Advances in Biological Chemistry
Vol.3 No.1(2013), Article ID:28161,6 pages DOI:10.4236/abc.2013.31019
PCR-based molecular markers linked to the leaf rust resistance gene Lr19 in different bread wheat cultivars
![]()
1Institute of Botany, ANAS, Baku, Azerbaijan
2Research Institute of Crop Husbandry, Ministry of Agriculture of Azerbaijan Republic, Baku, Azerbaijan
Email: huseynova-i@botany-az.org
Received 30 December 2012; revised 30 January 2013; accepted 5 February 2013
Keywords: Wheat; Brown Leaf Rust; Lr19 Gene; SCAR-Markers; PCR Analysis
ABSTRACT
61 varieties of wheat collected in the gene fund of the Research Institute of Crop Husbandry were screened using SCAR-markers associated with the gene of resistance to brown leaf rust, Lr19. As a result of PCR analysis using SCS123 marker the 737 bp locus was detected in 48 genotypes. The expected fragment of the 688 bp was detected in 53 genotypes using the SCS253 marker. The results obtained using both markers indicate that the Lr19 gene is present on 7D chromosomes of 45 genotypes. The existence of the Lr19 gene has not been proven only for 5 from the 61 analyzed wheat genotypes.
1. INTRODUCTION
Wheat is the most cultivated cereal crop, giving almost 30% of global grain production and providing food for more than a half the Earth’s population. In Azerbaijan, wheat is strategically important crop for covering needs of the population in food products. Therefore, it is cultivated annually in different regions of the republic at the area of over 580 thousands of hectares. The main factors limiting the yield of summer wheat in Azerbaijan are fungal, bacterial and viral diseases. One of the common and harmful diseases of cereal crops is brown leaf rust caused by basidium fungus Puccinia recondite f. sp. tritici [1-3].
Depending on the severity and duration of the infection, yield losses can reach 40% - 50% [4]. Brown leaf rust still remains the most harmful wheat disease in the world, despite of the advances in studying the nature of plant resistance, structure and variability of pathogen population and the success of practical breeding for resistance. Genetic method, i.e. creation of varieties resistant to infection, is the most cost effective, environmentally safe, and therefore, highly relevant method in the integrated system for protection of agricultural plants from these diseases [5,6].
The study of the genetic basis of plant resistance, the search for effective genes and their introduction into the culture of bread wheat significantly prevent the spread of the epiphytotic disease and stabilize the grain yield capacity. Development and deployment of cultivars with host genetic resistance is the most ecofriendly way to reduce the losses. Using hybridological analysis, it was established that wheat resistance to leaf rust pathogen is controlled by both dominant and recessive genes during their independent, complementary, polymeric, additive and epistatic actions and interactions. Using various genetic and biochemical approaches some attempts were made to study key genes responsible for resistance to wheat leaf rust pathogen [7,8]. In wheat, these genes are called “Lr” genes from the English “Leaf rust”. A number of effective Lr-genes of resistance to brown leaf rust pathogen is decreasing year by year. This is related to the fact that virulent biotypes and strains that can break this resistance appear in the pathogen as a result of sexual hybridization and other processes. A constant search for such genes is required. This approach is relevant and significant for breeding. To date, more than 65 leaf rust resistance (Lr) genes against the fungal pathogen Puccinia triticina have been described in common hexaploid wheat, tetraploid durum wheat and diploid wild wheat species [9]. Most of these leaf rust resistance genes condition a hypersensitive reaction and interact with the pathogen in a gene-for-gene fashion. Molecular biology tools in recent years led to the development of DNA markers that are a powerful tool for identification of the gene through binding with appropriate resistance genes. Molecular markers were identified for most of the resistance genes against brown leaf rust (Lr1, Lr3, Lr9, Lr10, Lr13, Lr14a, Lr16, Lr19, Lr20, Lr21, Lr22, Lr24, Lr25, Lr26, Lr28, Lr29, Lr32, Lr34, Lr35, Lr37, Lr39, Lr46, Lr47, Lr50, Lr51, Lr52, Lr57, Lr58) [10]. These works are carried out with the use of RAPD, SSR, SCAR, STS, and AFLP markers.
