Advances in Biological Chemistry
Vol. 3 No. 1 (2013) , Article ID: 28156 , 7 pages DOI:10.4236/abc.2013.31018
Identification of water-borne bacterial isolates for potential remediation of organophosphate contamination
![]()
Center for Life Sciences Technology, University of Houston, Houston, USA
Email: *riyer@uh.edu
Received 30 November 2012; revised 30 December 2012; accepted 12 January 2013
Keywords: Paraoxon Degradation; Methyl Parathion Degradation; Water Contamination; OP Pesticide
ABSTRACT
Three water-borne bacterial isolates were collected from the Houston metropolitan area. Each isolate was capable of growing upon carbon limited media inoculated with the organophosphorus (OP) compound paraoxon. All isolates were able to efficiently metabolize paraoxon and, to a lesser degree, methyl parathion to p-nitrophenol. 16S rDNA genome sequencing with universal bacterial primers identified the isolates as species belonging to the genera Aeromonas, Stenotrophomonas, or Exiguobacterium. All screened isolates harbor organophosphorus degradation (opd) genes that are approximately 99% similar over approximately 660 base pairs sequenced to one first isolated from Sphingobium fuliginis ATCC 27551 (formerly Flavobacterium sp. ATCC 27551). Additionally, two isolates KKWT11, identified as a putative Senotrophomonas maltophilia, and KKBO11, identified as a putative Exiguobacterium indicum, were found to possess genomic DNA that closely matched a metallobeta-lactamase that has been reported to function as a methyl parathion degradation (mpd) gene suggesting that both of these strains are prime candidates for wastewater remediation of a broad range of OP compounds.
1. INTRODUCTION
Organophosphorus (OP) compounds are potent nerve agents that act upon the enzyme acetylcholinesterase and are widely used as pesticides, fungicides, as well chemical warfare agents such as sarin and soman [1,2]. Common OP pesticides and insecticides include methyl parathion, parathion, paraoxon, chlorpyrifos, dianzon, malathion, and dichlorvos, malathion and chlorpyrifos and are used extensively throughout the developing world [3]. Organophosphates are degraded upon exposure to sunlight or water and are typically immobile in soil and do not typically represent hazards to underground water sources or farmland crops in the short term [4-6]. However, their acute toxicity to humans renders them extremely dangerous in any areas that are continuously treated with such compounds. Additionally, wastewater runoff often results in contamination of above ground water sources where it is extremely toxic to fish and other aquatic life [7,8]. In addition, self-poisoning through occupational exposure occurs regularly in agricultural settings including within the United States [9-12]. The toll on the developing world from organophosphate poisoning is tallied in the millions with approximately 200,000 deaths worldwide annually [9-12]. The growing use and accumulation of these toxic compounds in our food products, in the soils from which they are harvested and as potential runoff into nearby aquatic sources has fueled a growing interest in biotechnology that provides cheap, efficient OP pesticide detoxification to supplement both natural and expensive chemical methods already in use.
Efficient degradation of problematic OP compounds in the environment is typically tied to the presence of bacterial microorganisms that can metabolize OP compounds as sources of carbon, nitrogen or phosphorus [13]. In most cases the isolated species and strains found to possess such a genetic complement are soil microorganisms found in topsoil due to their immediate exposure to the pesticides added to farmland and gardens. While the bacterial genes encoding an OP degrading function are numerous [13], the two of the best described are (1) the opd gene and its homologues, originally isolated from plasmid DNA extracted from S. fuliginis and B. diminuta [14-17] and characterized as a parathion hydrolases (PHs) or organophosphorus hydrolases (OPHs) and (2) homologues of the methyl parathion degrading (mpd) genes that encode for metallo-beta-lactamases, denoted in the literature as a methyl parathion hydrolases or MPH. Unlike the majority of known opd genes, mpd gene homologues are found in both plasmid and chromosomal DNA. Among the first isolated putative MPH’s was originally taken from plasmid DNA in Pseudomona ssp. WBC-3 [18,19] and later was found in genomic DNA in Plesiomonas sp. strain M6 [20]. Both enzyme families work through similar degradative mechanisms, hydrolysis of the OP compound to p-nitrophenol and diethylthiophosphate or dimethylthiophosphate which may or may not be further reduced depending on capacity of the remediative organism or associating microbial population in the environment [14-19]. Currently opd genes encode hydrolytic enzymes with broad temperature and pH optima coupled the ability to at least partially degrade the widest range of organophosphate substrates yet demonstrated for any of the OP hydrolyzing enzyme [13,21], however, the mpd gene family exemplifies a separate OP degrading pathway that imparts a specialized ability to degrade and mineralize methyl parathion, methyl paraoxon and chlorpyrifos [20,22]. Further isolation and characterization efforts has found evidence that a subset of both of the above described OP genes can be flanked by insertion sequences and transposable elements, supporting a lateral transfer mechanism for widespread distribution of these genes [18,22,24-25]. In the latter case the mpd gene originally found in plasmid DNA from Pseudomona ssp. WBC-3 was shown to have migrated to the chromosomes of several Asiatic bacterial strains including Plesiomonas sp. strain M6 and is now found among species of the genera Pseudaminobacter, Achromobacter, Brucella, Ochrobactrum and Stentrophomonas [20,22,24].
