Journal of Biophysical Chemistry
Vol.2 No.3(2011), Article ID:6790,7 pages DOI:10.4236/jbpc.2011.23039
Indonesian avian influenza H274Y mutant neuraminidase homology models assessment
Departement of Physics, Mathematics and Science Faculty, University of Indonesia, Depok, Indonesia; *Corresponding Author: sigit.jaya.herlambang@gmail.com
Received 14 May 2011; revised 21 June 2011; accepted 1 July 2011.
Keywords: Neuraminidase; Model Assessment; Homology Model; Mutant; Avian Influenza
ABSTRACT
Five models of Indonesian H274Y mutant neuraminidase type 1 (N1) were generated from the template 3CKZ by homology modeling. The template has the best similarity percentage 97% with the model sequence. The models was evaluated to search the best model with DOPE, 3D-profiles and PROCHECK in a good rank. The results show model 3 as a potential model to be used in the simulation with the lowest DOPE score, highest verify-3D score and from Ramachandran plots we inferred that it also shared the 1st rank with model 4 based on the 99.4% of the residues found, without Glycine and Proline, at the most favoured and additionally allowed region of both model structures.
1. INTRODUCTION
The H5N1 avian influenza virus is a tremendous threat for a country with an economical background in poultry farming. If spread of the avian influenza virus (AIV) is left untreated, the poultry industry in a certain country could be disastrous. Indonesia is one of the countries that were impacted as a result of this pandemic. In poultry alone, it took down poultry in 31 of 33 provinces [1].
At the same time the AIV infected poultry, it also crossed species and was able to infect humans in the surrounding area of the infected poultry. Until 21st April 2010, there were 163 human cases with 135 deaths [2]. By this time, the virus had showed its virulence and the resulting high mortality rate. Recent infections of humans with H5N1 avian influenza viruses have rising the popularity of this type of influenza. The effective transmission of the viruses was a big help for the development of an influenza pandemic [3]. The toll taken by avian influenza virus was also correlated with the pathogenicity of this virus.
The ability of virus spread may have correlated with the two main glycoproteins, haemagglutinin (HA) and neuraminidase (NA) [4]. HA play an important role in the penetration of virus before it infect the host cells while NA takes the responsibility to make the matured virion release from the host cells after the replication [5]. The catalytic site of NA improved the rate of the cleaving process. The higher rate of the releasing process could determine the increase in the number of viruses that may infect other host cells.
From several studies, NAs are known to have the unique ability of rejecting inhibitor drug molecules after antigenic shift and antigenic drift even if it was caused by a single mutation [6-16]. It may also occur in Indonesian avian virus. In Indonesia, the research in this area is really hard to achieve even by its own native researchers. Since January 2007, the Government of Indonesia through the Ministry of Health has decided to withhold specimen (virus) sharing practice [17], including DNA (deoxyribonucleic acid) base codes or protein sequences, hindering further research on this topic. Therefore it is imperative to this research to generate NA structures based on sequences in an open influenza database then comparing the generated structures with other epidemic viruses NA sequences.
In this study we generated NA structures of Indonesian AIV that has a mutation at the active site. We then assessed the models that were constructed with the better approach using three different methods. One of the Indonesian H5N1 NA sequences, which have a single mutation in H274Y, was modeled by homology modeling. After the models were generated, the evaluation of models was examined to assess the overall stability of protein and the residues environment through verify 3D score and DOPE energy. The stereo chemical properties of the models were also evaluated through PROCHECK [18-20] that was provided online by SWISS EXPASY web [21].
2. MATERIALS AND METHODS
2.1. Materials
The NA amino acid sequence of Influenza A virus (A/Indonesia/560H/2006(H5N1)) was taken from National Center of Biotechnology Information (NCBI) [22] with the accession code ABW06159. The 449 residues long NA has one single mutation at the 274 residue position from Histidine into Tyrosine. This virus is known as one of Indonesian avian influenza virus that could infect humans.
The NA protein templates for homology modeling were selected from crystallized structures, with protein data bank code 3CKZ, stored in the protein data bank of Research Collaboratory for Structural Bioinformatics (RCSB) [23]. The crystal structures of NAs were selected by BLASTP 2.2.16 [24] provided by Swiss Expasy. The database is known to have a collection up to 55.000 sequences.
