Paper Menu >>
Journal Menu >>
 J. Biomedical Science and Engineering, 2009, 2, 70-75 Published Online February 2009 in SciRes. http://www.scirp.org/journal/jbise JBiSE 1 Computer-Assisted analysis of subcellular localization signals and post-translational modifications of human prion proteins Fatemeh Moosawi, Hassan Mohabatkar* Department of Biology, College of Sciences, Shiraz University, Shiraz, Iran, *Corresponding author to Fatemeh Moosawi (mohabat@shirazu.ac.ir), Telephone: +98711-6137426, Mobile: 09177157459, Fax: +98711-2280926. Received August 26th, 2008; revised December 19th, 2008; accepted December 7th, 2008 ABSTRACT In the present work, computational analyses were applied to study the subcellular localiza- tion and posttranslational modifications of hu- man prion proteins (PrPs). The tentative location of prion protein was determined to be in the nu- cleolus inside the nucleus by the following bio- informatics tools: Hum-PLoc, Euk-PLoc and Nuc-PLoc. Based on our results signal peptides with average of 22 base pairs in N-terminal were identified in human PrPs. This theoretical study demonstrates that PrP is post-translationally modified by: 1) attachment of two N-linked complex carbohydrate moieties (N181 and N197), 2) attachmet of glycosylphosphatidylinositol (GPI) at serine 230 and 3) formation of two di- sulfide bonds between “6–22” and “179–214” cysteines. Furthermore, ten protein kinase phosphorylation sites were predicted in human PrP. The above-noted phosphorylation was car- ried out by PKC and CK2. By using bioinfor- matics tools, we have shown that computation- ally human PrPs locate particularly into the nu- cleolus. Keywords: Prion protein; Subcellular localiza- tion; Signal peptides; Post-translational Modifi- cations; Bioinformatics 1. INTRODUCTION Prion diseases or transmissible spongiform encephalo- pathies (TSE) are fatal neurodegenerative conditions in humans and animals that originate spontaneously, ge- netically or by infection [1]. Human TSE diseases include sporadic, genetic, iatrogenic and variant Creutzfeldt- Jakob disease (CJD) and sporadic or familial fatal in- somnia. Animal counterparts are scrapie in sheep and goats, bovine spongiform encephalopathy, and chronic wasting disease of mule deer and elk [2,3]. The critical pathogenetic events in TSE diseases are conformational changes of the physiological host prion protein (PrPc) into an insoluble form (PrPsc) [4]. Prions are devoid of nucleic acid and seem to be com- posed exclusively of protein. The normal, cellular PrP is converted into PrPsc through a process whereby a por- tion of its α-helical and coil structure is refolded into β-sheet [5,6]. This structural transition is accompanied by profound changes in the physicochemical properties of the PrP [7]. At the molecular level, PrP is a sialogly- coprotein of 253 amino acids in human [8]. The C-terminal end is a signal peptide allowing the anchoring of the protein to the plasma membrane via a glycosyl- phosphatidylinositol residue in the early steps of matura- tion in the endoplasmic reticulum [9]. The ultimate des- tination is then the plasma membrane where PrP can be released by phospholipase or protease treatment [10]. Finally, PrP cycles between the plasma membrane and the endocytic pathway. During infection, PrPsc is thought to derive from PrPc after exposure to the plasma membrane [11]. It has been suggested that PrPc can bind a putative ‘‘protein X’’ that may function as a molecular chaperone in the formation of PrPsc [12]. The cellular role of the normal host protein PrP is still unknown. Oesch has characterized membrane associated proteins that interact with PrP [13,14]. Immunocyto- chemical studies reveal that PrPsc and PrPc are present, not only at the plasma membrane or in cytoplasmic compartments [15,16,17], but also in the nuclear com- partment, particularly into the