In this study we describe the isolation, identification and analysis of microbial isolates collected from both wastewater and potential drinking water sources throughout the Houston metropolitan area that were capable of metabolizing the OP pesticides paraoxon and methyl parathion. Identification of these bacterial organisms was conducted through 16S rDNA sequence analysis. Degradation efficiency of OP pesticides paraoxon and methyl parathion were investigated at the whole cell level and PCR was used to screen and confirm the presence of an organophosphorus degradation (opd) gene or methyl parathion degradation (mpd) gene or genes present in the isolates.
2. MATERIALS AND METHODS
2.1. Chemicals, Enzymes and Oligonucleotides
Paraoxon (O,O-diethyl p-nitrophenyl phosphate), methyl parathion and all other chemicals were obtained from Sigma (St. Louis, MO). Egg white lysozyme, DNase-free ribonuclease A and ampicillin were also obtained from Sigma (St. Louis, MO). Synthetic deoxyoligonucleotides were purchased from Sigma (St. Louis, MO). Proteinase K was acquired from Promega (Madison, WI).
2.2. Media for Bacterial Growth
The composition of all growth mediums used in the study has been tabulated in Table 1. Luria-Bertani medium (LB) was obtained from Difco Laboratories, Detroit, MI. The pH of the LB media was adjusted to 7.5 for the study.
2.3. Isolation of Paraoxon-Metabolizing Microbial Species from Water Samples
A total of three water samples were collected from sites located throughout the Houston-Sugar Land-Baytown metropolitan area. Each water sample was diluted 1:49 in LB medium for a total of 50 mL. The dilutions were kept for 2 d at 30˚C on a shaker. An aliquot (100 μL) from the LB culture was used to inoculate 3 mL of paraoxonsupplemented carbon-deficient minimal media (CDMM). The culture was incubated for 1 wk at 30˚C on a rotary shaker at 200 rpm. A 100 μL aliquot of the bacterial culture was transferred into 3 mL of fresh CDMM containing paraoxon and the incubation step was repeated. After five consecutive subcultivations, the three resultant isolates were placed on to CDMM agar plates containing paraoxon (100 μg∙mL−1). After overnight incubation at 30˚C, the isolates formed on the plates were transferred to glycerol-supplemented minimal media (GSMM) stocks and frozen at −80˚C.
2.4. Isolation of Total Cellular DNA
Bacterial glycerol stocks were used to inoculate 3 mL LB medium and incubated overnight at 37˚C and 200 rpm for approximately 24 h. A 2 mL aliquot of the resulting bacterial culture (OD = 0.6) was pelleted at 13,000 rpm for 3 min and the total cellular DNA was extracted using the CTAB (cetyltrimethylammonium bromide) method
Table 1. Composition of inoculation and growth media used in this study.

[26]. Cells were suspended in TE buffer containing lysozyme (50,000 units∙mL−1) and ribonuclease A (300 Kunitz units∙mL−1), and incubated for 1 h at 37˚C. Immediately prior to lysis, proteinase K (>30,000 units∙g−1) was added to a final concentration 0.25 mg∙mL−1. Cells were lysed with 0.5% (w/v) SDS for 1 h at 37˚C. 100 μL CTAB/NaCl were added to the lysed cells and incubated at 65˚C for 1 h. 500 μL phenol:chloroform:isoamyl alcohol (25:24:1) was then added and the mixture was spun down for 10 min at 13,000 rpm. The resultant aqueous phase was transferred to a clean tube, 500 μL chloroform:isoamyl alcohol (24:1) was added and the process was repeated for 10 min at 13,000 rpm. After transferring the aqueous phase to a new tube, 0.6 vol. cold isopropanol was added and the tubes were incubated at −20˚C overnight. The following day sample tubes were removed and spun down at 13,000 rpm for 15 min at 4˚C. The pellets were washed with 70% ethanol, respun and left to dry at room temperature for approximately 30 min after the supernatant had been decanted off. The DNA pellets were then suspended in 50 μL Tris_EDTA (TE) buffer pH 8.0 + RNAse (100 μg∙mL−1).