2.2. Methods
The sequences were then aligned with a few sequences from the template and other virus NAs. The sequences for other viruses were obtained from NCBI and were known to be collected when pandemics occurred (see Table 1). Those NAs were analyzed with multiple sequences alignment focusing on the consensus and trace residues to see the similar pattern of those viruses. A phylogenetic tree was analyzed using neighbor joining method [25], best tree mode and set a distance as Uncorrected (“p”). The parameters were set to simplify the analysis because the result would show the differences in percentage number of residue positions at which the sequences differ.
Building homology models were produced using MODELER [26] which is integrated in Accelrys Discovery Studio 2.1. The parameters during this process were a medium optimization level of models and cut overhangs to remove the terminal unaligned residues in model sequence for comparison reason with the template. The homology modeling generated 5 models that were produced by 3CKZ structure template.
The selected model was evaluated to verify its reliability with Discrete Optimized Protein Energy (DOPE) [27] method using MODELER and 3D-profiles [28,29]. The parameter for MODELER was set with DOPE-HR method, which is very similar to the DOPE method but obtained at higher resolution. The parameter for 3Dprofiles was set with smooth windows sized 10 and Kabsch-Sander algorithm for secondary structure method [30]. All models were sent to Swiss Expasy to be analyzed with PROCHECK to see the stereo chemical perspective of the model.
3. RESULTS
The comparison of H5N1 NA sequences with a H274Y mutation from Indonesia with other NA sequences from several pandemics had an objective to see the evolution link and relationship between them. From Figure 1, we could see that the NA from ABW06159 is in a different type with AAO46476 and AAT66420. The differences between them are up to 28% residues. This result showed that the H274Y virus may not correlate with the Asian and Hongkong flu pandemics.
Besides the template structure sequence, the closest sequence similarity was from the AAF77036 NA sequence which is a little bit closer than ADL39249. It could be inferred that the NA may have evolved from the 1918 Spanish flu virus. This is really interesting considering that the NA sequence which showed up with a 2-year separation has more differences than the one that emerged more than 90 years ago. Notice that the compared sequences are sequences that have caused major pandemics throughout the years, thus further investigation must be done with more sequences to seek the general relationship between every pandemic. However, the ABW06159 sequence has a close relationship with both of Spanish flu and the 2009 pandemic.
In Table 2, we provide the information of three different assessment methods of the generated models.

Table 1. The accession codes of Nas of influenza viruses from pandemics that occurred during the 20th and 21st centuries.
From the first score column, there are overall residues verify scores were taken using profiles-3d assessment method. The results show the ranges of score are from 192-201. All the models have a score above the high expected score which is only 175.54. This indicated that most residues of all models seem to be placed in an appropriate environment and are compatible for simulation. The verify score of template 3CKZ is exceeded only by model 3.