nucleolus [18]. The incon- sistency in the nuclear localization of PrP between non- infected and infected cells may account for important pathogenetical mechanisms [19]. The posttranslational modification of amino acids ex- pands the range of functions of the protein by attaching it to other biochemical functional groups such as acetate, phosphate lipids and carbohydrates. This occurs by al- tering the chemical nature of an amino acid or by making structural changes, like the formation of disulfide bridges. The molecular mechanism by which the PrPsc is formed and causes infectivity or neurodegeneration is not known. In an emerging view, post-translational modifications play roles in the transformation of PrPc to PrPsc [20]. Moreover, Post-translational modification of the scrapie prion protein is thought to account for the unusual fea- SciRes Copyright © 2009  F. Moosawi et al. / J. Biomedical Science and Engineering 2 (2009) 70-75 71 SciRes Copyright © 2009 JBiSE tures of this protein [21]. All eukaryotic cells are compartmentalized into sepa- rate membrane-bound organelles and require tightly regulated transport of proteins and lipids between these compartments. The function of a protein is closely cor- related with its subcellular location. With the rapid in- crease in new protein sequences entering into data banks, we are confronted with a challenge. Proteins are classi- fied, according to their subcellular locations, into the following 18 groups: cell wall, centriole, chloroplast, cyanelle, cytoplasm, cytoskeleton, endoplasmic reticulum, extracell, Golgi apparatus, hydrogenosome, lysosome, mitochondria, nucleus, peroxisome, plasma membrane, plastid, spindle pole body, and vacuole [22]. Determination of subcellular location of a protein is essential for understanding its biochemical function. These data are hard to obtain experimentally but have become especially significant since many protein se- quences are still lacking detailed functional information. To address this rarity of data, many computational analysis methods have been developed. However, these methods have varying levels of accuracy and perform differently based on the sequences that are presented to the underlying algorithm. Giving the huge number of uncharacterized protein sequences, computer-aided analy- sis of posttranslational modifications and translocation signals from amino acid sequence becomes a necessity. In this study, we have analyzed subcellular localiza- tion, signals peptides and posttranslational modifications of human PrPs. 2. MATERIALS AND METHODS 2.1. Amino Acids Sequence The sequences of human PrPs were obtained from http://www.ncbi.nlm.nih.gov/sites/Entrez and http//beta. uniprot.org. Accession numbers of human PrPs are shown in Table 1. 2.2. Consensus Sequence and Percentage of Different Amino Acids The consensus sequence was achieved by using Multalin 5.4.1 server and the percentage of different amino acids was calculated by expasy server. Table 1. Accession numbers of human PrPs AAD46098 AAG21693 AAO83636 AAC05365 AAC78725 AAV38303 AAB59442 AAB59443 AAA60182 AAO83635 A2A2V1 AAR21603 AAH12844 AAH22532 AAS80162 AAA19664 A1YVW6 ABM85428 ABD63004 ABM82244 ABL75508 BAA00011 CAG46836 CAD62016 CAB75503 CAM27320 CAA56283 CAI19053 CAI19053 CAA58442 CAG46869 EAX10449 EAX10450 NP_000302 NP_001073592 NP_001073591 NP_898902 NP_001073590 O75942 P04156 P23907 Q6FGR8 Q5QPB4 Q53YK7 Q6FGN5 Q540C4 Q6SES1 Q5U0K3 Q86XR1 2.3. Prediction of Signal Peptides Signal-CF server was employed to study the signal pep- tides. The web interface to the Signal-CF tool was acces- sible at http://www.chou.med.harvard.edu/shen. This server is called Signal-CF, where C stands for “coupling” and F for “fusion”, meaning that Signal-CF is formed by in- corporating the subsite coupling effects along a protein sequence and by fusing the results derived from many width-different scaled windows through a voting system. Signal-CF is featured by high success prediction rates with short computational time, and hence is particularly useful for the analysis of large-scale datasets [23]. 