Lr19 localized on the 7D chromosome is one of the few widely effective genes conferring resistance against brown leaf rust in wheat [8]. Foreign Lr19 gene demonstrated efficacy against all pathotypes of leaf rust in South Africa [10], India [11], Europe [12] and Canada [13]. The Lr19 translocation is associated with deleterious agronomic effects and as a result modified forms of the translocation have been derived by different researchers in an attempt to remove the genes responsible. It was reported that Lr19 was associated with increases in grain yield. Aerial biomass was also increased when Lr19 was introgressed, although differences were not associated with improved light interception (indirectly measured) or radiation use efficiency (RUE). The physiological basis of the increased biomass and the mechanisms causing increased number of grains per spike, in terms of dynamic of floret development, are not completely understood [14].
Lr19 translocation originally produce by Sharma and Knott [15] when they transform leaf rust resistance genes 7e l1 chromosome of Thinopyrum ponticum a long arm of chromosome 7 D of common wheat [16]. HeurtaEspino and Singh [17] reported first virulence in Puccini a Triticinan to Lr-19 and it is an effective source of leaf rust resistance worldwide. The cut-of point of Lr19 translocation is located in the middle of long arm of chromosome 7D and find that the distal half of 7D was replaced by Thinopyrum Chromatinv [18]. During meiosis Thinopyrum segment 7DL does not pair with homologous wheat segment, complicating attempts to study linkage relationship or to recombine its genes [16,19,20].
Despite the virulence for the Lr19 gene, there are reports that in the last decade it has demonstrated high efficacy in wheat cultivation areas [17,21]. High efficacy of the Lr19 gene in Asia, Australia and Europe indicates that this gene can be used in combination with other Lr genes for long-term resistance to leaf rust all over the world [22,23].
On this basis, the objective of this study was to determine the presence of the Lr19 gene in different wheat genotypes using SCAR markers.
2. MATERIALS AND METHODS
61 bread wheat genotypes (Triticum aestivum L.) from the genefund of the Research Institute of Crop Husbandry served as a research objects. Plants were grown in field conditions. SCAR markers were used for the screening.
2.1. Extraction of Plant DNA
DNA extraction was carried out using the CTAB method with some modifications [24]. Fresh plant tissue as a fragment of leaf was minced in liquid nitrogen and suspended in 1000 µl of CTAB extraction buffer (100 mM Tris-HCl, pH 8.0; 20 mM EDTA, pH 8.0; 1.4 mM NaCl; 40 mM b-mercaptoethanol), pre-warmed in a water bath at 60˚C. Homogenization was completed by intense Vortex shaking. Then 400 ml of chloroform (99.8%) were added into each tube and the tubes were gently mixed. Next the tubes were placed in a water bath and incubated for 10 min at 60˚C. After incubation, the tubes were centrifuged in an Eppendorf type benchtop centrifuge (15,000 × g) for 10 min at room temperature. After centrifugation the supernatant was carefully selected (taking care not to capture sediment particles) and transferred to clean 1.5 ml tubes and 600 ml of cold isopropanol were added, then mixed well and left at room temperature for 3 - 5 minutes. At this stage we can observe the dispersed DNA precipitate. The tube contents were centrifuged at room temperature in the Eppendorf type benchtop centrifuge (15,000 × g) for 10 min.
The precipitate was washed several times with 70% ethanol, dried in a thermostat at 56˚C for 5 minutes and dissolved in TE buffer (10 mM Tris-HCl, pH 8; 1 mM EDTA). Samples were left in a refrigerator at 4˚C for the complete dissolution of the DNA in a buffer.
2.2. DNA Quantification
After dissolution of the DNA the quantity was determined by optical density (OD) at λ = 260 using the ULTROSPEC 3300 PRO spectrophotometer (“AMERSHAM”, USA). Purity of the genomic DNA was determined by the ratio of absorptions at A260/A280. Quality of the DNA was checked on the basis of performance of the extracted DNA samples in 0.8% agarose gel stained with 10 mg/mL of ethidium bromide in 1 × TBE (Tris base, Boric acid, EDTA) buffer. The gel was developed and photographed under ultraviolet light using “Gel Documentation System UVITEK” (UK).
