Advances in Bioscience and Biotechnology
Vol.3 No.8(2012), Article ID:25840,11 pages DOI:10.4236/abb.2012.38148
Detection of human papillomavirus L1 gene DNA fragments in postmortem blood and spleen after Gardasil® vaccination—A case report
![]()
Milford Hospital and Milford Molecular Laboratory, Milford, Connecticut, USA
Email: shlee01@snet.net
Received 15 September 2012; revised 22 October 2012; accepted 26 November 2012
Keywords: Gardasil®; Postmortem; HPV; DNA; Aluminum; Vaccine
ABSTRACT
A same-nested PCR was used to re-amplify the amplicon of a hypervariable region of the HPV-16 L1 gene DNA in the postmortem blood and splenic tissue obtained at autopsy of a formerly healthy teenage girl who suffered a sudden unexpected death in sleep 6 months after 3 intramuscular injections of a quadrivalent HPV vaccine, Gardasil®. A full autopsy analysis revealed no cause of death. The HPV-16 gene DNA detected in the postmortem materials was similar to the HPV-16 gene DNA fragments in Gardasil® in that both were in non-B-conformation, requiring nondegenerate GP6 and MY11 primers to re-amplify the PCR amplicon for detection and to generate a template useful for direct DNA sequencing. A sequence excised from the base-calling DNA sequencing electropherogram was analyzed by Basic Local Alignment Search Tool (BLAST) alignment and a 45 - 60 base sequence fully matched with a standard hypervariable region of the HPV-16 L1 gene retrieved from the National Center for Biotechnology Information database validated the correct genotyping for HPV- 16 L1 gene DNA. These naked non-proliferating HPV- 16 L1 gene DNA fragments appeared to be in the macrophages of the postmortem blood and spleen, and were protected from degradation by binding firmly to the particulate aluminum adjuvant used in vaccine formulation. The significance of these HPV DNA fragments of a vaccine origin found in postmortem materials is not clear and warrants further investigation.
1. INTRODUCTION
Virus-like particles (VLPs) of human papillomavirus (HPV) are irregularly shaped 30 - 50 nm structures composed of self-assembled HPV major capsid L1 protein pentamers manufactured by a DNA recombinant technology [1,2]. Genotype-specific VLPs for HPV-16, -18, -11 and -6 are used as the active ingredient of a quadrivalent HPV vaccine Gardasil® which has been shown to reduce the incidence of cervical intraepithelial neoplasia grade 2 and grade 3 in vaccinated women [3], and is approved as a vaccine against cancer of the uterine cervix [4]. HPV VLPs adsorbed to particulate amorphous aluminum hydroxyphosphate sulfate (AAHS) adjuvant are exceptionally effective in eliciting production of neutralizing antibodies against HPV-16 in clinical trials [5-7]. The recent revelation that Gardasil® does contain recombinant HPV L1 gene DNA fragments [8,9] which appear to be firmly bound to AAHS nanoparticles [9] may offer a plausible explanation for the high immunogenicity of Gardasil® because co-delivery of both a DNA component and a protein component of a vaccine with aluminum phosphate salts may have the advantage of stimulating a potent and multivalent immune response [10].
On the other hand, the estimated rate of anaphylaxis in the young women receiving Gardasil® vaccination is also high and has been reported to be 5 to 20 times those identified in comparable school-based vaccination programs, using the Brighton case definition of anaphylaxis for diagnostic certainty [11]. A number of cases of possibly immune-based inflammatory neurodegenerative disorders involving the central nervous system, known as acute disseminated encephalomyelitis, following Gardasil® injections have been reported in world literature [12-18]. Among 12,424 reported adverse events following Gardasil® vaccination from June 1, 2006 through December 31, 2008, there were 32 deaths with a mean age of 18 years old, who died 2 to 405 days after the last Gardasil® injection [19]. Medical records and autopsy reports on 20 of the 32 deaths were available for review and confirmed there were 4 unexplained deaths and 6 cardiac-related deaths [19]. No investigative work was attempted to confirm or to exclude any link of a death to Gardasil® vaccination although there was disproportional reporting of syncope among the Gardasil® recipients [19].
The parents of a formerly healthy New Zealand young woman who suffered a sudden unexpected death in sleep 6 months after Gardasil® vaccination requested testing for the presence of HPV L1 gene DNA in the post-mortem samples of their deceased daughter collected at the time of autopsy. Some of the consultants to the parents suggested that if residual HPV L1 gene DNA which is known to be present in the Gardasil® vaccine [8,9] were present in the postmortem samples, there might be a potential link between the residual HPV DNA and the unexplained death of their daughter. This paper reports the experience in developing a method for the detection and validation of minute quantities of HPV-16 L1 gene DNA in the postmortem blood and spleen obtained at autopsy. The data reported in this paper were extracted from a full report which was submitted to the Wellington coronial court at a public inquest held on August 8-9, 2012.
The parents of the deceased have granted their permission to the author to publish the data contained in this report.
2. MATERIALS AND METHODS
2.1. Postmortem Samples
An 18-year-old healthy woman living with her parents was found dead in bed. The only relevant medications that she received before death were three injections of Depo Provera over a period of four years and three doses of intramuscular injections of the HPV vaccine, Gardasil®, in the last year of her life with the last dose of Gardasil® vaccination given six months prior to her demise. There was no history of alcohol or drug abuse. According to the documents presented at the inquest, the patient experienced temperament changes shortly after the first dose of Gardasil® injection, started to have dizziness spells, pins-and-needles feelings in her hands, memory lapses and abdominal pains after the second injection, and developed intermittent weak arm, frequent tiredness requiring daytime naps, increased pins-and-needles feelings in hands causing things to drop from hands, appetite increase with no weight gain, night sweats, loss of ability to use common objects, intermittent chest pain and sudden unexpected “racing heart”. A full autopsy analysis revealed no anatomical, histological, toxicological, genetic or microbiological findings that might be linked to a potential cause of death.
At the parents’ request and by order of the coronial court, DNA samples of the postmortem blood and splenic tissue of the deceased were prepared by Dr. Donald Love at Auckland Hospital Molecular Genetics Laboratory to be tested for the presence of HPV L1 gene DNA. According to Dr. Love, the commercial Gentra® Puregene® Blood Kit (Qiagen) was used to extract the DNA from the nucleated cell fraction of the unfixed blood and splenic tissue which were obtained at the time of autopsy and had been stored at –80˚C. The purified DNA was finally dissolved in TE buffer at the concentration of 0.5 µg of DNA per µL, speedvac dried in plastic tubes, and sent to the author’s laboratory for analyses. Based on the second edition of Gentra® Puregene® Handbook (September 2007), the DNA yield with this method of preparation is about 6 pg DNA per human diploid cell. Therefore, 0.5 µg of DNA was equivalent to the amount of DNA extracted from ~80,000 nucleated cells (500 ng/6pg). A few split samples of the dried DNA are stored at Auckland Hospital for possible future investigation by independent laboratories at the order of the coronial court.