2.4. Prediction of Subcellular Localization Several computational tools for predicting the subcellu- lar localization of a protein are available. In this study, Hum-PLoc, Euk-PLoc and Nuc-PLoc have been utilized to study the localization of prion protein sequences. Hum-PLoc is a server that analyzes the subcellular lo- calization of human proteins among the following 12 loca- tions: centriole, cytoplasm, cytoskeleton, endoplasmic re- ticulum, extracell, Golgi apparatus, lysosome, microsome, mitochondrion, nucleus, peroxisome, and plasma mem- brane [24]. The web interface to this tool is present at http://www URL: http://www.chou.med.harvard.edu/shen. Euk-PLoc is available as a web-server at http://202. 120.37.186/bioinf/euk. A new benchmark dataset is con- structed that covers the following 18 localizations: cell wall, centriole, chloroplast, cyanelle, cytoplasm, cy- toskeleton, endoplasmic reticulum, extracell, Golgi ap- paratus, hydrogenosome, lysosome, mitochondria, nu- cleus, peroxisome, plasma membrane, plastid, spindle pole body, and vacuole [25]. A new classifier, called Nuc-PLoc, has been devel- oped that can be exploited to recognize nuclear proteins among the following nine subnuclear locations: chroma- tin, heterochromatin, nuclear envelope, nuclear matrix, nuclear pore complex, nuclear speckle, nucleolus, nucleo- plasm and nuclear promyelocytic leukemia (PML) body. As a user-friendly web-server, Nuc-PLoc is accessible at http://chou.med.harvard.edu/ bioinf/Nuc-PLoc [25]. 2.5. Analysis of Posttranslational Modifica- tions N-myristoylation, N-glycosylation, protein kinase C, casein kinase II and Serine, threonine, tyrosine phos- phorylation sites were predicted. Expasy which is avail- able at www.expasy.ch/tools was applied for this purpose. Big-PI server was utilized to study the glycosylphos- phatidylinositol (GPI) anchor signal [26]. The web server http://clavius.bc.edu /~clotel ab/DiANNA was chosen for prediction of disulfide bonds [27]. 3. RESULTS 3.1. Sequences and Signal Peptides Number of amino acids and molecular weight of human consensus sequence of prion protein were 253 and 27661.1 respectively. The consensus sequence of human PrPs is shown in Figure 1. 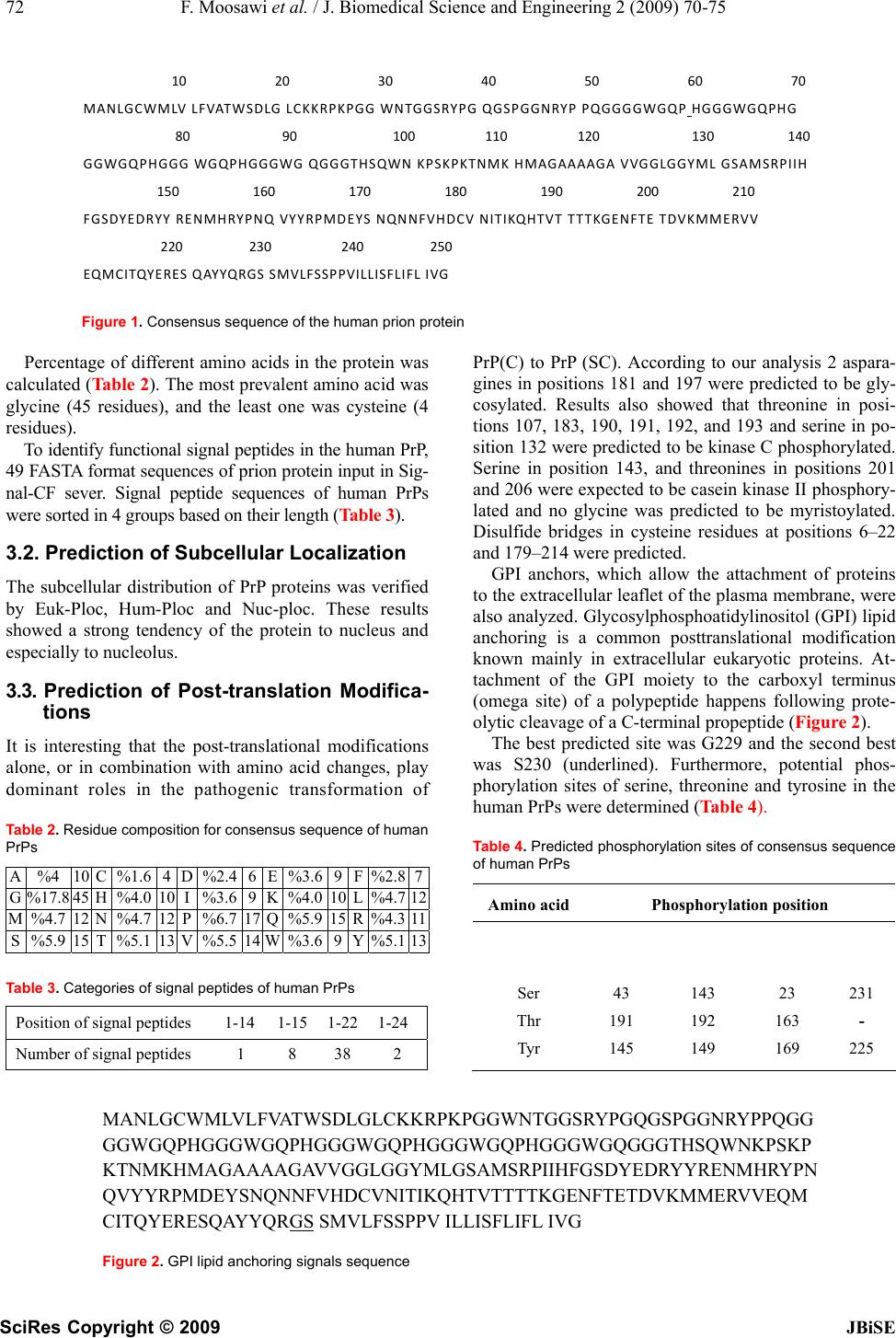 72 F. Moosawi et al. / J. Biomedical Science and Engineering 2 (2009) 70-75 SciRes Copyright © 2009 JBiSE Figure 1. Consensus sequence of the human prion protein Percentage of different amino acids in the protein was calculated (Table 2). The most prevalent amino acid was glycine (45 residues), and the least one was cysteine (4 residues). To identify functional signal peptides in the human PrP, 49 FASTA format sequences of prion protein input in Sig- nal-CF sever. Signal peptide sequences of human PrPs were sorted in 4 groups based on their length (Table 3). 3.2. Prediction of Subcellular Localization The subcellular distribution of PrP proteins was verified by Euk-Ploc, Hum-Ploc and Nuc-ploc. These results showed a strong tendency of the protein to nucleus and especially to nucleolus. 3.3. Prediction of Post-translation Modifica- tions It is interesting that the post-translational modifications alone, or in combination with amino acid changes, play dominant roles in the pathogenic transformation of Table 2. Residue composition for consensus sequence of human PrPs A %4 10 C %1.6 4 D %2.46 E %3.6 9 F%2.87 G %17.8 45 H %4.0 10 I %3.69 K %4.0 10 L%4.7 12 M %4.7 12 N %4.7 12 P %6.7 17 Q %5.9 15 R %4.311 S %5.9 15 T %5.1 13 V %5.514 W %3.6 9 Y%5.113 Table 3. Categories of signal peptides of human PrPs Position of signal peptides 1-14 1-15 1-221-24 Number of signal peptides 1 8 38 2 PrP(C) to PrP (SC). According