2.5. 16S rDNA Sample Identification
The 16S rDNA gene was amplified by PCR from cellular DNA using universal 16S rDNA-specific primers (Forward: AGAGTTTGATCMTGGCTCAG, Reverse: GGTTACCTTGTTACGACTT) [27,28] with PCR 2x Master Mix (Promega, Madison, WI). PCR conditions were set as follows: initial denaturation at 94˚C for 4.5 min, 32 cycles consisting of denaturation at 94˚C for 0.5 min, annealing at 52˚C for 0.5 min, and extension at 72˚C for 1 min, and final elongation at 72˚C for 4 min. PCR products were purified using a QIAquick PCR Purification Kit (Qiagen) and cloned into the pJET1.2/blunt cloning vector using the Clone Jet PCR Cloning Kit and a sticky-end cloning protocol supplied by the manufacturer (Fermentas). Approximately 5 μL of the ligation mixture was used directly to transform chemically competent DH5alpha cells at 42˚C for 40 s. Following a 1 h incubation period in LB media, 125 μL aliquots were plated onto LB-ampicillin-agar plates and grown overnight. Transformants were grown overnight in liquid LB media and harvested for plasmid DNA using a Qiaprep Spin Miniprep Kit (Qiagen). Minipreps for each isolate were sequenced bi-directionally by the dye-terminator method with plasmid specific primers supplied with the kit from the manufacturer (Fermentas) by SeqWright, Inc. (Houston, TX). DNA alignments were made using ClustalX 2.0.12, and sequences were manually edited when necessary to remove gaps and undetermined bases pairs. Corrected sequences were then compared to highly similar sequences in the NCBI database to determine the genus and species of each bacterial isolate.
2.6. Detection of Organophosphorus Degradation Genes by PCR Amplification
Total cellular DNA from each bacterial isolate was used as a template to probe for the presence of an organophosphorus degradation (opd) gene first isolated in Sphingobium sp. ATCC 27551. PCR amplification of the opd gene region was conducted using the following primer set (F196-CGCGGTCCTATCACAATCTC and R840-CTTCTAGACCAATCGCACTG) with PCR 2x Master Mix (Promega, Madison, WI) [29]. Amplification of a putative methyl parathion hydrolase from cellular DNA was conducted with the following primer sets:
(5’-CGTCTATCACCAGCGCATC-3’ and 5’ CACGCATAGGCCTTACCC-3’) and 5’-GAATTCATATGCCCCTGAAGAAC-3’, and reverse, 5’-GAATTCTCGAGCTTGGGGTTGACGACCG-3’) with PCR 2x Master Mix (Promega, Madison, WI). The first mpd primer set targets a conserved region of a putative chromosomal mpd gene encoding for a metallo-beta-lactamase protein often found in Stenotrophomonas and Xanthamonas sp. The second set directly targets the mpd gene sequence of Plesiomonas sp. M6and P. putida WBC-3 [22]. PCR conditions for all three templates were set as follows: initial denaturation at 94˚C for 5 min, 35 cycles consisting of denaturation at 94˚C for 1 min, annealing at 56˚C or 60˚C for 1 min, and extension at 72˚C for 1 min, and final elongation at 72˚C for 5 min. Products were purified using a QIAquick PCR Purification Kit (Qiagen) and checked through electrophoresis on a 1.2% agarose gel. Subsequent cloning, transformation, and sequencing of PCR products were the same as described for 16S rDNA identification in Section 2.5.