2.2. Low Temperature PCR
The traditional heat-resistant Taq DNA polymerase could not generate a useful nested PCR amplicon from a minute quantity of target HPV DNA in the postmortem materials to be used as a template for direct DNA sequencing. As a result, a LoTemp® PCR with a highly processive HiFi® DNA polymerase system programmed at thermocycling steps not to exceed 85˚C was selected for this study. The general method used to detect HPV L1 gene DNA by heminested (nested) LoTemp® PCR amplification with the GP/MY degenerate consensus primers and validation with direct automated DNA sequenceing for genotyping has been described in detail elsewhere for clinical samples [20-25] and for detecting residual HPV DNA fragments in the Gardasil® vaccine [9]. Each primary PCR consisted of 1 μL of reconstituted DNA solution in molecular grade water containing about 0.5 μg human DNA, 2 μL of water, 1 μL of 10 μM forward primer, 1 μL of 10 μM reverse primer, and 20 μL of ready-to-use LoTemp® PCR master mix with HiFi® DNA polymerase (www.hifidna.com) in a total volume of 25 μL. The thermocycling steps for the LoTemp® PCR system were programmed for an initial heating at 85˚C for 10 min, followed by 30 cycles, each set at 85˚C for 30 sec, 40˚C (low stringency PCR) or 50˚C (high stringency PCR) for 30 sec, and 65˚C for 1 min. The final extension was 65˚C for 10 min (30-cycle Lo-temp program). One μL of each reconstituted sample was placed in a separate PCR tube with a β-globin primer pair for human genomic DNA amplification to assure specimen adequacy.
2.3. HPV-16 L1 Gene DNA Detected by Same-Nested PCR
Transferring of PCR products was accomplished by a micro-glass rod to eliminate the need for micropipetteting to avoid aerosol contamination [25]. The nested PCR mixture consisted of 3 μL of water, 1 μL of 10 μM forward primer, 1 μL of 10 μM reverse primer, and 20 μL of ready-to-use LoTemp® PCR mix with HiFi® DNA polymerase in a total volume of 25 μL. The thermo-cycling steps for the nested PCR were identical to those for primary PCR.
A “same-nested” PCR was introduced for re-amplification of a target region of an HPV L1 gene in the postmortem samples in this case. To perform a same-nested PCR, the primary PCR and the subsequent same-nested PCR(s) were conducted with an identical pair of PCR primers, or the subsequent same-nested PCR was conducted with a pair of the same primers having a few new bases added to the 3’end for one or for both of the primers which had been used in the prior PCR. As a result, all same-nested PCR products were terminated by the first pair of PCR primers used to initiate the primary PCR. The same-nested PCR protocol was found to be necessary to amplify the HPV-16 L1 gene DNA fragments in the postmortem materials in this case and the HPV-16 L1 gene DNA fragments in the Gardasil® vaccine [26].
After completion of the primary and the nested PCR, a 5 µL aliquot of the PCR products was pipetted out from each tube and mixed with 2 µL loading fluid for electrophoresis in a 2% agarose gel containing ethidium bromide. The gel was examined under UV light for various PCR product bands in the agarose gel.
2.4. Direct DNA Sequencing of Nested PCR Amplicon
For DNA sequencing, a trace of the positive nested PCR product was transferred directly with a micro-glass rod from the positive nested PCR tube into a 20 µL volume of a cycle sequencing reaction mixture consisting of 14.5 µL water, 3.5 µL of 5× buffer, 1 µL of BigDye Terminator 1.1 (Applied Biosystems) and 1 µL of 10 µM sequencing primer. After thermal cycling according to the manufacturer’s recommendation, the reaction mixture was loaded in an automated ABI 3130 four-capillary Genetic Analyzer for sequence analysis. Alignment analysis of a 45 - 60 base sequence in the hypervariable region of the L1 gene excised from the computer-generated base-calling electropherogram was performed against various standard HPV genotype sequences stored in the GenBank, using the on-line BLAST (Basic Local Alignment Search Tool) system to validate the specific HPV genotyping.
2.5. Oligonucleotides Used as Primers
The PCR and DNA sequencing primers used and referred to in this report with their DNA sequences are listed as follows.
Degenerate HPV L1 gene primers:
MY09 = 5’-CGTCCMARRGGAWACTGATC-3’
MY11 = 5’-GCMCAGGGWCATAAYAATGG-3’
Key to degenerate nucleotides: M = (A + C), R = (A + G), W = (A + T), Y = (C + T)
Non-degenerate HPV L1 gene primers:
GP5 = 5’-TTTGTTACTGTGGTAGATAC-3’
GP6 = 5’-GAAAAATAAACTGTAAATCA-3’
GP6+ = 5’-GAAAAATAAACTGTAAATCATATTC- 3’
MY11 (1) = 5’-GCACAGGGACATAACAATGG-3’
MY11 (2) = 5’-GCACAGGGACATAATAATGG-3’
MY11 (3) = 5’-GCACAGGGTCATAACAATGG-3’
MY11 (4) = 5’-GCACAGGGTCATAATAATGG-3’
MY11 (5) = 5’-GCCCAGGGACATAACAATGG-3’
MY11 (6) = 5’-GCCCAGGGACATAATAATGG-3’
MY11 (7) = 5’-GCCCAGGGTCATAACAATGG-3’
MY11 (8) = 5’-GCCCAGGGTCATAATAATGG-3’
HPV16MY11+ = 5’-GCACAGGGCCACAATAATGGCAT-3’
The choice of an appropriate combination of PCR primers at different stages was crucial to generate a relatively pure template useful for direct DNA sequencing. Lengthening a PCR primer increased the specificity of target DNA amplification at the expense of sensitivity in this case. The purified full-length HPV-16 plasmid DNA purchased from American Type Culture Collection, diluted to a concentration of 1 copy of HPV-16 L1 gene DNA per µL in TE buffer, was used as the positive control. At the latter theoretical concentration, about 50% of the primary PCRs and 100% of the same-nested PCRs would generate a PCR amplicon of ~190 bp in size at agarose gel electrophoresis when 1 µL of the HPV-16 positive control was used as the template and the degenerate consensus 20-base GP6/MY11 primer pair as the same-nested PCR primers.