to our analysis 2 aspara- gines in positions 181 and 197 were predicted to be gly- cosylated. Results also showed that threonine in posi- tions 107, 183, 190, 191, 192, and 193 and serine in po- sition 132 were predicted to be kinase C phosphorylated. Serine in position 143, and threonines in positions 201 and 206 were expected to be casein kinase II phosphory- lated and no glycine was predicted to be myristoylated. Disulfide bridges in cysteine residues at positions 6–22 and 179–214 were predicted. GPI anchors, which allow the attachment of proteins to the extracellular leaflet of the plasma membrane, were also analyzed. Glycosylphosphoatidylinositol (GPI) lipid anchoring is a common posttranslational modification known mainly in extracellular eukaryotic proteins. At- tachment of the GPI moiety to the carboxyl terminus (omega site) of a polypeptide happens following prote- olytic cleavage of a C-terminal propeptide (Figure 2). The best predicted site was G229 and the second best was S230 (underlined). Furthermore, potential phos- phorylation sites of serine, threonine and tyrosine in the human PrPs were determined (Table 4). Table 4. Predicted phosphorylation sites of consensus sequence of human PrPs Amino acidPhosphorylation position Ser Thr Tyr 43 191 145 143 192 149 23 163 169 231 - 225 MANLGCWMLVLFVAT WSDLGLCKKRPKPGGW NTGGSRYPGQGSPGGN RYPPQ GG GGWGQPHGGGWGQPH GGGWG QPHG GGWG QPHG GGWGQGG GTHSQWNK PSKP KTNMKHMAGAAAAGAVVGGL GGY MLGSAMSRPIIH FGSDYE DRYYRENMHRYP N QVYYRPMDEYSNQNNF VHDCVNITIKQHTVTTT TKGENFTET DVKMMERVVEQM CITQYERESQAYYQRGS SMVLFSSPPV ILLISFLIFL IVG Figure 2. GPI lipid anchoring signals sequence 10203040506070 MANLGCWMLVLFVATWSDLGLCKKRPKPGGWNTGGSRYPGQGSPGGNRYPPQGGGGWGQPHGGGWGQPHG 8090100110120130140 GGWGQPHGGGWGQPHG GGWGQGG GTHS QWNKPSKPKTNMKHMAGAAAAGAVVGGLGGYMLGSAMSRPIIH 150160170180190200210 FGSDYEDRYYRENMHRYPNQVYYRPMDEYSNQNNFVHDCVNITIKQHTVT TTTKGENFTETDVKMMERVV 220230240250 EQM C ITQYE R E S QAYYQRGSSMVLFSSPPVILLISFLIFLIVG  F. Moosawi et al. / J. Biomedical Science and Engineering 2 (2009) 70-75 73 SciRes Copyright © 2009 JBiSE 4. DISCUSION The goal of this investigation was to apply bioinformat- ics methods to study the subcellular localizations, signal peptides and posttranslational modifications of human PrPs. 4.1. Identification of Signal Peptides in PrP Based on our results, there were signal peptides with average 22 bp in N-terminal of human PrPs. According to a survey, conducted by Alexandre and his colleagues, PrP does not contain a nuclear localization signal and that, in normal conditions, PrP cannot be released in the cytosolic compartment remaining membrane bound till its degradation and in infected cells, PrP can interact with a molecule to form a complex able to be released in the cytosol and then targeted enters the nucleus [18]. However in another investigation, the presence of two independent nuclear localization signals in the N-termi- nal region of PrP was observed. The first signal included residues 23–28, and the second one included residues 101–106 of PrP [29]. Protein signals have become crucial tools for re- searchers to construct new drugs which are expected to enter a particular organelle to correct a specific defect. For example, by adding a specific tag to a desired protein, one can tag it for excretion, making it much easier to harvest. To use such a tool successfully, first one has to identify the signal sequences. Since the number of nas- cent protein sequences entering databanks is rapidly in- creasing, it is time consuming and expensive to identify the signal peptides entirely by experiments [28]. 