After overall verify scores were obtained, we looked further into specific residue verify score to find a negative verify score residue. The negative verify score show poor compatibility of the residue. From the Table 3, we could see three residues that has negative verify score (highlighted in yellow). They are E412-C (partially buried), L412-D (buried) and T413 (partially buried). These occurrences varies, in model 1 and 2, they do not occur. In model 3 there are three residues with a negative value. Negative values of residues are also found in model 4 at residue T413, and in model 5 at L412-D and T413. This is interesting that the model with highest overall verify score has more residues with negative verify score than the other models. And moreover, the model 4 and 5, which has negative verify score residues, possess a rela-
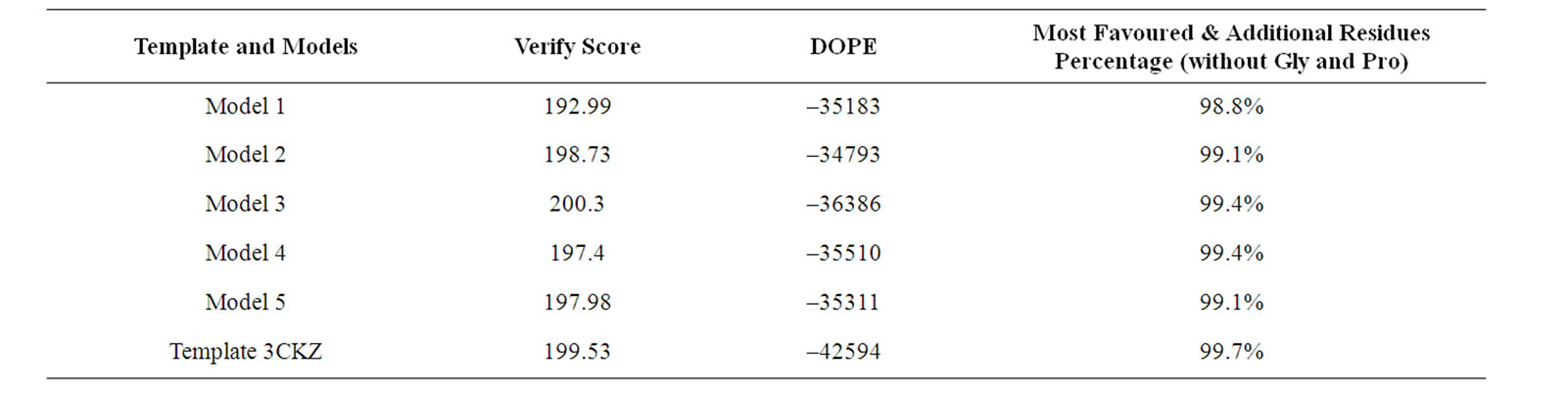
Table 2. The assessment Scores of 5 homology models.
Table 3. The comparisons of specific residue verify score of 5 models from the residues which has a problem.
tively good overall verify score. Another wondering fact is that those three residues had the same secondary structure (a-helix) in all models generated. We overlooked the negative verify score residues problems based on argument that the ratio of the number of negative verify score residues and overall residues is very low (3/385), and the specific Ramachandran plot (not shown) did not put these three residues in the disallowed region.
The next column in Table 2 contained DOPE-HR scores that relate with the stability of molecules. The molecule model which has the most stable structure (showed from its energy) related with the ability of the structure to take the temperature increment which related with the kinetic energy applied when molecular dynamoics simulation is conducted. The higher DOPE score of the molecule, the more unstable they are. The DOPE scores of our models are varies in range –36386 to –34793 and all of them had satisfied the spatial restraints. Even the DOPE score for the template 3CKZ is lowest than the models, the model 3 is the model which has a most negative score compared by other models generated in this experiment. It indicates that model 3 is relatively the most stable and reliable model to be simulated from an energetic perspective.
The third column in Table 2 shows the percentage of residues which are placed in most favoured and additionally allowed regions. The template 3CKZ has the best percentage and over all models structures. For the models generated by homology modeling, the rank is divided by three levels. The first rank is occupied by model 3 and 4, the second rank is occupied by model 2 and 5, and the last rank is occupied only by model 1. All models have not any residue which placed in the disallowed region and have a percentage of residues which placed in generously allowed region below 2%. This indicated that the stereo-chemistry of all models were good enough. However, model 3 and 4 are more preferable to be used for simulation. But if we look back at two other assessment methods, we would able to see that model 3 is the most reliable structure to be simulated with molecular dynamics simulation.
4. DISCUSSION
In this study, the experiment used homology modeling which compared structure of template which had similar sequence with H274Y mutant neuraminidase sequence from the virus circulated in Indonesia. Five models have been successfully generated. From the template 3CKZ which the homology models were generated, we found that a few residues are double numbered in one position and some skipped numbers. The residues that were double numbered are at position 169, 345 and 412, meanwhile the skipped residues number are at position 307, 334, 337, 338, 343, 393 and 434 (N1 residues numbering). The double numbered residues do not give any contribution to the homology models we have generated since its does not mean there are two residues in one position, but only the next residue has numbered, with the same number and the addition alphabet after the number, with its previous residue. The residues that double numbered in one position are 169 and 169-A, 345 and 345-A, and 412, 412-A, 412-B, 412-C and 412-D.