3. RESULTS
3.1. Genomic DNA Interfered with Re-Amplification of Target HPV DNA
To detect minute quantities of HPV L1 gene DNA fragments in the whole blood DNA, 1 µL of undiluted reconstituted sample containing 0.5 µg of human genomic DNA was used to start each primary PCR. The general protocol of MY09/MY11 degenerate primer amplification followed by degenerate consensus GP6/MY11 heminested PCR amplification, which has proved highly successful in detecting HPV DNA and in preparing templates for direct DNA sequencing for testing HPV DNA in clinical specimens [25] and in Gardasil® samples [9, 26], generated numerous overlapping PCR product bands at agarose gel electrophoresis when the same protocol was used for testing the postmortem materials. None of such PCR products could be used for direct DNA sequencing. When the 8 individual non-degenerate MY11 primers, i.e. MY11 (1), MY11 (2), MY11 (3), MY11 (4), MY11 (5), MY11 (6), MY11 (7), or MY11 (8), were paired with a GP6 primer to perform 8 individual parallel same-nested PCRs, numerous PCR products of various bp sizes were generated by the same-nested PCR as shown in a gel electrophoresis (Figure 1). Only one of these bands, ~190 bp in size, suggestive of a possible HPV DNA amplicon, although light in intensity, was obtained by PCR with the MY11 (1)/GP6 primer pair (Figure 1, lane 1). DNA sequencing of this same-nested PCR product, using GP6 nucleotide as the sequencing primer, showed an electropherogram of mixed DNA fragments, possibly including an HPV-16 L1 gene DNA sequence by visual analysis (Figure 2). Heminested PCR using a pair of MY09/GP5 primers did not generate an amplicon compatible with an HPV DNA PCR product at gel electrophoresis.
3.2. Second Same-Nested PCR with Elongated Primer for HPV-16 L1 DNA Amplification
To generate an amplicon suitable for DNA sequencing validation, a second same-nested PCR was performed, using a pair of HPV16MY11+/GP6 primers for re-amplification (Figure 3). A two-directional sequencing carried out on this new same-nested PCR amplicon, using the GP6 and the HPV16MY11+ oligonucleotide as the sequencing primer, respectively, showed a typical segment of HPV-16 L1 gene DNA (Figures 4 and 5).
3.3. Co-Amplification of Human Genomic DNA by HPV DNA Primers
In order to characterize the non-target PCR products resulting from a GP6/MY11 primer nested PCR amplification, the amplicon illustrated in lane 2 of Figure 1 was sequenced from its both ends, using the MY11 (2) and GP6 oligonucleotide as the sequencing primer, respectively. The base-calling electropherograms of DNA sequencing confirmed that the amplicon observed in lane 2, Figure 1 represented a 318-bp human genomic DNA sequence terminated by a GP6 primer and an MY11 primer (Figures 6 and 7). The 20-base GP6 and MY11 HPV DNA primers were apparently able to anneal to multiple sites of the human genome with various degrees of complementary base matches and were capable of initiating non-target DNA PCR amplifications in a samenested PCR setting.
3.4. Selection of Non-Degenerate MY11/GP6 Primers to Prepare Template for DNA Sequencing
Based on the above experiments, a same-nested PCR
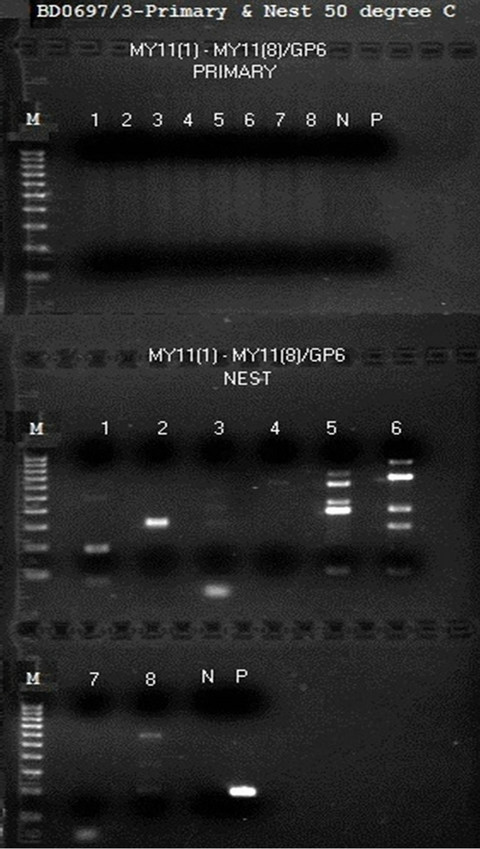
Figure 1. Same-nested PCR amplification with individual nondegenerate MY11 primers pairing with the GP6 primer for detection of HPV L1 gene DNA in a postmortem blood sample. This is a gel electrophoresis showing various same-nested PCR products with different primer pairs. Both the primary PCR and the same-nested PCR were performed in a 25 µL volume containing 20 µL of LoTemp® PCR master mix with HiFi® DNA polymerase, 1 µL of 10 µM GP6 primer and 1 µL of 10 µM of 1 of the 8 non-degenerate MY11 primers labeled MY11 (1) to MY11 (8) with their individual sequences described in the Materials and Methods section. For thermocycling, after an initial heating for 10 min at 85˚C, a 30-cycle amplification with 85˚C for 30 sec, 50˚C for 30 sec, and 65˚C for 1 min was programmed, with a final extension at 65˚C for 10 min for both primary and nested PCRs. The results show that only the MY11 (1)/GP6 primer pair generated a possible HPV DNA amplicon of ~190 bp in lane 1. Numerous nonspecific PCR products were generated when GP6 paired with other non-degenerate MY11 primers. Note: BD0697/3 = Batch No. assigned by Auckland Hospital. Molecular ruler 100 - 1000 bp on the left; N = negative water control; P = HPV-16 plasmid DNA amplified by a pair of degenerate consensus GP6/MY11 primer pair.

Figure 2. DNA sequencing electropherogram on a possible HPV DNA amplicon. Using GP6 as the sequencing primer, the sequencing electropherogram of the ~190 bp MY11 (1)/GP6 same-nested PCR amplicon shown in lane 1, Figure 1, suggested an HPV-16 L1 gene DNA fragment. However, the coexistent interfering nonspecific PCR products prevented a base-calling for validation.