4.2. Determinants of Subcellular localization of PrP Protein subcellular localization prediction has been widely studied (reviewed in [30,31]). Available servers differ in many aspects including the computational method, the type and diversity of protein characteristics, the localization coverage, the target organism(s) and the reliability. Servers can be grouped into 4 general classes based upon the protein characteristics that are considered: amino acid composition and order based predictors [32,33,34], sorting signal predictors [35,36], homology based predictors [37,38] and hybrid methods that inte- grate several sources of information to predict localiza- tion [39,40]. Nowadays, the importance of developing a powerful high-throughput tool to predict protein subcel- lular location has become obvious [41]. In the present study, the tentative location of prion protein was determined to be in the nucleolus inside the nucleus by bioinformatics tools, Hum-PLoc, Euk-PLoc and Nuc-PLoc. There are different opinions regarding the subcellular localization of PrP. Stahl and his col- leagues considered a signal peptide at the C-terminus of prion protein allowing the anchoring of the protein to the plasma membrane via a glycosylphosphatidylinositol residue in the early steps of maturation in the endoplas- mic reticulum [8]. Oesch has characterized membrane- associated proteins that interact with PrP [12,13]. More recently, it has been shown that the 37-kDA laminin re- ceptor interacts with PrP [42]. A number of cellular proteins, among them the nuclear lectin CBP35, was identified that bound to the predicted RNA stem-loop structure of PrP RNA. CBP35 could also be detected in purified infections prions, [43]. Moreover, the presence of PrP in the nucleus and its subnuclear location in the nucleolus has been reported [17,44]. In addition, Gu and his coworkers demonstrated that nuclear accumulation of PrP fragments was mediated by nuclear localization signals in the N-terminal domain of PrP that became functional under certain conditions and might contribute to the pathogenesis of certain prion disorders [29]. 4.3. Post-translational Modifications of Consensus Sequence of PrP Our analysis shows that PrP is post-translationally modi- fied by the attachment of two N-linked complex carbo- hydrate moieties (N181 and N197) and a GPI anchor at serine 230 as well as by the formation of a disulfide bond between 6-22 and 179-214 cysteins. Glycosylation is one of the most complex and ubiqui- tous post-translational modifications of proteins in eu- karyotic cells. It is a dynamic enzymatic process in which saccharides are attached to proteins or lipoproteins, usually on serine (S), threonine, asparagine, and trypto- phan residues. Glycosylation, like phosphorylation, is clinically important because of its role in a wide variety of cellular, developmental and immunological processes, including protein folding, protein trafficking and local- ization, cell-cell interactions, and epitope recognition [45,46,47,48,49,50]. The number of glycosylation sites in our work is in agreement with the results obtained by molecular cloning of a PrP cDNA [20]. It has already been shown also that addition of one or two N-glycans causes retention of the N-terminal PrP fragment in the endoplasmic reticulum in a partially aggregated form, and a small amount is secreted into the medium. Pres- ence of two glycans in the N-terminal fragment is more conducive to proper folding and secretion into the medium than one glycan, which largely remains in the ER [29]. In GPI anchors, a hydrophobic phosphatidylinositol group is linked to a residue at or near the C-terminus of a protein through a carbohydrate-containing linker. GPI anchor addition is both structurally and functionally re- lated to another important post-translational modification, prenylation, in which hydrophobic farnesyl or geranyl- geranyl moieties are added to C-terminal cysteine resi- dues of target proteins. Additionally, GPI anchors pro- teins to the cell membrane [51]. Although we determined the nucleus as the tentative location for prion proteins, this fact also should be taken in mind that according to a previous study GPI anchor and the N-glycans function in a complicated way to reduce the tendency of PrP for lo- calization in nucleus [30].  