2.7. Degradation of Paraoxon and Methyl Parathion by Bacterial Isolates
Aliquots of each isolate and the associated negative control strain were taken from glycerol stock, inoculated into duplicate 150 mL LB media flasks and placed in an incubator at 37˚C, 200 rpm for approximately 24 h (OD = 1). Cells were then pelleted at 2500 rpm for 10 min at 4˚C. The supernatant was poured off and the pellet was washed three times with 0.05 M potassium phosphate buffer (pH = 7.0) at 2500 rpm for 10 min at 4˚C. A total of 25 mL of 0.05 M potassium phosphate buffer was added to the pellet and the mixture was suspended on a rotary shaker at 0˚C over a period of 1 h. The cell suspension was transferred to a clean 250 mL flask to which paraoxon or methyl parathion was added to a final concentration of 100 μg∙mL−1. No additional supplements or metal cofactors (such as CoCl2) were added to the buffer. Each mixture was incubated at 30˚C, 120 rpm for 144 h and absorbance readings of each sample were taken at 405 nm with a Tecan i-control plate spectrophotometer
(set for the BD Falcon35-3072 plate definition and a well diameter of 6000 μm) and 96-well CytoOne plate to measure breakdown to p-nitrophenol every 24 h. Absorbance values were then converted to μM using the extinction coefficient for p-nitrophenol (18,000 M−1∙cm−1). Aliquots taken from 0.05 M potassium phosphate buffer inoculated with the negative control strain and a final concentration of 100 μg∙mL−1 of paraoxon or methyl parathion were used to blank each sample for p-nitrophenol production. Escherichia coli strain JM109 lacking either an opd or mpd gene was used as the negative control. Statistical analysis was used to generate confidence intervals using the standard deviation between replicates of each isolate and an alpha value set at 0.5.
3. RESULTS AND DISCUSSION
Three water-borne bacterial strains (2 gram-positive, 1 gram-negative) that grew on paraoxon and methyl parathion supplemented CDMM plates were collected from the Houston metropolitan area (Table 2). Identification of the species collected in the study was conducted by probing the 16S rDNA by PCR using universal bacterial primers as previously described [33,34]. Sequencing results (Table 3) identify the three isolates as Aeromonas hydrophilia (KKSB11), Stenotrophomonas maltophilia (KKWT11) and Exiguobacterium indicum (KKBO11).
All isolates were screened for OP degradation genes through PCR amplification from total cellular DNA extracted from each isolate. For each isolate an expected band at 663 bp was seen denoting the presence of a Sphingobium-like opd gene (Figure 1(a)). Sequencing results for the 663 bp fragment revealed that the gene present in all three isolates is approximately 99% identical at the nucleotide level to the plasmid-borne gene found in Sphingobium and later in Brevundimonasdi
Table 2. Location of each water isolate collected for this study.
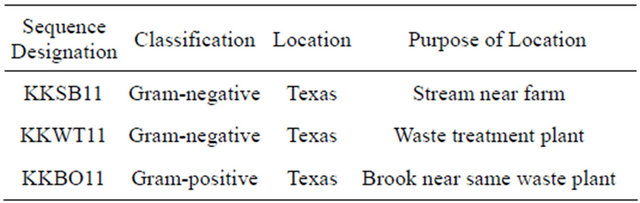
Table 3. Results of 16S rDNA analysis using the NCBI BLAST tool to search for highly similar sequences. The max identity details the highest percent identity for a set of aligned segments to the subject sequence.
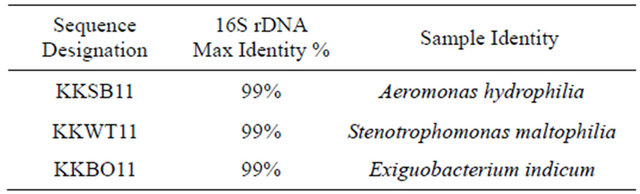
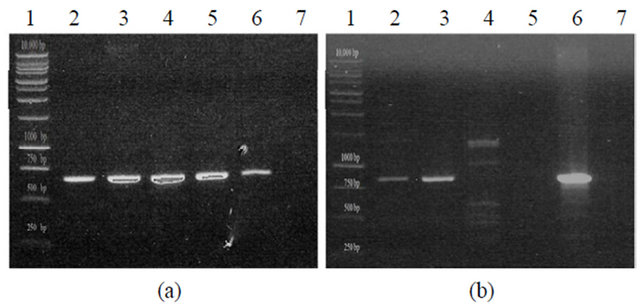
Figure 1. PCR amplification of organophosphorus degradation genes from bacterial isolates. (a) Putative organophosphorus hydrolase (Expected product size of 663 bp). KKBO11_A/B represent two different genomic preps of the same isolate. Lane 1: Promega 1KB Ladder, Lane 2: KKSB11, Lane 3: KKWT11, Lane 4: KKBO11_A, Lane 5: KKBO11_B, Lane 6: (+) Control, Lane 7: (-) Control; (b) Putative methyl parathion hydrolase (Expected product size of 820 bp). Lane 1: Promega 1KB Ladder, Lane 2: KKWT11, Lane 3: KKBO11, Lane 4: KKSB11_A (Annealing temp of 56˚C), Lane 5: KKSB11_B (Annealing temp of 60˚C, Lane 6: (+) Control, Lane 7: (-) Control.