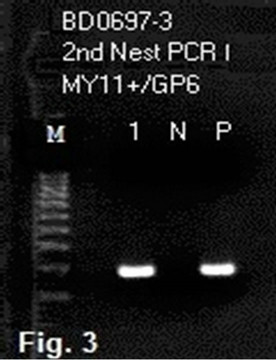
Figure 3. Second nested PCR to generate an amplicon for direct DNA sequencing. The 1st samenested PCR product shown in lane 1, Figure 1, was further amplified by a pair of HPV16MY11+ and GP6 primers in a 2nd nested PCR. This gel electrophoresis shows a clean band of HPV amplicon in lane 1. Note: Lane 1 = 2nd nested PCR amplicon using the 1st nested PCR product described in lane 1, Figure 1 as the template; N = negative water control; P = PCR product from the P control in 1st nested PCR was used as the template.
with a pair of 20-base non-degenerate MY11 (1)/GP6 primers was chosen for the initial detection of HPV-16 L1 gene DNA fragments in all batches of DNA extraction on this case. It turned out that only about 1/3 of the same-nested PCRs with this protocol showed a positive amplicon of HPV-16 L1 gene DNA, eventually validated by DNA sequencing. The non-target human genomic DNA fragments which were co-amplified in the samenested PCR varied considerably. For example, in a splenic DNA extract, when four 1 µL aliquots of the reconstituted DNA sample were used for a parallel same-nested PCR experiment, all using the non-degenerate MY11 (1)/GP6 primer pair for amplification, and two of the PCRs were cycled under a low stringency condition with a 40˚C annealing temperature while two under a high stringency condition with a 50˚C annealing temperatureonly one of the PCRs generated a ~190 bp HPV DNA amplicon. Even in the latter nested PCR, there was a heavy ~500 bp product which was the result of co-amplification of non-target DNA in a heminested PCR setting (Figure 8, lane 4). A 3rd nested PCR using a nondegenerate elongated HPV16MY11+/GP6+ primer pair was needed to selectively amplify the target HPV DNA (Figure 9) in preparation of an amplicon suitable to be used for DNA sequencing (Figure 10) to confirm that HPV-16 L1 gene DNA was also present in the splenic tissue obtained at autopsy.
4. DISCUSSION
A same-nested PCR in which one identical pair of nondegenerate primers selected from the well-characterized degenerate consensus 20-base GP6/MY11 primer group, referred to as the MY11 (1)/GP6 primers in this report, was needed to detect the HPV-16 L1 gene DNA fragments present in the postmortem blood and the splenic tissue obtained at autopsy of a teenage girl who suffered a sudden unexpected death in sleep 6 months after Gardasil® vaccination. Since the human genomic samples contain numerous DNA fragments which are substantially complementary to the base sequences of the HPV PCR primers, co-amplification of non-target DNAs of the human genome invariably occurs in the same-nested PCR settings when PCR amplicons are re-amplified with the same primer(s). These human genomic DNA segments may act as primer-binding PCR inhibitors even when the partially matched primer-binding does not generate PCR amplicon bands visible at agarose gel electrophoresis. The same-nested PCR procedure has the effects of reducing the concentration of inhibitors carried over from the original sample by simple dilution so that the chance of obtaining a target DNA amplicon is significantly increased [25]. However, additional same-nested PCR with an elongated 23-base HPV16MY11+ primer and an elongated 25-base HPV16GP6+ primer may be needed to selectively generate a specific 184-bp HPV-16 PCR amplicon to be used as the template for direct DNA sequencing.
The elongated HPV16MY11+/HPV16GP6+ primers cannot be used to generate a PCR amplicon directly from the HPV-16 DNA template in the postmortem materials in this case. In comparison, the 184-bp L1 gene DNA
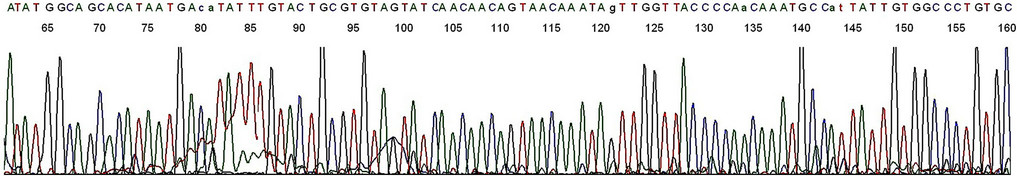
Figure 4. Base-calling electropherogram of DNA sequencing of an HPV PCR amplicon using GP6 as the sequencing primer. The 2nd nested PCR products described in lane 1, Figure 3, was used as the template for sequencing. The last 23 bases are the HPV16MY11+ primer-binding site.
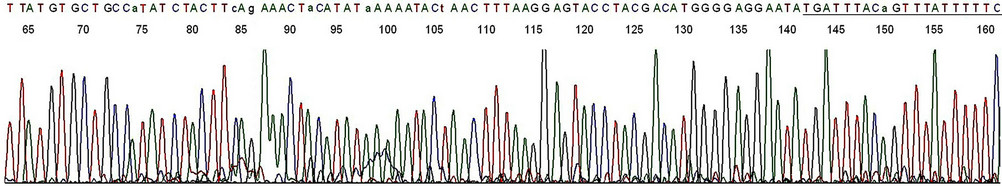
Figure 5. Base-calling electropherogram of DNA sequencing of an HPV PCR amplicon using HPV16MY11+ as the sequencing primer. The 2nd nested PCR products described in lane 1, Figure 3, was used as the template for sequencing. The last 20 bases are the GP6 primer-binding site (underlined). A composite two-directional 184-base sequence derived from the base-calling electropherograms depicted in Figures 4 and 5 is 100% matched with that of the standard HPV-16 L1 gene DNA as follows: GCACAGGGCCACAATAATGGCATTTGTTGGGGTAACCAACTATTTGTTACTGTTGTTGATACTACACGCAGTACAAATATGTCATTATGTGCTGCCATATCTACTTCAGAAACTACATATAAAAATACTAACTTTAAGGAGTACCTACGACATGGGGAGGAATATGATTTACAGTTTATTTTTC-3’ (HPV-16 genome Locus ID AF125673, location 6582-6765, direction 5’-3’, retrieved from the National Center for Biotechnology Information database).

Figure 6. Base-calling electropherogram of DNA sequencing of a non-target PCR amplicon, using MY11 (2) as the sequencing primer. The nested PCR amplicon shown in lane 2, Figure 1, was used as the template for sequencing. The 20 bases of GP6 primer-binding site are in the end as for the HPV-16 L1 gene DNA sequence shown in Figure 5.