74 F. Moosawi et al. / J. Biomedical Science and Engineering 2 (2009) 70-75 SciRes Copyright © 2009 JBiSE In our study, 10 protein kinases phosphorylation sites were predicted in the human PrPs. The addition of a phosphate molecule to a polar R group of an amino acid residue can turn a hydrophobic portion of a protein into a polar and extremely hydrophilic part. In this way, it can introduce a conformational change in the structure via interaction with other hydrophobic and hydrophilic resi- dues in the protein. Moreover, phosphorylation may modulate PrP biological activity. Regarding the bonding states of cysteine also, it has been found out that it plays important functional and structural roles in proteins. Par- ticularly, disulfide bond formation is one of the most important factors influencing the three-dimensional fold of proteins [52]. In conclusion, this study can help in better under- standing of signal peptides of prion proteins. Generally, our results indicate the role that bioinformatics can play in analysis of proteins modification and localization. ACKNOWLEDGMENT Support of this study by Shiraz University is acknowledged. REFERENCES [1] G. G. Kovacs and H. Budka, (2008) Prion Diseases: From Pro- tein to Cell Pathology. The American Journa of Pathology, 172(3): 555-565. [2] R. G. Will, J. W. Ironside, M. Zeidler, S. N. Cousens, K. Estibiro, A. Alperovitch, S. Poser, M. Pocchiari, A. Hofman and P. G. Smitch, (1996) A new variant of Creutsfeldt-Jakob disease in the UK. Lancet, 347: 921-925. [3] M. E. Bruce, (2006) New variant Creutsfeldt-Jakob disease and bovine spongiform encephalopathy. Nature medicine, 6: 258- 259. [4] I. Loredana, N. Beatriz, Z. Andrea, C. Franco, A. Raquel, S. Marco, B. Simona, I. Marcello, L. Quanguo, V. Vito, L. Mei, F. Franco, C. Salvatore, F. Antonio and P. Maurizio, (2006) Scrapie infectivity is quickly cleared in tissues of orally-infected farmed fish. BMC Veterinary Research, 2: 21. [5] D. A. Lysek, C. Schorn, L. G. Nivon, V. Esteve-Moya, B. Chris- ten, L. Calzolai, C. Von Schroetter, F. Fiorito, T. Herrmann, P. Guntert and K. (2005) Wuthrich, Prion protein NMR structure of cats, dogs, pigs and sheep. Proceedings of the National Academy of Sciences of the United States of America, 102: 640-645. [6] K. M. Pan, M. Baldwin, J. Nguyen, M. Gasset, A. Serban, D. Groth, I. Mehlhorn, Z. Huang, R. J. Fletterick and F. E. Cohen, et al. (1993) Conversion of alpha-helices into beta-sheets fea- tures in the formation of the scrapie prion proteins. Proceedings of National Academy of Sciences of the United States of Amer- ica, 90(23): 10962-10966. [7] B. P. Stantley, (1998) Proceedings of the National Academy of Sciences of the United States of America, 95: 13363-13383. [8] H. A. Kretzschmar, L. E. Stowring, D. Westaway, W. H. Stub- blebine, S. B. Prusiner and S. J. DeArmond, (1986) Molecular cloning of a human prion protein cDNA. DNA, 5: 315–324. [9] N. Stahl, D. R. Borchelt, K. Hsiao and S. B. Prusiner, (1987) Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell, 51: 229-240. [10] N. Stahl, D. R. Borchelt and S. B. Prusiner, (1990) Differential release ofcellular and scrapie prion proteins fromcellular mem- branes by phosphatidylinositol-specific phospholipase C. Biochemistry, 29: 5405-5412. [11] B. Caughey and G. J. Raymond, (1991) The scrapie-associated form of PrP is made from a cell surface precursor that is both protease and phospholipase-sensitive. The Journal of biological chemistry, 266: 18217–18223. [12] G. C. Telling, M. Scott, J. Mastrianni, R. Gabizon, M. Torchia, F. E. Cohen, S. J. DeArmond and S. B. Prusiner, (1995) Prion propagation in mice expressing human and chimeric PrP trans- genes implicates the