969. http://dx.doi.org/10.1603/029.102.0529
- J. Diaz-Montano, J. C. Reese, W. T. Schapaugh and L. R. Campbell, “Chlorophyll Loss Caused by Soybean Aphid (Hemiptera: Aphididae) Feeding on Soybean,” Journal Economic Entomology, Vol. 100, No. 5, 2007, pp. 1657- 1662. http://dx.doi.org/10.1603/0022-0493(2007)100[1657:CLCBSA]2.0.CO;2
- A. C. P. Juhász, D. L. B. Moraes, B. O. Soares, S. Pimenta, H. O. Rabello and M. D. V. Resende, “Parâmetros Genéticos e Ganho com a Seleção para Populações de Pinhão Manso (Jatropha curcas),” Pesquisa Florestal Brasileira, Vol. 30, No. 61, 2010, pp. 25-35 2010.
- F. B. Abreu, M. D. V. Resende, J. L. Anselmo, H. M. Saturnlno, J. A. M. Brenha and F. B. Freitas, “Variabilldade Genética entre Acessos de Pinhão-Manso na Fase Juvenil,” Magistra, Vol. 21, No. 1, 2009, pp. 36-40.
- J. T. Farias Neto, M. S. P. Oliveira, A. A. Muller, O. L. Nogueira and D. F. S. P. Anaissi, “Variabilidade Genética em Progênies Jovens de Açaizeiro,” Cerne, Vol. 11, No. 4, 2005, pp. 336-341.
- R. F. Missio, A. M. Silva, L. A. S. Dias, M. L. T. Moraes and M. D. V. Resende, “Estimates of Genetic Parameters and Prediction of Additive Genetic Values in Pinus kesya Progenies,” Crop Breeding Applied Biotechnology, Vol. 5, 2005, pp. 394-401.
- R. B. Costa, M. D. V. Resende, A. Z. Contini, F. L. H. Rego, R. A. R. Roa and W. J. Martins, “Avaliação Gené- tica de Indivíduos de Erva-Mate (Ilex Paraguariensis st. hil.) na Região de Caarapó, MS, Pelo Procedimento REML/BLUP,” Ciência Florestal, Vol. 15, No. 4, 2005, pp. 371-376.
- R. B. Costa, M. D. V. Resende, P. S. Gonçalves, J. F. Chichorro and R. A. R. Roa, “Variabilidade Genética e Seleção Para Caracteres de Crescimento da Seringueira,” Bragantia, Vol. 67, No. 2, 2008, pp. 299-305. http://dx.doi.org/10.1590/S0006-87052008000200005
- R. F. Missio, L. A. S. Dias, M. L. T. Moraes and M. D. V. Resende, “Selection of Pinus caribaea var. Bahamensis Progenies Based on the Predicted Genetic Value,” Crop Breeding Applied Biotechnology, Vol. 4, 2004, pp. 399- 407.
- G. C. Sharma, M. G. Jotwani, B. S. Rana and N. G. P. Rao, “Resistance to the Sorghum Shoot Fly, Atherigona soccata (Rondani) and Its Genetic Analysis,” Journal Entomology Research, Vol. 1, No. 1, 1997, pp. 1-12.
- R. Vencovsky, “Herança Quantitative,” In: E. Paterniani and G. P. Viegas, Eds., Melhoramento e Produção do Milho. 2nd Edition, Fundação Cargill, Campinas, 1987, pp. 137-214.
NOTES
*Corresponding author.