2.3. DNA Amplification
Polymerase chain reaction was performed by Williams et al. [25] using SCAR markers. DNA amplification was performed in a 25 µl reaction mixture volume, containing 10 × buffer, 20 ng of the genomic DNA, 0.2 µM primer, 200 µM of each of the following: dATP, dCTP, dGTP and dTTP, 2.5 mM МgCl2, and 0.2 units of Taq-polymerase in the incubation buffer. Two SCAR primers—SCS123 and SCS253 (Eurofins mwg operon)— were used for the test (Table 1). PCR was performed in the “Applied Biosystems 2720” thermal cycler (Singapore) under the following conditions: 1 cycle—3 minutes at 94˚C; 38 cycles—1 min at 94˚C, 1 min at 60 and 63˚C (for SCS123 and SCS253 respectively), 2 minutes at
Table 1. Nucleotide sequence of the SCAR primers used for the DNA amplification.
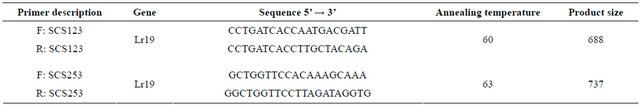
72˚C; the final elongation cycle was performed at 72˚C for 10 min, then kept at 4˚C.
The reaction products were separated by electrophoresis in a 1.2% agarose gel in the HR-2025-High Resolution (“IBI SCIENTIFIC” US) horizontal electrophoresis system with addition of ethidium bromide and documented using “Gel Documentation System UVITEK”. Dimensions of amplified fragments determined with respect to 1kb DNA marker. Statistical analysis included binary matrix compilation for each of the primers, in which “presence” (+) or “absence” (−) of fragments with equal molecular weight on the electrophoregram were noted.
3. RESULTS AND DISCUSSION
DNA samples from wheat (Triticum aestivum L.) from genefund of Research Institute of Crop Husbandry were screened using two SCAR molecular markers bound to a known Lr19 gene of resistance to brown leaf rust. SCAR markers are polymorphic and amplified unique bands linked to the Lr19 gene [8]. This gives a possibility of using these markers in marker assisted breeding for Lr19 gene.
Figure 1 reflects the PCR profiles performed using SCS123 F/R molecular marker (5’CCTGATCACCAATGACGATT3’/5’CCTGATCACCTTGCTACAGA3’).
This marker must lead to the amplification of fragments of 688 bp in size. As a result of PCR test with this primer the locus of the 688 bp region was detected only in 48 genotypes. This is approximately 79% of all investigated genotypes.
Fragment linked to the SCS123F/R marker was not synthesized in the following genotypes—Pirshahin-1, Pactole, 8th WWEERYT (32 №), 3 RBWYT (521 №), 3 RBWYT (536 №), 11th IWWYT-R (9816 №), S5, 16th FAWWON-IR (90), 16th FAWWON-IR (47), S1, Nurlu- 99 Kyrmyzygyul-1, 12th FAWWON № 97 (130/21).