interaction of cellular PrP with another pro- tein. Cell, 83: 79-90. [13] B. Oesch, D. B. Teplow, N. Stahl, D. Serban, L. E. Hood and S. B. Prusiner, (1990) Identification of cellular proteins binding to the scrapie prion. Biochemistry, 29: 5848–5855. [14] B. Oesch, (1994) Characterization of PrP binding proteins. Phi- losophical transactions of the Royal Society of London. Series B, Biological, 343: 443–445. [15] B. Caughey, K. Neary, R. Buller, D. Ernst, L. Perry, B. Chesebro and R. Race, (1990) Normal and scrapie-associated forms of prion protein differ in their sensitivities to phospholipase and proteases in intact neuroblastoma cells. Journal of virology, 64: 1093-1101. [16] A. Taraboulos, M. Rogers, D. R. Borchelt, M. P. McKinley, M. Scott, D. Serban and S. B. Prusiner, (1990) Acquisition of prote- aseresistance by prion proteins in scrapie-infected cells does not- require asparagine-linked glycosylation. Proceedings of the Na- tional Academy of Sciences of the United States of America, 87: 8262-8266. [17] M. P. McKinley, A. Taraboulos, L. Kenaga, D. Serban, A. Stieber, S. J. DeArmond, S. B. Prusiner and N. Gonatas, (1991) Ultra- structural localization of scrapie prion proteins in cytoplasmic vesicles of infected cultured cells. Laboratory investigation, 65: 622-630. [18] K. Pfeifer, M. Bachmann, H. C. Schroder, J. Forrest and W. E. G. Muller, (1993) Kinetics of expression of prion protein in unin- fected and scrapieinfected N2 (a) mouse neuroblastoma cells. Cell biochemistry and function, 11: 1-11. [19] J. Alexandre, M. Franck, M. Jean, C. Bertrand, P. D. Jean and D. Dominique, (1998) Search for a Nuclear Localization Signal in the Prion Protein. Molecular and Cellular Neuroscience, 11: 127-133. [20] L. Otvos and M. Cudic, (2002) Post-translational modifications in prion proteins. Current protein and peptide science, 3(6): 643-652. [21] T. Haraguchi, S. Fisher, S. Olofsson, T. Endo, D. Groth, A. Tar- entino, D. R. Borchelt, D. Teplow, L. Hood, and A. Burlingame, et al. (1989) Asparagine-linked glycosylation of the scrapie and cellular prion proteins. Archives of biochemistry and biophysics, 274(1): 1-13. [22] H. B. Shen, J. Yang and K. C. Chou, (2007) Euk-PLoc: an en- semble classifier for large-scale eukaryotic protein subcellular location prediction. Amino Acids, 33(1): 57-67. [23] K. C. Chou and H. B. Shen, (2007) Signal-CF: a subsite-coupled and window fusing approach for predicting signal peptides. Bio- chemical and biophysical research communications, 357(3): 633-640. [24] K. C. Chou, and H. B. Shen, (2006) Hum-PLoc: a novel ensem- ble classifier for predicting human protein subcellular localiza- tion. Biochemical and biophysical research communications, 347(1): 150-157. [25] H. B. Shen, and K. C. Chou, (2007) Nuc-PLoc: a new web-server for predicting protein subnuclear localization by fusing PseAA composition and PsePSSM. Protein engineering, design and selection, 20(11): 561-567. [26] B. Eisenhaber, G. Schneider, M. Wildpaner and F. Eisenhaber, (2004) A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to ge- nome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosac- charomyces pombe. Journal of molecular biology, 337(2): 243-253. [27] F. Ferrè, and P. Clote, (2005) Clote. DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Research, 33: 230-232. [28] K. C. Chou, (2001) Using subsite coupling to predict signal  F. Moosawi et al. / J. Biomedical Science and Engineering 2 (2009) 70-75 75 SciRes Copyright © 2009 JBiSE peptides. Protein Engineering, 2: 75-79. [29] Y. Gu, J. Hinnerwisch, R. Fredricks, S. Kalepu, R. S. Mishra and N. Singh, (2002) Identification of cryptic nuclear localization sig- nals in the prion protein. Neurobiology of Disease, 12: 133-149. [30] Z. P. Feng, (2002) An overview on predicting the subcellular location of a protein. In silico biology, 2: 291-303. [31] A. Reinhardt and