minuta. Isolates were also tested for the presence of an mpd gene using two different sets of primers. All strains were found to lack the transposable Plesiomonas sp. M6 mpd gene variant, though both the KKWT11 and KKBO11 strains were both found to harbor individual metallo-beta-lactamase proteins that could potentially act as a methyl parathion hydrolase in OP degradation (Figure 1(b)). The putative mpd gene found in the KKWT11 strain was found to be closely related to other known S. maltophilia methyl parathion hydrolases sharing approximately 90% - 95% sequence homology with strains S. maltophilia K279a and D457. The mpd gene sequenced from E. indicum is distantly related with little sequence homology with S. maltophilia or other known metallo-beta-lactamases outside of the priming regions.
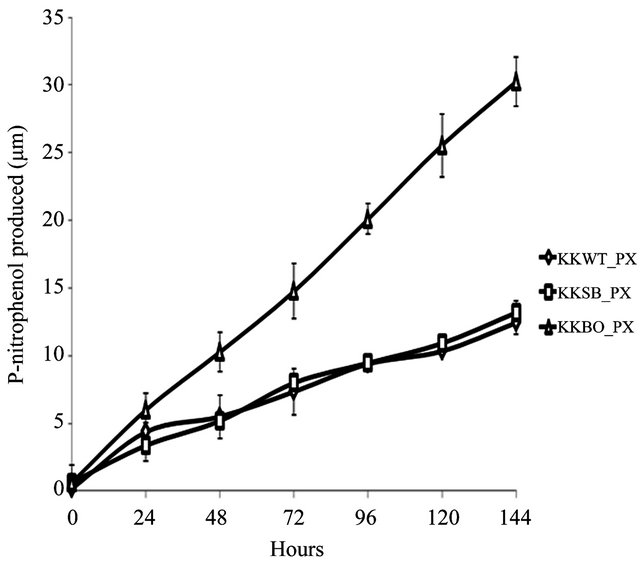
The ability to metabolize OP pesticides paraoxon and methyl parathion to one of its primary metabolites, pnitrophenol, was tested over a period of 144 h. Zero hour represents a time approximately 15 min after inoculation with either pesticide. All three samples were found to be capable of degrading paraoxon to p-nitrophenol (Figure 2(a)) with varying degrees of efficiency. The E. indicum (KKBO11) isolate shows a rapid rise in p-nitrophenol production after 24 h that continued steadily for the duration of the experiment. The p-nitrophenol productionrate of the two remaining isolates S. maltophilia (KKWT11) and A. hydrophilia (KKSB11) are slower, but also remained very consistent over each 24 h time period. Methyl parathion degradation rates were considerably lower and more dynamic in comparison (Figure 2(b)). In general, the KKWT11 isolate demonstrated a sharp rise in p-nitrophenol production for 24 h that quickly tapered off and did not increase further over the course of the study and in fact decreased over the last 48 hours. Both
 (a)
(a)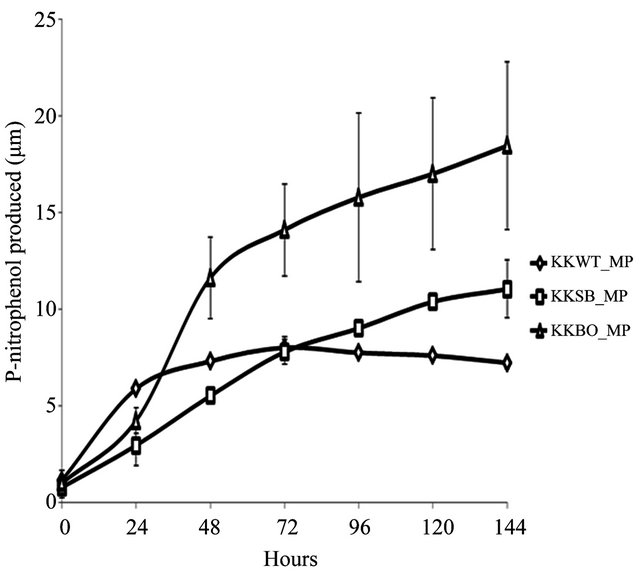 (b)
(b)
Figure 2. Degradation of paraoxon and methyl parathion to p-nitrophenol by bacterial isolates. All isolates were blanked to the negative control for both studies. Error bars are set at a 95% confidence level. (a) Starting concentration of paraoxon was 100 μg∙mL−1. Absorbance readings were taken at 405 nm to detect the concentration of p-nitrophenol in the media over a period of 144 h. E. coli strain JM109 was used as a negative control; (b) Starting concentration of methyl parathion was 100 μg∙mL−1. Absorbance readings were taken at 405 nm to detect the concentration of p-nitrophenol in the media over a period of 144 h. E. coli strain JM109 was used as a negative control.