The skipped residues number problem is different. The investigation for skipped residues was executed with the align-and-superimpose protein protocol. In this special occasion, main-chain atoms and secondary structures were used to trace the proteins and superimposed them. The results showed that the skipped residues had narrow spaces (in range 0.23 to 1.41 Å) which would not fit even with one amino acid (Figures 2(a), 2(b), 2(c), 2(d) and 2(e)). The models generated (in blue) have been refined during homology modeling. Sequence alignment (see Figure 3) shows there is no gap in the position of the skipped residues number. Figure 3 is the sequence alignment of Indonesian influenza virus H274Y mutant NA and template 3CKZ. If there are no gaps in the sequence were aligned, then the crystals structure itself which has a problem.
Investigation further was done by checking other N1 crystal structure 2HU0 which was obtained from rcsb. org and found that there is no gap between position 306 and 308 (see Figure 4). The others skipped numbered positions are also not showing the broken peptide bond in 2HU0 (not showed in Figure 4). In the Figure 4 showed only residues at positions 306 and 308 for evidence that the broken bond between two residues do not occur in 2HU0 structure. All investigation results indicate that the spaces in the skipped residue numbers might be the broken peptide bond that occurred during the x-ray crystallographic process.
The PROCHECK main structure assessment did not contain glycine and proline residues in the same Ramachandran plots of general residues because they have special properties. They were provided in specific residue Ramachandran plots. The glycine and proline which placed in unfavourable conformation was put on the Table 4 below. The percentage of most favoured and additional residues in overall structure of the models, and with glycine and proline Ramachandran plots may have a relationship with the verify score where model 1 has the lowest verify score. Even if it is have not occurred in the template structure 3CKZ which has 3 glycine in unfavourable conformation where one of unfavourable glycine is the one with a broken peptide bond.
Figure 5 is the Ramachandran plot of model 3 that
 (a)
(a) (b)
(b) (c)
(c) (d)
(d) (e)
(e)
Figure 2. The superimposed lost residues show very narrow space (pointed out by arrows): (a) between N306 and Q308; (b) between G333 and S335, and between C336 and G339; (c) between S342 and S344; (d) between V392 and K394; (e) between E433 and S435.

Figure 3. Sequences alignment of indonesian virus and template 3CKZ (Number 1 in this figure is number 83 in N1 amino acid position).

Table 4. The list of gly and pro placed in the disallowed region.
shows the residues positioned at categorized regions which are determined by Phi and Psi angles. There are actually four different regions. Most favoured region, additional allowed region, generously allowed region and disallowed region. The stereo-chemistry of model 3 was fit in to the preferably good category. There are no residues placed in the disallowed region. The values of PROCHECK result were compared with other molecules model assessment and show that our resulting model is in the same order. The amylase, ninjurin and lycopene [31-33] models evaluation showed about 87.3%, 87.3% and 88.2% respectively, residues were in the most favoured region while our best model has about 88%. The closest experiment was done with nucleocapsid of H1N1

Figure 4. The structure of amino acids at position 306 and 308 from 2HU0 which do not show the broken peptide bond.
Figure 5. Ramachandran plot of the best model has 88% of its residues (without gly and pro) in the most favoured residues. those in generously allowed regions are labeled.
with percentage of residue in favoured region in range 76.3% - 87.4% [34].
5. CONCLUSIONS
Homology models generated from homology modeling may contain errors since it is only a method to predict how molecules are shaped based on its secondary structure by comparing with the template that was crystallized. The unique perspective of each assessment method that were used here may be an advantage to selecting the best model to simulate. The preferably good structure chosen is model 3 which have the best rank at verify-3D and DOPE score and shared the 1st rank with model 4 as the percentage of the residues which found at the most favoured and additionally allowed region of both models based on their Ramachandran plots.
6. ACKNOWLEDGEMENTS
We would like to express gratitude towards Ding Ming Chee of Accelrys Singapore for the Accelrys Discovery Studio 2.1 trial sent to us.