T. Hubbard, (1998) Using neural networks for prediction of the subcellular location of proteins. Nucleic acids research, 26: 2230-2236. [32] S. Hua and Z. Sun, (2001) Support vector machine approach for protein subcellular localization prediction. Bioinformatics, 17: 721-728. [33] K. C. Chou, (2001) Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins, 43: 246-255. [34] J. D. Bendtsen, H. Nielsen, G. Heijne and S. Brunak, (2004) Improved prediction of signal peptides: SignalP 3.0. Journal of molecular biology, 340: 783-795. [35] O. Emanuelsson, H. Nielsen, S. Brunak and G. Heijne, (2000) Predicting subcellular localization of proteins based on their N- terminal amino acid sequence. Journal of molecular biology, 300: 1005-1016. [36] E. M. Marcotte, I. Xenarios, A. M. Der Bliek and D. Eisenberg, (2000) Localizing proteins in the cell from their phylogenetic profiles. Proceedings of the National Academy of Sciences of the United States of America, 97: 12115-12120. [37] Z. Lu, D. Szafron, R. Greiner, P. Lu, D. S. Wishart, B. Poulin, J. Anvik, C. Macdonell and R. Eisner, (2004) Predicting subcellu- lar localization of proteins using machine-learned classifiers. Bioinformatics, 20: 547-556. [38] K. Nakai and M. Kanehisa, (1992) A knowledge base for pre- dicting protein localization sites in eukaryotic cells. Genomics, 14: 897-911. [39] A. Drawid and M. Gerstein, (2000) A Bayesian system integrat- ing expression data with sequence patterns for localizing pro- teins: comprehensive application to the yeast genome. Journal of molecular biology, 301: 1059-1075. [40] M. S. Scott, D. Y. Thomas, and M. T. Hallett, (2004) Predicting subcellular localization via protein motif co-occurrence. Genome research, 14: 1957-1966. [41] Y. D. Cai, and K. C. Chou, (2004) Predicting subcellular local- ization of proteins in a hybridization space. Bioinformatics, 20(7): 1151-1156. [42] R. Rieger, F. Edenhofer, C. I. Lasme´zas, and S. Weiss, (1997) The human 37-kDA laminin receptor precursor interacts with the prion protein in eukaryotic cells. Nature medicine, 3: 1383-1388. [43] W. E. Müller, U. Scheffer, S. Perovic, J. Forrest and H. C. Schröder, (1997) Interaction of prion protein mRNA with CBP35 and other cellular proteins Possible implications for prion replication and age-dependent changes. Archives of ger- ontology and geriatrics , 25(1): 41-58. [44] J. F. Bazan, R. J. Fletterick, M. P. McKinley and S. B. Prusiner, (1987) Pre-dicted secondary structure and membrane topology of the scrapie prion protein. Protein engineering, 2: 125-135. [45] R. Haltiwanger and J. Lowe, (2004) role of glycosylation in development. Annual Review of Biochemistry, 73: 491-537. [46] P. Mentesana and J. Konopka, (2001) Mutational analysis of the role of N glycosylation in alpha-factor receptor function. Bio- chemistry, 40(32): 9685-9694. [47] K. Pilobello and L. Mahal, (2007) Deciphering the glycocode: the complexity and analytical challenge of glycomics. Current opinion in chemical biology, 11(3): 300-305. [48] S. Miyamoto, (2006) Clinical applications of glycomic ap- proaches for the detection of cancer and other diseases. Current opinion in molecular therapeutics, 8: 507-513. [49] R. Gupta and S. Brunak, (2002) Prediction of glycosylation across the human proteome and the correlation to protein func- tion. Pacific Symposium on Biocomputing, 310-322. [50] C. W. Von der Lieth, A. Bohne-Lang, K. K. Lohmann and M. Frank, (2004) Bioinformatics for glycomics: Status, methods, requirements and perspectives. Briefings in Bioinformatics, 5(2): 164-178. [51] B. Eisenhaber, P. Bork and F. Eisenhaber, (1999) Prediction of Potential GPI-modification Sites in Protein Sequences. Journal of molecular biology, 292: 741-758. [52] S. M. Muskal, S. R. Holbrook and S. H. Kim, Prediction of the disulfide-bonding state of cysteine in proteins. Protein Engi- neering 1990, 3(8): 667-672. |