gram-positive isolates, KSB11 and KBO11, also demonstrated sharp rises in p-nitrophenol present in the medium after 24 - 48 h then slower production for the remainder of the study. Of these remaining two isolates KBO11 had a noticeably greater amount of p-nitrophenol present in the medium by the end of the study.
These results suggest that the overall degradative capacity of each isolate was reliant upon the organism’s gene complement, opd and/or mpd, its tolerance of the stringent media conditions that lacked all other sources of carbon, nitrogen, phosphorus, or any enzymatic cofactors as well as other secondary metabolic pathways that may further break down the primary metabolic byproducts including p-nitrophenol. The promiscuous opd gene that was found in all three isolates would appear to be the principal contributor to the KKSB11 isolate’s capacity to metabolize OP compounds and as such it degrades the substrate paraoxon with greater efficacy then methyl parathion. Accelerated degradation of paraoxon and methyl parathion by the KKBO11 isolate is likely due to the presence of more available enzyme for OP degradation from both its opd and mpd gene complements as the opd gene is still capable of acting upon methyl parathion though with reduced efficacy. The final isolate, KKWT11, also possessed two different genes for OP degradation but both degradative curves are notably different from that of the others. P-nitrophenol levels in the medium were found to decreases lightly over time when methyl parathion was the sole carbon source and suggests a different metabolic pathway may be at work utilizing the p-nitrophenol as a secondary source of carbon.
4. CONCLUSION
Pesticide runoff into above ground water sources is comparatively rare and finding multiple organisms in aquatic reservoirs with the capacity to efficiently degrade OP compounds through shared degradation genes was unexpected. The ubiquitous nature of the Sphingobium sp. opd gene found in each of these water-borne isolates demonstrates that potential sources of drinking water around the Houston metropolitan area have been and are being exposed to OP compounds through runoff or leaching of inoculated soil from nearby farmland, gardens, or are being contaminated from runoff out of water treatment plants. Given that both hydrolases identified in this study together cover a broad range of substrates these isolated strains could be very useful in future bioremediative efforts for a variety of different compounds. Furthermore, taking into account its tolerance in harsh environmental conditions, its genetic complement for OP degradation even in the limiting absence of enzymatic cofactors, the isolate identified as a strain of E. indicum emerges as very strong candidate for bioaugmentation of contaminated wastewater and other aquatic environments.
5. ACKNOWLEDGEMENTS
Partial funding for this project was provided by the Texas Workforce Commission (Meeting Industries’ Critical Workforce Initiative) grant. The authors wish to thank Kevin Smith, biotechnology lab manager, and the Fall 2010 BTEC 3100 undergraduate class for assistance with sample preparation, Kristy Kirkman, undergraduate biotechnology major, for her help in compiling pesticide degradation data and Ashish Damania, undergraduate bioinformatics major, for his help in compiling sequence data.