The second SCAR marker linked to the studied Lr19 gene of resistance to brown leaf rust was SCS253 F/R (5’ GCTGGTTCCACAAAGCAAA 3’/5’ GGCTGGTTCCTTAGATAGGTG 3’). Amplification products with the use of this marker are detected in the 737 bp region. As can be seen from Figure 2, the expected fragment in the 737 bp region was synthesized in only in 53 of 61 genotypes, in other words, in approximately 87% of all inves-
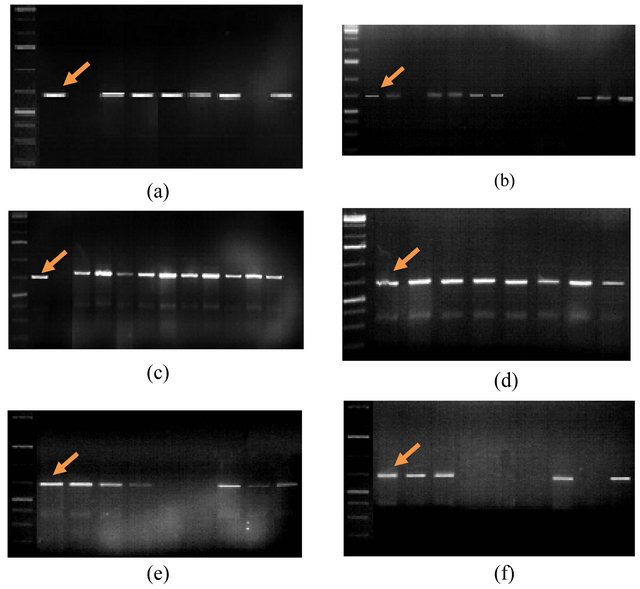
Figure 1. PCR-profiles of Triticum aestivum L. plants induced by SCS123F/R primer (5’CCTGATCACCAATGACGATT3’/ 5’CCTGATCACCTTGCTACAGA3’). Arrow indicates the 688 bp. 1st line on all figures is molecular weight marker: 100 bp. (a) 2nd line: Akinchi-84, 3rd: Pirshahin-1, 4th: Guneshli, 5th: Dagdash, 6th: FARAN Dole, 7th: Renan, 8th: Avreka, 9th: Pactole, 10th: 38 IBWSN (129 №); (b) 2nd line: 10 SAWVT (11 №), 3rd: 3rd FAWWON (117 №), 4th: 8th WWEERYT (32 №), 5th: 4th RWVT-LRCA (89 №), 6th: 14th FAWWON (86 №), 7th: 8th WON-SA (65 №), 8th: 9th WON-SA (27 №), 9th: 3 RBWYT (521 №), 10th: 12th FAWWON №97 (130/21), 11th: 3 RBWYT (536 №), 12th: 3 RBWYT (518 №), 13th: 39 IBWSN (113 №), 14th: 14 SAWYT (49 №); (c) 2nd line: 39 IBWSN (97 №), 3rd - 11st IWWYT-R (9816 №), 4th: S5, 5th: Mirbashir-128, 6th: Yegane, 7th: Zirve-80, 8th: Fatima, 9th: Aran, 10th: Azeri, 11th: Murov, 12th: Murov-2, 13th: Saba, 14th: Tereggi; (d) 2nd line: Beyaz, 3rd: Shafag, 4th: KSI-13, 5th: Pirshahin, 6th: Ugur, 7th: Parzivan-1, 8th: Parzivan-2, 9th: Sheki-1; (e) 2nd line: 16th FAWWON-IR (61), 3rd: 16th FAWWON-IR (46), 4th: 16th FAWWON-IR (52), 5th: 29 ES WVT (7), 6th: 16th FAWWON-IR (90), 7th: 16th FAWWON-IR (47), 8th: 29 ES WVT (26), 9th: 29 ES WVT (38), 10th: 29 ES WVT (30); (f) 2nd line: 16 SAWWVT (29), 3rd: 16 SAWWVT (34), 4th: Azamatli-95, 5th: Nurlu-99, 6th: Gyrmyzy gul-1, 7th: S1, 8th: Ruzi-84, 9th: Tale-38, 10th: 3 RBWYT (510 №).
tigated genotypes. Fragments specific for the SCS 253F/R SCAR marker were not amplified in the following genotypes—3 RBWYT (521 №), Zirve-80, Gyrmyzy gul-1, S1, Azamatli-95, Tale-38, Ruzi-84 and 12th FAW-
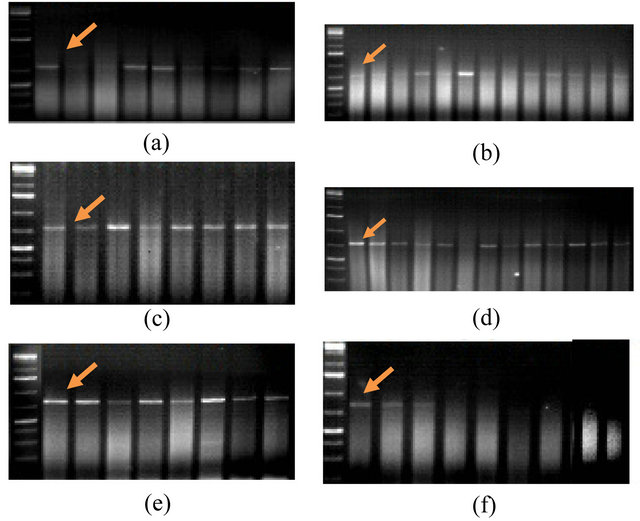
Figure 2. PCR-profiles of Triticum aestivum L. plants induced by the SCS253F/R primer (5’GCTGGTTCCACAAAGCAAA3’/5’ GGCTGGTTCCTTAGATAGGTG3’). Arrow indicates the 737 bp. 1st line on all figures is molecular weight marker - 100 bp. (a) 2nd line: Akinchi-84, 3rd: Pirshahin-1, 4th: Guneshli, 5th: Dagdash, 6th: FARAN Dole, 7th: Renan, 8th: Avreka, 9th: Pactole, 10th: 38 IBWSN (129 №); (b) 2nd line: 10 SAWVT (11 №), 3rd: 3rd FAWWON (117 №), 4th: 8th WWEERYT (32 №), 5th: 4th RWVT-LRCA (89 №), 6th: 14th FAWWON (86 №), 7th: 8th WON-SA (65 №), 8th: 9th WON-SA (27 №), 9th: 3 RBWYT (521 №), 10th: 3 RBWYT (510 №), 11th: 3 RBWYT (536 №), 12th: 3 RBWYT (518 №), 13th: 39 IBWSN (113 №), 14th: 14 SAWYT (49 №); (c) 