template in the HPV-16 plasmid DNA control and in the HPV-16 DNA isolated from clinical cervicovaginal cytology samples is always successfully amplified by the 20-base degenerate consensus GP6/MY11 primer pair and by the elongated HPV16MY11+ /HPV16GP6+ primer pair under identical same-nested PCR conditions. Unlike the L1 gene in the HPV-16 plasmid DNA and in the HPV-16 isolates from clinical cervicovaginal samples, the HPV-16 L1 gene DNA fragments found in the postmortem blood and splenic samples cannot be amplified under low stringency PCR condition and lacks a useful MY09 primer-binding site for PCR amplification. These variances in PCR amplification characteristics indicate that there are topological conformational changes in the HPV-16 L1 gene DNA fragments in the postmortem samples. Similar topological non-B conformational changes in HPV L1 gene DNA fragments bound to the AAHS particles in the Gardasil® vaccine have been demonstrated by a low temperature (LoTemp®) PCR catalyzed by a highly processive DNA polymerase with proof-reading function [26]. Aluminum, unlike other metals, is known to stabilize and destabilize portions of a dsDNA molecule at different pHs, cause intrastrand cross-links, and create a “non-cooperative melting profile” for the bound
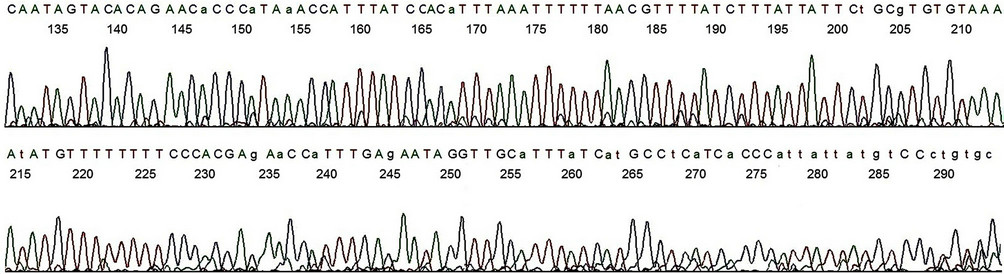
Figure 7. Base-calling electropherogram of DNA sequencing of a non-target PCR amplicon, using GP6 as the sequencing primer. The nested PCR amplicon shown in lane 2, Figure 1, was used as the template for sequencing. The 20 bases of MY11 (2) primer-binding site are in the end of the electropherogram as for the HPV-16 L1 gene DNA shown in Figure 4. A composite two-directional inter-primer sequence derived from the base-calling electropherograms depicted in Figures 6 and 7 shows a human genomic DNA sequence flanked by a GP6 primer-binding site (underlined small letters) and by an MY11 primer-binding site (underlined capitalized letters) as follows:
gcactgggacataataatggGTGATGAGGCATGATAAATGCAACCTATTCTCAAATGGTTCTGGTGGGAAAAAAAACATATTTTACACACGCAGAATAATAAAGATAAAACGTTAAAAAATTTAAATGTGGATAAATGGTTTATGGGTGTTCTGTGTACTATTGCTACTTTTCTAAAAGTTAAACATTGTTTCAAAAAACATAAGGTTTTTTCACACTAGAAATTCTTTTTATATGCCATTTATATTGTTTACAAAATATTCCGCCTTATATATGTATCATAACTTATAGACTATTTTGTGATTTACAGTTTATTTTTC.
DNA molecule [27].
Gardasil® is a quadrivalent vaccine which is known to contain residual recombinant HPV L1 gene DNA fragments [8]. As vaccine excipient, HPV L1 gene DNA fragments of all four genotypes, namely HPV-16, -18, -11 and -6, are expected to be present in any vaccine lot. However, previous studies have shown that only the L1 gene DNA of HPV-11 or HPV-18, or a combination of both was successfully amplified by a pair of degenerate consensus GP6/MY11 PCR primers [9], and that specially modified non-degenerate GP6/MY11 primers were needed to amplify the HPV-16 L1 gene DNA fragments bound to the insoluble fraction of the vaccine [26], indicating conformational changes taking place when naked HPV DNA is bound to the AAHS adjuvant during vaccine formulation. The topological conformational changes in the bound DNA may be genotype-related. The current study shows that only HPV-16 L1 gene DNA was detected 6 months after last vaccination, further suggesting that the non-B-conformation has protected the HPV-16 L1 gene DNA fragments from being degraded by various nucleases in the human body. Unprotected foreign DNA fragments in B conformation introduced into peripheral blood of a mammalian host are known to be degraded and eliminated within 48 hours [28].
HPV-16 is a virus which only infects human mucosal epithelial cells. HPV-16 DNA may be detected in the plasma of patients with invasive squamous cervical cancer harboring the same genotype of virus, but not in the control subjects without cervical cancer [29]. HPV-16 DNA has been reported to be present in peripheral blood mononuclear cells from human immunodeficiency virus (HIV)-infected pediatric patients and even from healthy blood donors [30]. However, unlike the HPV-16 L1 gene DNA fragments found in the Gardasil® vaccine and in the postmortem materials in this autopsy case, the HPV- 16 L1 gene DNA in those reported clinical samples is always in B conformation which is readily amplified by a pair of degenerate or consensus PCR primers from both ends defined by the MY09 and MY11 binding sites [30].
The HPV-16 L1 gene DNA fragments detected in the postmortem blood and splenic tissue in this case are presumably in minute quantities and in the nucleated cells, probably macrophages. Naked viral and bacterial DNA fragments firmly bound to insoluble aluminum salts can be carried into tissue macrophages through phagocytosis to initiate a series of DNA-related immune reactions [31-34]. Intramuscular injection of free HPV-16 L1 plasmid DNA in BALB/C mice without adjuvant has been known to induce a strong CD8 T cell response [35], indicating that under certain conditions non-replicating HPV L1 gene DNA can activate the immune system. However, to be detectable 6 months after intramuscular injection, the naked foreign DNA in the host must be in a stabilized physical condition, either by remaining bound to the AAHS nanoparticles or by integration into the human genome through hitherto poorly understood mechanisms [36-40].
The presence of HPV-16 L1 gene DNA fragments of a vaccine origin indicates possible co-existence of other companion microbial DNA, such as DNA fragments of the plasmid pGAL110 and yeast cells which are used in the vaccine production by the manufacturer [2]. A potential consequence of these viral and microbial DNA fragments with their unmethylated CpG motifs in macro-
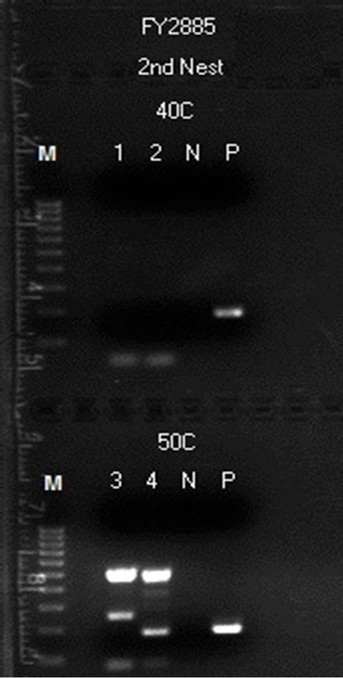
Figure 8. Detection of HPV L1 gene DNA in a postmortem splenic sample by a second same-nested PCR amplification with a nondegenerate HPV16MY11+/GP6 primer pair. Description: Four parallel same-nested PCRs with the MY11 (1)/GP6 primer pair, each started with 0.5 µg of human genomic DNA extracted from spleen, were performed, two under low stringency condition with 40˚C annealing temperature and two under high stringency condition with 50˚C annealing temperature. No HPV amplicon was obtained in the 1st same-nested PCRs. Then the second same-nested PCRs each with an HPV16MY- 11+/GP6 primer pair were performed. As depicted in this gel electrophoresis, an intense HPV DNA amplicon band of ~190 bp in size was generated and shown in lane 4 in one of the two second nested PCRs under high stringency condition only. However, due to coamplification of human genomic DNA which generated a large amount of high molecular weight PCR products, it was impossible to use this material as the template for DNA sequencing.