REFERENCES
- Eddleston, M., Karalliedde, L., Buckley, N., Fernando, R., Hutchinson, G., Isbister, G., Konradsen, F., Murray, D., Piola, J.C., Senanayake, N., Sheriff, R., Singh, S., Siwach, S.B. and Smit, L. (2002) Pesticide poisoning in the developing world—A minimum pesticides list. Lancet, 360, 1163-1167. doi:10.1016/S0140-6736(02)11204-9
- Marrs, T.C. (2007) Toxicology of organophosphate nerve agents. In: Marrs, T.C., Maynard R.L. and Sidell F.R., Eds., Chemical Warfare Agents: Toxicology and Treatment, John Wiley & Sons Ltd., Chichester, 191-221. doi:10.1002/9780470060032.ch8
- World Health Organization in collaboration with the United Nations Environmental Programme (1990) Public Impact of Pesticides Used In Agriculture. WHO, Geneva.
- Cáceres, T., Megharaj, M., Venkateswarlu, K., Sethunathan, N. and Naidu, R. (2010) Fenamiphos and related organophosphorus pesticides: Environmental fate and toxicology. Reviews of Environmental Contamination and Toxicology, 205, 117-162. doi:10.1007/978-1-4419-5623-1_3
- Ragnarsdottir, V.K. (2000) Environmental fate and toxicology of organophosphate pesticides. Journal of the Geological Society of London, 157, 859-876. doi:10.1144/jgs.157.4.859
- Vagi, M.C., Petsas, A.S., Kostopoulou, M.N. and Lekkas, T.D. (2010) Adsorption and desorption processes of the organophosphate pesticides, dimethoate and fenthion, onto three Greek agricultural soils. International Journal of Environmental Analytical Chemistry, 90, 369-389. doi:10.1080/03067310903194980
- Coupe, R.H., Manning, M.A., Foreman, W.T., Goolsby, D.A. and Majewski, M.S. (2000) Occurrence of pesticides in rain and air in urban and agricultural areas of Mississippi. April-September 1995. Science of the Total Environment, 248, 227-240. doi:10.1016/S0048-9697(99)00545-8
- Schipper, P.N., Vissers, M.J., van der Linden, A.M. (2008) Pesticides in groundwater and drinking water wells: Overview of the situation in the Netherlands. Water Science and Technology, 57, 1277-1286. doi:10.2166/wst.2008.255
- Aardema, H., Meertens, J.H., Ligtenberg, J.J., PetersPollman, O.M., Tulleken, J.E. and Zijlstra, J.G. (2008) Organophosphorus pesticide poisonings: Cases and developments. Netherlands Journal of Medicine, 66, 149- 153.
- Buckley, N.A., Karalliedde, L., Dawson, A., Senanayake, N. and Eddleston, M. (2004) Where is the evidence for treatments used in pesticide poisoning? Is clinical toxicology fiddling while the developing world burns? Journal of Toxicology—Clinical Toxicology, 42, 113-116. doi:10.1081/CLT-120028756
- Eddleston, M., Eyer, P., Worek, F., Mohamed, F., Senarathna, L., von Meyer, L., Juszczak, E., Hittarage, A., Azhar, S., Dissanayake, W., Sheriff, M.H., Szinicz, L., Dawson, A.H. and Buckley, N.A. (2005) Differences between organophosphorus insecticides in human selfpoisoning: A prospective cohort study. Lancet, 366, 1452- 1459. doi:10.1016/S0140-6736(05)67598-8
- Karalliedde, L. and Senanayake, N. (1989) Organophosphorus insecticide poisoning. British Journal of Anaesthesia, 63, 736-750. doi:10.1093/bja/63.6.736
- Singh, B.K. and Walker, K.A. (2006) Microbial degradation of organophosphorus compounds. FEMS Microbiology Reviews, 30, 428-471. doi:10.1111/j.1574-6976.2006.00018.x
- Serdar, C.M., Gibson, D.T., Munnecke, D.M. and Lancaster, J.H. (1982) Plasmid involvement in parathion hydrolysis by Pseudomonas diminuta. Applied and Environmental Microbiology, 44, 246-249.
- Harper, L.L., McDaniel, C.S., Miller, C.E. and Wild, J.R. (1988) Dissimilar plasmids isolated from Pseudomonas diminuta MG and Flavobacterium sp. (ATCC 27551) contain identical opd genes. Applied and Environmental Microbiology, 54, 2586-2589.
- Mulbry, W.W., Kearney, P.C., Nelson, J.O. and Karns, J.S. (1987) Physical comparison of parathion hydrolase plasmids from Pseudomonas diminuta and Flavobacterium sp. Plasmid, 18, 173-177. doi:10.1016/0147-619X(87)90046-1
- McDaniel, C.S., Harper, L.L. and Wild, J.R. (1988) Cloning and sequencing of a plasmid-borne gene (opd) encoding for a phosphotriesterase. Journal of Bacteriology, 170, 2306-2311.