REFERENCES
- Sedyaningsih, E.R., Isfandari, S., Soendoro, S., Supari, S.F. (2008) Towards mutual trust, transparency and equity in virus sharing mechanism: The avian influenza case of Indonesia. Annals Academy of Medicine Singapore, 37, 482-488
- World Health Organization (2006) Acian influenza (“bird flu”)—Fact sheet. Global Alert and Response. http://www.who.int/csr/disease/avian_influenza/avianinfluenza_factsheetJan2006/en/index.html
- Lowen, A.C. and Palese, P. (2007) Influenza virus transmission: Basic science and implications for the use of antiviral drugs during a pandemic. Infectious Disorders— Drug Targets, 7, 318-328. doi:10.2174/187152607783018736
- Mitnaul, L.J., Matrosovich, M.N., Castrucci, M.R., Tuzikov, A.B., Bovin, N.V., Kobasa, D. and Kawaoka, Y. (2000) Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. Journal of Virology, 74, 6015-6020. doi:10.1128/JVI.74.13.6015-6020.2000
- Garman, E. and Laver, G. (1996) The structure, function and inhibitors. Trends in Cell Biology, 67-71.
- Russell, R.J., Haire, L.F., Stevens, D.J., Collins, P.J., Lin, Y.P., Blackburn, G.M., Hay, A.J., Gamblin, S.J. and Skehel, J.J. (2006) The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature, 443, 45-49. doi:10.1038/nature05114
- Chachra, R. and Rizzo, R.C. (2008) Origins of resistance conferred by the R292K neuraminidase mutation via molecular dynamics and free energy calculations. Journal of Chemical Theory and Computation, 4, 1526-1540. doi:10.1021/ct800068v
- McKimm-Breschkin, J.L., Sahasrabudhe, A., Blick, T.J., McDonald, M., Colman, P.M., Hart, G.J., Bethell, R.C. and Varghese, J.N. (1998) Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to neu5ac2en-derived inhibitors. The Journal of Virology, 72, 2456-2462.
- Mishin, V.P., Hayden, F.G. and Gubareva, L.V. (2005) Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrobial Agents and Chemotherapy, 49, 4515-4520. doi:10.1128/AAC.49.11.4515-4520.2005
- Sheu, T.G., Deyde, V.M., Okomo-Adhiambo, M., Garten, R.J., Xu, X., Bright, R.A., Butler, E.N., Wallis, T.R., Klimov, A.I. and Gubareva, L.V. (2008) Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrobial Agents and Chemotherapy, 52, 3284-3292. doi:10.1128/AAC.00555-08
- Wetherall, N.T., Trivedi, T., Zeller, J., Hodges-Savola, C., McKimm-Breschkin, J.L., Zambon, M. and Hayden, F.G. (2003) Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: Report of the neuraminidase inhibitor susceptibility network. Journal of Clinical Microbiology, 41, 742-750. doi:10.1128/JCM.41.2.742-750.2003
- McKimm-Breschkin, J., Trivedi, T., Hampson, A., Hay, A., Klimov, A., Tashiro, M., Hayden, F. and Zambon, M. (2003) Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrobial Agents and Chemotherapy, 47, 2264-2272. doi:10.1128/AAC.47.7.2264-2272.2003
- Yen, H., Ilyushina, N.A., Salomon, R., Hoffmann, E., Webster, R.G. and Govorkova, E.A. (2007) Neuraminidase inhibitor-resistant recombinant A/Vietnam/1203/04 (H5N1) influenza viruses retain their replication efficiency and pathogenicity in vitro and in vivo. Journal of Virology, 81, 12418-12426. doi:10.1128/JVI.01067-07
- Meijer, A., Lackenby, A., Hungnes, O., Lina, B., Van der Werf, S., Schweiger, B., Opp, M., Paget, J., Van de Kassteele, J., Hay, J. and Zambon, M. (2009) Oseltamivirresistant influenza virus A (H1N1), Europe, 2007-2008 season. Emerging Infectious Diseases, 15, 552-560. doi:10.3201/eid1504.081280
- Monto, A.S., McKimm-Breschkin, J.L., Macken, C., Hampson, A.W., Hay, A., Klimov, A., Tashiro, M., Webster, R.G., Aymard, M., Hayden, F.G. and Zambon, M. (2006) Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrobial Agents and Chemotherapy, 50, 2395-2402. doi:10.1128/AAC.01339-05
- Tamura, D., Mitamura, K., Yamazaki, M., Fujino, M., Nirasawa, M., Kimura, K., Kiso, M., Shimizu, H., Kawakami, C., Hiroi, S., Takahashi, S., Hata, M., Minagawa, H., Kimura, Y., Kaneda, S., Sugita, S., Horimoto, T., Sugaya, N. and Kawaoka, Y. (2009) Oseltamivir-resistant influenza A viruses circulating in Japan. Journal of Clinical Microbiology, 47, 1424-1427. doi:10.1128/JCM.02396-08
- Supari, S.F. (2008) It’s time for the world to change: In the spirit of dignity, equity, and transparency. In: Cardiyan, H.I.S., Alex Tri Tjansono Widodo P.T., Eds., Divine Hand behind Avian Influenza, Sulaksana Watinsa Indonesia, Jakarta.