2nd line: Beyaz, 3rd: Shafag, 4th: KSI-13, 5th: Pirshahin, 6th: Ugur, 7th: Parzivan-1, 8th: Parzivan-2, 9th: Sheki-1; (d) 2nd line: 39 IBWSN (97 №), 3rd: 11st IWWYT-R (9816 №), 4th: S5, 5th: Mirbashir-128, 6th: Yegane, 7th: Zirve-80, 8th: Fatima, 9th: Aran, 10th: Azeri, 11th: Murov, 12th: Murov-2, 13th: Saba, 14th: Tereggi; (e) 2nd line: 16th FAWWON-IR (61), 3rd: 16th FAWWON-IR (46), 4th: 16th FAWWON-IR (52), 5th: 16th FAWWON-IR (90), 6th: 16th FAWWON-IR (47), 7th: 29 ES WVT (7), 8th: 29 ES WVT (26), 9th: 29 ES WVT (38), 10th: 29 ES WVT (30); (f) 2nd line: 16 SAWWVT (29), 3rd: 16 SAWWVT (34), 4th: Nurlu-99, 5th: Azamatli-95, 6th: Tale-38, 7th: Ruzi-84, 8th: 12th FAWWON №97 (130/21), 9th: S1, 10th: Gyrmyzy gul-1.
WON № 97 (130/21).
Comparative analysis of PCR profiles obtained with the use of both SCAR markers demonstrates (Table 2) that the results are the same in 82% of genotypes: specific amplification fragments were identified in 45 genotypes with the use of both SCS123F/R and SCS253F/R markers, which indicates that the Lr19 gene of resistance to brown leaf rust is present on 7D chromosomes of these genotypes. The existence of the Lr19 gene has not been proven in 5 of 61 genotypes used, because specific fragments amplified with any of the applied markers were not identified in these genotypes.
The results obtained with different markers did not match in 18% of genotypes. After using the SCS123F/R marker, nine genotypes (Pirshahin-1, Pactole, 8th WW-
Table 2. Results of the PCR tests using SCAR markers SCS123F/R and SCS253F/R.*


*Note: [+]—the presence of the expected locus, [-]—the absence of this locus.
EERYT (32 №), 3 RBWYT (536 №), 11th IWWYT-R (9816 №), S5, 16th FAWWON-IR (90), 16th FAWWONIR (47), Nurlu-99) did not match, i.e. fragments in the 688 bp region specific for the SCS123F/R marker were not synthesized in these genotypes, on the contrary, the 737 bp fragments, linked with the SCS253F/R marker, were amplified. And this kind of mismatch was detected in three genotypes (Zirve-80, Azamatli-95, Ruzi-84) with the use of the SCS253F/R marker, in other words, amplification products specific for the SCS253F/R marker were absent in these genotypes, on the contrary, synthesis of PCR profiles specific for the SCS123F/R marker has successfully been performed.
The absence of marker components with the Lr19 gene in these samples may be due to an incomplete linkage of the marker and the gene [26].
Attention is drawn to the fact that resistance and high sensitivity to brown leaf rust are observed among the genotypes in which amplification products have not been revealed, thus indicating the absence of the Lr19 gene.