phages [41-46] is to cause release of various cytokines, including tumor necrosis factor (TNF), a recognized myocardial depressant [47-51]. TNF-induced hypotensive shock is a documented observation among animals
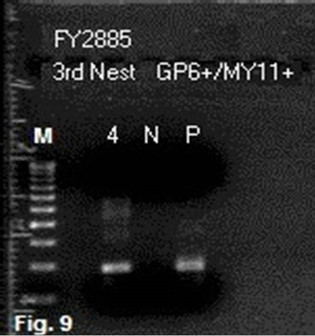
Figure 9. Third same-nested PCR amplification with two elongated primers to prepare HPV-16 DNA template from splenic DNA for sequencing. The second samenested PCR products of the splenic DNA depicted in lane 4, Figure 8, were selectively amplified by a pair of HPV16MY-11+ and GP6+ primers to obtain a target HPV- 16 amplicon for DNA sequencing. Note: M = molecular ruler; 4 = re-amplification of the lane 4 PCR products depicted in Figure 8 with two elongated primers; N = negative water control; P = HPV-16 plasmid DNA control.
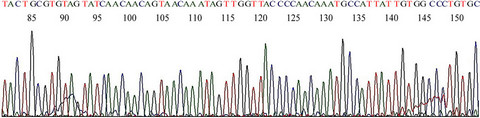
Figure 10. Base-calling electropherogram of DNA sequencing on HPV PCR amplicon from postmortem splenic DNA. This is a typical HPV-16 L1 gene DNA sequence using GP6+ as the sequencing primer and the #4 PCR amplicon described in Figure 9 as the template.
[52,53] and humans [54,55]. To answer the question whether the quantity of these persistent viral or microbial DNA fragments can stimulate the macrophages to release enough TNF to generate a significant pathophysiological impact following Gardasil® vaccination needs expanded research.
5. CONCLUSION
Detection of HPV-16 L1 gene DNA fragments in non-Bconformation in postmortem blood and spleen from a person who died suddenly and unexpectedly 6 months after quadrivalent HPV vaccination has not been previously reported and warrants further investigation.
6. ACKNOWLEDGEMENTS
This study was commissioned and sponsored by SANE VAX, Inc. for a future payment not to exceed one US dollar. The author thanks Ms. Veronica S. Vigliotti and Ms. Jessica S. Vigliotti for donating their extremely valuable technical and professional time to assist completion of this study.
REFERENCES
- Mach, H., Volkin, D.B., Troutman, R.D., Wang, B., Luo, Z., Jansen, K.U. and Shi, L. (2006) Disassembly and reassembly of yeast-derived recombinant human papillomavirus virus-like particles (HPV VLPs). Journal of Pharmaceutical Sciences, 95, 2195-2206. doi:10.1002/jps.20696
- Bryan, J.T. (2007) Developing an HPV vaccine to prevent cervical cancer and genital warts. Vaccine, 25, 3001- 3006. doi:10.1016/j.vaccine.2007.01.013
- The Future II Study Group (2007) Quadrivalent vaccine against human papillomavirus to prevent high-grade cervical lesions. The New England Journal of Medicine, 356, 1915-1927. doi:10.1056/NEJMoa061741
- Merck & Co., Inc. (2006) Gardasil®[Quadrivalent Human Papillomavirus Types 6, 11, 16, 18 Recombinant Vaccine]. Merck Product Document 9883616. http://www.merck.com/product/usa/pi_circulars/g/gardasil/gardasil_pi.pdf
- Einstein, M.H., Baron, M., Levin, M.J., Chatterjee, A., Edwards, R.P., Zepp, F., Carletti, I., Dessy, F.J., Trofa, A.F., Schuind, A., Dubin, G. and HPV-010 Study Group (2009) Comparison of the immunogenicity and safety of Cervarix and Gardasil human papillomavirus (HPV) cervical cancer vaccines in healthy women aged 18 - 45 years. Human Vaccines, 5, 705-719. doi:10.4161/hv.5.10.9518
- Giuliano, A.R., Lazcano-Ponce, E., Villa, L., Nolan, T., Marchant, C., Radley, D., Golm, G., McCarroll, K., Yu, J., Esser, M.T., Vuocolo, S.C. and Barr, E. (2007) Impact of baseline covariates on the immunogenicity of a quadrivalent (types 6, 11, 16, and 18) human papillomavirus virus-like-particle vaccine. Journal of Infectious Diseases, 196, 1153-1162. doi:10.1086/521679
- Villa, L.L., Ault, K.A., Giuliano, A.R., Costa, R.L., Petta, C.A., Andrade, R.P., Brown, D.R., Ferenczy, A., Harper, D.M., Koutsky, L.A., Kurman, R.J., Lehtinen, M., Malm, C., Olsson, S.E., Ronnett, B.M., Skjeldestad, F.E., Steinwall, M., Stoler, M.H., Wheeler, C.M., Taddeo, F.J., Yu, J., Lupinacci, L., Railkar, R., Marchese, R., Esser, M.T., Bryan, J., Jansen, K.U., Sings, H.L., Tamms, G.M., Saah, A.J. and Barr, E. (2006) Immunologic responses following administration of a vaccine targeting human papillomavirus types 6, 11, 16, and 18. Vaccine, 24, 5571-5583. doi:10.1016/j.vaccine.2006.04.068
- US Food and Drug Administration (2011) FDA information on gardasil-presence of DNA fragments expected, no safety risk. http://www.fda.gov/BiologicsBloodVaccines/Vaccines/ApprovedProducts/ucm276859.htm
- Lee, S.H. (2012) Detection of human papillomavirus (HPV) L1 gene DNA possibly bound to particulate aluminum adjuvant in the HPV vaccine Gardasil®. Journal of Inorganic Biochemistry, 117, 85-92. doi:10.1016/j.jinorgbio.2012.08.015
- Kwissa, M., Lindblad, E.B., Schirmbeck, R. and Reimann, J. (2003) Codelivery of a DNA vaccine and a protein vaccine with aluminum phosphate stimulates a potent and multivalent immune response. Journal of Molecular Medicine, 81, 502-510. doi:10.1007/s00109-003-0452-9
- Brotherton, J.M., Gold, M.S., Kemp, A.S., McIntyre, P.B., Burgess, M.A., Campbell-Lloyd, S. and New South Wales Health HPV Adverse Events Panel (2008) Anaphylaxis following quadrivalent human papillomavirus vaccination. Canadian Medical Association Journal, 179, 525-533. doi:10.1503/cmaj.080916
- Sutton, I., Lahoria, R., Tan, I., Clouston, P. and Barnett, M. (2009) CNS demyelination and quadrivalent HPV vaccination. Multiple Sclerosis Journal, 15, 116-119. doi:10.1177/1352458508096868
- Wildemann, B., Jarius, S., Hartmann, M., Regula, J.U. and Hametner, C. (2009) Acute disseminated encephalomyelitis following vaccination against human papilloma virus. Neurology, 72, 2132-2133. doi:10.1212/WNL.0b013e3181aa53bb