- Fiser, R.K. and Sali, A. (2000) Modeling of loops in protein structures. Protein Science, 9, 1753-1773. doi:10.1110/ps.9.9.1753
- Laskowski, R.A., MacArthur, M.W., Moss, D.S. and Thornton, J.M. (1993) PROCHECK: A program to check the stereochemical quality of protein structures. Journal of Applied Crystallography, 26, 283-291. doi:10.1107/S0021889892009944
- Morris, A.L., MacArthur, M.W., Hutchinson, E.G. and Thornton, J.M. (1992) Stereochemical quality of protein structure coordinates. Proteins: Structure, Function, and Bioinformatics, 12, 345-364. doi:10.1002/prot.340120407
- Arnold, K., Bordoli, L., Kopp, J. and Schwede, T. (2006) The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics, 22, 195-201. doi:10.1093/bioinformatics/bti770
- Influenza Virus Sequence Database. http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi?go=database
- Protein Data Bank. http://www.pdb.org/pdb/home/home.do
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389-3402. doi:10.1093/nar/25.17.3389
- Saitou, N. and Nei, M. (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406-425
- Sali, A., Pottertone, L., Yuan, F., Van Vlijmen, H. and Karplus, M. (1995) Evaluation of comparative protein modeling by MODELLER. Proteins, 23, 318-326. doi:10.1002/prot.340230306
- Shen, M.-Y. and Sali, A. (2006) Statistical potential for assessment and prediction of protein structures. Protein Science, 15, 2507-2524. doi:10.1110/ps.062416606
- Lüthy, R., Bowie, J.U. and Eisenberg, D. (1992) Assessment of protein models with three-dimensional profiles. Nature, 356, 83-85. doi:10.1038/356083a0
- Ramachandran, G.N., Ramakrishnan, C. and Sasisekharan, V. (1963) Stereochemistry of polypeptide chain configurations. Journal of Molecular Biology, 7, 95. doi:10.1016/S0022-2836(63)80023-6
- Kabsch, W. and Sander, C. (1983) Dictionary of protein secondary structure: Pattern recognition of hydrogenbonded and geometrical features. Biopolymers, 22, 2577- 2637. doi:10.1002/bip.360221211
- Patel, A., Dewangan, R., Khatri, S., Choubey, J., Gupta, S.K. and Verma, M.K. (2009) Identification of insilico 3D structure of amylase (Drosophila melanogaster) and comparative computational studies. Journal of Engineering and Technology Research, 1, 39-45.
- Khatri, S., Patel, S., Choubey, J., Gupta, S.K. and Verma, M.K. (2010) Insilico 3D structure prediction of cell membrane associated protein ninjurin (homosapiens). Current Research Journal of Biological Sciences, 2, 1-5.
- Satpathy, R., Guru, R.K., Behera, R. and Priyadarshini, A. (2010) Homology modelling of lycopene cleavage oxygenase: The key enzyme of bixin production. Journal of Computer Science & Systems Biology, 3, 59-61.
- Singh, S., Kumar, A., Patel, A., Tripathi, A., Kumar, D., and Verma, D. (2010) Silico 3D structure prediction and comparison of nucleocapsid protein of H1N1. Journal of Modelling and Simulation of Systems, 1, 108-111.
Abbreviations
Amino Acid: G = Glycine; A = Alanine; I = Isolucine; F = Phenylalanine; L = Leucine; V = Valine; P = Proline; W = Tryphtophan; S = Serine; D = Aspartat Acid; M = Methionine; C = Cysteine; E = Glutamic Acid; K = Lysine; H = Histidine; R = Arginine; N = Asparagine; Q = Glutamine; T = Threonine; Y = Tyrosine;