Gyrmyzy gul-1 wheat genotype in field conditions also demonstrates high susceptibility to the brown rust pathogen and is completely affected by the Puccinia recondite f. sp. tritici fungus. The genotypes called 3 RBWYT (521 №), S1 and Tale-38 in field conditions are estimated as moderately resistant to this disease. It is interesting that the 12th FAWWON № 97 (130/21) genotype actually demonstrates high resistance to this harmful disease, despite the absence of the Lr19 gene. Apparently, the resistance of this genotype may be caused by other Lr-genes.
The wheat cultivars are of different types and become susceptible to different types of rust because it has narrow genetic bases for resistance. The evolution rates of pathogens are very fast and rapid. So, it is necessary to find out new and better sources for resistance. The genetic resistance is important to control many phytopathogenic epidemics. The wheat production has been dependent on the use and development of rust resistance genotypes having well characterized and diverse genes. It is also concluded that, in wheat certain and different combinations of genes give long lasting and better resistance for rust diseases than given by any individual genes [16,27].
This work requires the continuation of the study. However, the material studied by us is a valuable source for wheat breeding for leaf rust resistance. Understanding the genetic regulation of resistance allows avoiding the possible use of the same donor gene in selection and developing the programs of rational use of high-effective genes of resistance. The obtained results can be used in breeding and genetic programs on creation of forms resistant to leaf rust pathogen populations in Azerbaijan. Thus, information about the existence of effective Lrgenes in adapted varieties that can be used as donors for resistance, and usage of these distinct genes or by pyramiding of different resistance genes in the genotype can significantly improve the efficiency of breeding of resistant varieties [12], thus assisting to avoid the creation of varieties that are genetically homogeneous [3].
REFERENCES
- Singh, R.P., Huerto-Espino, J. and Rajaram, S. (2000) Achieving near-immunity to leaf and stripe rusts in wheat by combining slow rusting resistance genes. Acta Phytopathologica et Entomologica Hungarica, 35, 133-139.
- Oelke, L.M. and Kolmer, J.A. (2004) Characterization of leaf rust resistance in hard red spring wheat cultivars. Plant Disease, 88, 1127-1133. doi:10.1094/PDIS.2004.88.10.1127
- Mebrate, S.A., Oerke, E.C., Dehne, H.W. and Pillen, K. (2008) Mapping of the leaf rust resistance gene Lr38 on wheat chromosome arm 6DL using SSR markers. Euphytica, 162, 457-466. doi:10.1007/s10681-007-9615-z
- McIntosh R.A., Wellings C.R. and Park R.F. (1995) Wheat rusts: An atlas of resistance genes. CSIRO Publications, Melbourne, 208. doi:10.1007/978-94-011-0083-0
- Marasas, C.N., Smale, M. and Singh, R.P. (2003) The economic impact of productivity maintenance research: Breeding for leaf rust resistance in modern wheat. Agricultural Economics, 29, 253-263. doi:10.1111/j.1574-0862.2003.tb00162.x
- Woxniak-Strzembicka, A. (2003) Wirulencja populacji Puccinia recondita f. sp. tritici w Polsce wlatach 1998-2001. Biuletyn Instytutu Hodowli i Aklimatyzacji Roslin, 230, 109-117.
- Vanzetti, L.S., Campos, P., Demichelis, M., Lombardo, L.A., Aurelia, P.R., Vaschetto, L.M., Bainotti, C.T. and Helguera, M. (2011) Identification of leaf rust resistance genes in selected Argentinean bread wheat cultivars by gene postulation and molecular markers. Electronic Journal of Biotechnology, 14, 3. doi:10.2225/vol14-issue3-fulltext-14
- Gupta, S.K., Charpe, A., Prabhu, K.V. and Haque, Q.M. (2006) Identification and validation of molecular markers linked to the leaf rust resistance gene Lr19 in wheat. Theoretical and Applied Genetics, 113, 1027-1036. doi:10.1007/s00122-006-0362-7
- McIntosh, R.A., Dubcovsky, J., Rogers, W.J., Morris, C., Appels, R. and Xia, X.C. (2011) Catalogue of gene symbols for wheat: 2011 supplement. http://www.shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2011.pdf
- Prins, R., Marais, G.F., Pretorius, Z.A., Janse, B.J.H. and Marais, A.S. (1997) A study of modified forms of the Lr19 translocation of common wheat. Theoretical and Applied Genetics, 95, 424-430. doi:10.1007/s001220050579
- Tomar, S.M.S. and Menon, M.K. (1998) Adult plant response of nearisogenic lines and stocks of wheat carrying specific Lr genes against leaf rust. Indian Phytopathology, 51, 61-67.