- Mendoza Plasencia, Z., González López, M., Fernández Sanfiel, M.L. and Muñiz Montes, J.R. (2010) Acute disseminated encephalomyelitis with tumefactive lesions after vaccination against human papillomavirus. Neurologia, 25, 58-59. doi:10.1016/S0213-4853(10)70023-2
- Chang, J., Campagnolo, D., Vollmer, T.L. and Bomprezzi, R. (2011) Demyelinating disease and polyvalent human papilloma virus vaccination. Journal of Neurology, Neurosurgery, and Psychiatry, 82, 1296-1298. doi:10.1136/jnnp.2010.214924
- DiMario Jr., F.J., Hajjar, M. and Ciesielski, T. (2010) A 16-year-old girl with bilateral visual loss and left hemiparesis following an immunization against human papilloma virus. Journal of Child Neurology, 25, 321-327. doi:10.1177/0883073809349322
- Balamoutsos, G., Bouktsi, M., Paschalidou, M., Tascos, N. and Milonas, I. (2009) A report of five cases of CNS demyelination after quadrivalent human papilloma virus vaccination: Could there be any relationship? www.guthyjacksonfoundation.org/services/download.php?2297.pdf+374
- Rossi, M., Bettini, C. and Pagano, C. (2011) Bilateral papilledema following human papillomavirus vaccination. Journal of Medical Cases, 2, 222-224.
- Slade, B.A., Leidel, L., Vellozzi, C., Woo, E.J., Hua, W., Sutherland, A., Izurieta, H.S., Ball, R., Miller, N., Braun, M.M., Markowitz, L.E. and Iskander, J. (2009) Postlicensure safety surveillance for quadrivalent human papillomavirus recombinant vaccine. The Journal of the American Medical Association, 302, 750-757. doi:10.1001/jama.2009.1201
- Lee, S.H., Vigliotti, V.S., Vigliotti, J.S. and Pappu, S. (2007) Routine human papillomavirus genotyping by DNA sequencing in community hospital laboratories. Infectious Agents and Cancer, 2, 11. doi:10.1186/1750-9378-2-11
- Lee, S.H., Vigliotti, V.S. and Pappu, S. (2009) Human papillomavirus (HPV) infection among women in a representative rural and suburban population of the United States. International Journal of Gynecology & Obstetrics, 105, 210-214. doi:10.1016/j.ijgo.2009.01.019
- Lee, S.H., Vigliotti, V.S. and Pappu, S. (2009) Molecular tests for human papillomavirus (HPV), Chlamydia trachomatis and Neisseria gonorrhoeae in liquid-based cytology specimen. BMC Women’s Health, 9, 8. doi:10.1186/1472-6874-9-8
- Lee, S.H., Vigliotti, V.S., Vigliotti, J.S. and Pappu, S. (2009) Validation of human papillomavirus genotyping by signature DNA sequence analysis. BMC Clinical Pathology, 9, 3. doi:10.1186/1472-6890-9-3
- Lee, S.H., Vigliotti, V.S. and Pappu, S. (2010) Signature sequence validation of human papillomavirus type 16 (HPV-16) in clinical specimens. Journal of Clinical Pathology, 63, 235-239. doi:10.1136/jcp.2009.069401
- Lee, S.H. (2012) Guidelines for the use of molecular tests for the detection and genotyping of human papilloma virus from clinical specimens. Methods in Molecular Biology, 903, 65-101. doi:10.1007/978-1-61779-937-2_5
- Lee, S.H. (2013) Topological conformational changes of human papillomavirus (HPV) DNA bound to an insoluble aluminum salt—A study by low temperature PCR. Advances in Biological Chemistry, in press.
- Karlik, S.J., Eichhorn, G.L., Lewis, P.N. and Crapper, D.R. (1980) Interaction of aluminum species with deoxyribonucleic acid. Biochemistry, 19, 5991-5998. doi:10.1021/bi00567a008
- Schubbert, R., Renz, D., Schmitz, B. and Doerfler, W. (1997) Foreign (M13) DNA ingested by mice reaches peripheral leukocytes, spleen, and liver via the intestinal wall mucosa and can be covalently linked to mouse DNA. Proceedings of the National Academy of Sciences of the United States, 94, 961-966. doi:10.1073/pnas.94.3.961
- Shimada, T., Yamaguchi, N., Nishida, N., Yamasaki, K., Miura, K., Katamine, S. and Masuzaki, H. (2010) Human papillomavirus DNA in plasma of patients with HPV16 DNA-positive uterine cervical cancer. Japanese Journal of Clinical Oncology, 40, 420-424. doi:10.1093/jjco/hyp193
- Bodaghi, S., Wood, L.V., Roby, G., Ryder, C., Steinberg, S.M. and Zheng, Z.M. (2005) Could human papillomaviruses be spread through blood? Journal of Clinical Microbiology, 43, 5428-5434. doi:10.1128/JCM.43.11.5428-5434.2005
- Marichal, T., Ohata, K., Bedoret, D., Mesnil, C., Sabatel, C., Kobiyama, K., Lekeux, P., Coban, C., Akira, S., Ishii, K.J., Bureau, F. and Desmet, C.J. (2011) DNA released from dying host cells mediates aluminum adjuvant activity. Nature Medicine, 17, 996-991. doi:10.1038/nm.2403
- Gherardi, R.K., Coquet, M., Cherin, P., Belec, L., Moretto, P., Dreyfus, P.A., Pellissier, J.F., Chariot, P. and Authier, F.J. (2001) Macrophagic myofasciitis lesions assess long-term persistence of vaccine-derived aluminium hydroxide in muscle. Brain, 124, 1821-1831. doi:10.1093/brain/124.9.1821
- Exley, C., Swarbrick, L., Gherardi, R.K. and Authier, F.J. (2009) A role for the body burden of aluminium in vaccine-associated macrophagic myofasciitis and chronic fatigue syndrome. Medical Hypotheses, 72, 135-139. doi:10.1016/j.mehy.2008.09.040
- Gherardi, R.K. and Authier, F.J. (2012) Macrophagic myofasciitis: Characterization and pathophysiology. Lupus, 21, 184-189. doi:10.1177/0961203311429557
- Caulfield, M.J., Shi, L., Wang, S., Wang, B., Tobery, T.W., Mach, H., Ahl, P.L., Cannon, J.L., Cook, J.C., Heinrichs, J.H. and Sitrin, R.D. (2007) Effect of alternative aluminum adjuvants on the absorption and immunogenicity of HPV16 L1 VLPs in mice. Human Vaccines, 3, 139-145. doi:10.4161/hv.3.4.4309
- Würtele, H., Little, K.C. and Chartrand, P. (2003) Illegitimate DNA integration in mammalian cells. Gene Therapy, 10, 1791-1799. doi:10.1038/sj.gt.3302074
- Milot, E., Belmaaza, A., Wallenburg, J.C., Gusew, N., Bradley, W.E. and Chartrand, P. (1992) Chromosomal illegitimate recombination in mammalian cells is associated with intrinsically bent DNA elements. European Molecular Biology Organization Journal, 11, 5063-5070.