- Wei, M., Zhang, J.J., Liu, H., Wang, S.J., Fu, H. and Zhou, N.Y. (2009) A transposable class 1 composite transposon carrying mph (methyl parathion hydrolase) from Pseudomonas sp. Strain WBC-3. FEMS Microbiology Letters, 292, 85-91. doi:10.1111/j.1574-6968.2008.01468.x
- Liu, H., Zhang, J.J., Wang, S.J., Zhang, X.E. and Zhou, N.Y. (2005) Plasmid-borne catabolism of methyl parathion and p-nitrophenol in Pseudomonas sp. Strain WBC- 3. Biochemical and Biophysical Research Communications, 334, 1107-1114. doi:10.1016/j.bbrc.2005.07.006
- Cui, Z.L., Li, S.P. and Fu, G.P. (2001) Isolation of methyl parathion-degrading strain M6 and cloning of the methyl parathion hydrolase gene. Applied and Environmental Microbiology, 67, 4922-4925. doi:10.1128/AEM.67.10.4922-4925.2001
- Cycoń, M., Wójcik, M. and Piotrowska-Seget, Z. (2009) Biodegradation of the organophosphorus insecticide diazinon by Serratia sp. and Pseudomonas sp. and their use in bioremediation of contaminated soil. Chemosphere, 76, 494-501. doi:10.1016/j.chemosphere.2009.03.023
- Yang, C., Liu, N., Guo, X. and Qiao, C. (2005) Cloning of mpd gene from a chlorpyrifos-degrading bacterium and use of this strain in bioremediation of contaminated soil. FEMS Microbiology Letters, 265, 118-125. doi:10.1111/j.1574-6968.2006.00478.x
- Siddavattam, D., Khajamohiddin, S., Manavathi, B., Pakala, S.B. and Merrick, M. (2003) Transposon-like organization of the plasmid-borne organophosphate degradation (opd) gene cluster found in Flavobacterium sp. Applied and Environmental Microbiology, 69, 2533- 2539. doi:10.1128/AEM.69.5.2533-2539.2003
- Zhang, R., Cui, Z.L., Jiang, J., He, J., Gu, X. and Li, S.P. (2005) Diversity of organophosphorus pesticide-degrading bacteria in a polluted soil and conservation of their organophosphorus hydrolase genes. Canadian Journal of Microbiology, 51, 337-343. doi:10.1139/w05-010
- Zhang, R., Cui, Z.L., Zhang, X, Jiang, J., Gu, J.D. and Li, S.P. (2006) Cloning of the organophosphorus pesticide hydrolase gene clusters of seven degradative bacteria isolated from a methyl parathion contaminated site and evidence of their horizontal gene transfer. Biodegradation, 17, 465-472. doi:10.1007/s10532-005-9018-6
- Wilson, K. (1990) Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.2. In: Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K., Eds., Current Protocols in Molecular Biology, John Wiley & Sons, Inc., New York.
- Baker, G.C., Smith, J.J. and Cowan, D.A. (2003) Review and re-analysis of domain-specific 16S primers. Journal of Microbiological Methods, 55, 541-555. doi:10.1016/j.mimet.2003.08.009
- Frank, J.A., Reich, C.I., Sharma, S., Weisbaum, J.S., Wilson, B.A. and Olson, G.J. (2008) Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Applied and Environmental Microbiology, 74, 2461-2470. doi:10.1128/AEM.02272-07
- Sprenger. W.W., Dijkstra, A., Zwart, G.J., Agterveld, M.P., Noort, P.C. and Parson, J.R. (2003) Competition of a parathion-hydrolyzing Flavobacterium with bacteria from ditch water in carbon-, nitrateand phosphate-limited continuous cultures. FEMS Microbiology Ecology, 43, 45-53. doi:10.1111/j.1574-6941.2003.tb01044.x
ABBREVIATIONS
OP, organophosphorusopd, organophosphorus degradation genePH, parathion hydrolaseOPH, organophosphorus hydrolasempd, methyl parathion degradation geneMPH, methyl parathion hydrolaseCDMM, carbon-deficient minimal media)GSMM (glycerol-supplemented minimal mediaCTAB, cetyltrimethylammonium bromide.
NOTES
*Corresponding author.