- Mesterhazy, A., Bartos, P., Goyeau, H., Niks, R.E., Csosz, M., Andersen, O., Casulli, F., Ittu, M., Jones, E., Manisterski, J., Manninger, K., Pasquini, M., Rubiales, D., Schachermayr, G., Strzembicka, A., Szunics, L., Todorova, M., Unger, O., Vanco, B., Vida, G. and Walther, U. (2000) European virulence survey for leaf rust in wheat. Agronomie, 20, 793-804. doi:10.1051/agro:2000104
- McCallum, B.D. and Seto-Goh, P. (2003) Physiologic specialization of wheat leaf rust (Puccinia triticina) in Canada in 2000. Canadian Journal of Plant Pathology, 25, 91-97. doi:10.1080/07060660309507053
- Miralles, D.J., Resnicoff, E. and Carretero, R. (2007) Yield Improvement associated with Lr19 translocation in wheat. In: Spiertz, J.H.J., Struik, P.C. and van Laar, H.H., Eds., Scale and Complexity in Plant Systems Research: Gene-Plant-Crop Relationships, Springer, Dordrecht, 171-178. doi:10.1007/1-4020-5906-X_14
- Sharma, D. and Knott, D.R. (1966) The transfer of leaf rust resistance from Agropyron to triticum by irradiation. Canadian Journal of Genetics and Cytology, 8, 137-143.
- Sehgal, S.A., Tahir, R.A., Anwar, Z., Abbas, G., Kausar Nawaz Shah, M. and Zaman Khan Khattak, J. (2012) Molecular genetic characterization of rust in wheat genotypes. Asian Journal of Agricultural Sciences, 4, 337-340.
- Huerta-Espino, J. and Singh, R.P. (1994) First report of virulence for wheat leaf rust gene Lr19 in Mexico. Plant Disease, 78, 640. doi:10.1094/PD-78-0640C
- Knott, D.R. (1980) Mutation of a gene for yellow pigment link to Lr-19 in wheat. Canadian Journal of Genetics and Cytology, 22, 651-654.
- Kim, N.S., Amstrong, K. and Knott, D.R. (1993) Molecular detection of lophopyrum chromatin in wheat lophopyrum recombinants and their use in the physical mapping of chromosome 7D. Theoretical and Applied Genetics, 85, 561-567. doi:10.1007/BF00220914
- Marais, G.F. and Marais, A.S. (1990) The assignment of a thinopyrum disticum (thunb) love derived translocation to the long arm of wheat chromosome 7D using endopeptidase polymorphism. Theoretical and Applied Genetics, 79, 182-186. doi:10.1007/BF00225949
- Sibikeev, S.N., Kruprov, V.A., Voronina, S.A. and Elesin V.A. (1996) First report of leaf rust pathotypes virulent on highly effective Lr-genes transferred from Agropyron species to bread wheat. Plant Breeding, 115, 276-278. doi:10.1111/j.1439-0523.1996.tb00917.x
- Roelfs, A.P. (1988) Genetic control of phenotypes in wheat stem rust. Annual Review of Phytopathology, 26, 351-367. doi:10.1146/annurev.py.26.090188.002031
- Pink, D. (2002) Strategies using genes for non-durable disease resistance. Euphytica, 124, 227-236. doi:10.1023/A:1015638718242
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research, 8, 4321-4325. doi:10.1093/nar/8.19.4321
- Williams, J.G., Kubelik, K.J., Livak, J.A. and Tingey, S.V. (1990) DNA polymorphisms amplified by arbitrary primers are useful genetic markers. Nucleic Acids Research, 18, 6531-6535. doi:10.1093/nar/18.22.6531
- Tirishkin, L.G. (2006) Genetic control of the effective juvenile brown rust resistance of collection Triticum aestivum L. wheat samples. Genetics, 42, 377-384.
- Dyck, P.L. and Sambroski, D.J. (1982) The inheritance of resistance to puccinia in a group of common wheat cultivars. Canadian Journal of Genetics and Cytology, 24, 2733-2783.