- Doerfler, W., Schubbert, R., Heller, H., Kämmer, C., Hilger-Eversheim, K., Knoblauch, M. and Remus, R. (1997) Integration of foreign DNA and its consequences in mammalian systems. Trends in Biotechnology, 15, 297- 301. doi:10.1016/S0167-7799(97)01061-5
- Bergen, J.M., Park, I.K., Horner, P.J. and Pun, S.H. (2008) Nonviral approaches for neuronal delivery of nucleic acids. Pharmaceutical Research, 25, 983-998. doi:10.1007/s11095-007-9439-5
- Lechardeur, D., Verkman, A.S. and Lukacs, G.L. (2005) Intracellular routing of plasmid DNA during non-viral gene transfer. Advanced Drug Delivery Reviews, 57, 755- 767. doi:10.1016/j.addr.2004.12.008
- Sparwasser, T., Miethke, T., Lipford, G., Erdmann, A., Häcker, H., Heeg, K. and Wagner, H. (1997) Macrophages sense pathogens via DNA motifs: Induction of tumor necrosis factor-alpha-mediated shock. European Journal of Immunology, 27, 1671-1679. doi:10.1002/eji.1830270712
- Häcker, H., Mischak, H., Miethke, T., Liptay, S., Schmid, R., Sparwasser, T., Heeg, K., Lipford, G.B. and Wagner, H. (1998) CpG-DNA-specific activation of antigen-presenting cells requires stress kinase activity and is preceded by non-specific endocytosis and endosomal maturation. European Molecular Biology Organization Journal, 17, 6230- 6240.
- Häcker, G., Redecke, V. and Häcker, H. (2002) Activation of the immune system by bacterial CpG-DNA. Immunology, 105, 245-251.
- Yoshida, H., Nishikawa, M., Yasuda, S., Mizuno, Y. and Takakura, Y. (2008) Cellular activation by plasmid DNA in various macrophages in primary culture. Journal of Pharmaceutical Sciences, 97, 4575-4585. doi:10.1002/jps.21302
- Boccaccio, G.L., Mor, F. and Steinman, L. (1999) Noncoding plasmid DNA induces IFN-gamma in vivo and suppresses autoimmune encephalomyelitis. International Immunology, 11, 289-296. doi:10.1093/intimm/11.2.289
- Fukuhara, Y., Naoi, T., Ogawa, Y., Nishikawa, M. and Takakura, Y. (2007) Plasmid DNA uptake and subsequent cellular activation characteristics in human monocyte-derived cells in primary culture. Journal of Pharmaceutical Sciences, 96, 1576-1584. doi:10.1002/jps.20816
- Parrillo, J.E., Burch, C., Shelhamer, J.H., Parker, M.M., Natanson, C. and Schuette, W. (1985) A circulating myocardial depressant substance in humans with septic shock. Septic shock patients with a reduced ejection fraction have a circulating factor that depresses in vitro myocardial cell performance. Journal of Clinical Investigation, 76, 1539-1553. doi:10.1172/JCI112135
- Kumar, A., Paladugu, B., Mensing, J., Kumar, A. and Parrillo, J.E. (2007) Nitric oxide-dependent and independent mechanisms are involved in TNF-alpha induced depression of cardiac myocyte contractility. American Journal of Physiology—Regulatory, Integrative, and Comparative Physiology, 292, R1900-R1906. doi:10.1152/ajpregu.00146.2006
- Cauwels, A., Van Molle, W., Janssen, B., Everaerdt, B., Huang, P., Fiers, W. and Brouckaert, P. (2000) Protection against TNF-induced lethal shock by soluble guanylate cyclase inhibition requires functional inducible nitric oxide synthase. Immunity, 13, 223-231. doi:10.1016/S1074-7613(00)00022-4
- Cauwels, A. and Brouckaert, P. (2007) Survival of TNF toxicity: Dependence on caspases and NO. Archives of Biochemistry and Biophysics, 462, 132-139. doi:10.1016/j.abb.2007.01.021
- Cauwels, A., Janssen, B., Waeytens, A., Cuvelier, C. and Brouckaert, P. (2003) Caspase inhibition causes hyperacute tumor necrosis factor-induced shock via oxidative stress and phospholipase A2. Nature Immunology, 4, 387-493. doi:10.1038/ni914
- Weinberg, J.R., Wright, D.J. and Guz, A. (1988) Interleukin-1 and tumour necrosis factor cause hypotension in the conscious rabbit. Clinical Science (London), 75, 251- 255.
- Turner, C.R., Esser, K.M., Wheeldon, E.B., Slivjak, M. and Smith, E.F. III (1989) Cardiovascular and pulmonary effects of human recombinant tumor necrosis factor in the conscious rat. Circulatory Shock, 28, 369-384.
- Chapman, P.B., Lester, T.J., Casper, E.S., Gabrilove, J.L., Wong, G.Y., Kempin, S.J., Gold, P.J., Welt, S., Warren, R.S., Starnes, H.F., Sherwin, S.A., Old, L.J. and Oettgen, H.F. (1987) Clinical pharmacology of recombinant human tumor necrosis factor in patients with advanced cancer. Journal of Clinical Oncology, 5, 1942-1951.
- Brouckaert, P., Ameloot, P., Cauwels, A., Everaerdt, B., Libert, C., Takahashi, N., Van Molle, W. and Fiers, W. (1994) Receptor-selective mutants of tumour necrosis factor in the therapy of cancer: Preclinical studies. Circulatory Shock, 43, 185-190.

