American Journal of Plant Sciences
Vol. 4 No. 3 (2013) , Article ID: 29003 , 6 pages DOI:10.4236/ajps.2013.43077
Characterization of Nuclear and Chloroplast Microsatellite Markers for Falcaria vulgaris (Apiaceae)
![]()
Department of Biology and Microbiology, South Dakota State University, Brookings, SD, USA.
Email: *Madhav.Nepal@sdstate.edu
Received January 10th, 2013; revised February 8th, 2013; accepted February 18th, 2013
Keywords: Falcaria vulgaris; Invasive Species; Microsatellite; trnL Intron; trnL-trnF Intergenic Spacer; Sickleweed
ABSTRACT
Falcaria vulgaris (sickleweed) is native to Eurasia and a potential invasive plant of the United States. No molecular markers have been developed so far for sickleweed. Characterization of molecular markers for this plant would allow investigation into its population structure and biogeography thereby yielding insights into risk analysis and effective management practices of the plant. In order to characterize the molecular markers, DNA samples were collected from eight populations in Iowa, Nebraska and South Dakota. Nuclear microsatellite markers developed for other Apiaceae taxa were screened and tested for inter-generic transferability to sickleweed. The chloroplast trnL intron and trnL-F intergenic spacer regions were sequenced and the sequences were used to design primers to amplify the microsatellites present within each region. We characterized eight polymorphic microsatellite markers for sickleweed that included six nuclear and two chloroplast markers. Our result showed inter-generic transferability of six nuclear microsatellite markers from Daucus carota to F. vulgaris. The markers we characterized are useful for population genetic study of F. vulgaris.
1. Introduction
Sickleweed (Falcaria vulgaris Bernh.; Apiaceae) is native to Europe and Asia [1] and was introduced to the United States in early 1920s [2]. It has been reported from 35 counties across 16 states in the United States and exhibits disjunct distribution in the Midwestern and Eastern USA [3]. Sickleweed exhibits some characteristics of invasive plant species including the production of a large number of seeds, effective seed dispersal mechanism whereby the seeds attached to the stem are carried away by the wind as the stems break at the nodes after the plant senescence, and the ability of the plant to reproduce asexually through root sprouting. These characteristics are perhaps facilitating its emergence as an aggressive weed in the Midwest [4]. Continuous increase in area coverage of sickleweed in Fort Pierre National Grassland (FPNG) and Buffalo Gap national Grassland (BGNG) of South Dakota has attracted attention of ecologists in the Midwest. The plant has also been listed as potential invasive plant by Nebraska Invasive Species Council [5].
Both nuclear and chloroplast DNA markers are commonly used for the genetic analysis of invasive plant populations particularly to predict the invasiveness of the introduced species, identify the source populations and help to design effective control programs for invasive species [6]. Microsatellite markers are one of the most preferred molecular markers because they are co-dominant, hyper-variable and are highly reproducible [7,8]. However, development of novel microsatellite markers is expensive and quite laborious task [9]. Cross-species transferability of microsatellite markers and identification of chloroplast microsatellites using universal chloroplast markers for sequencing chloroplast region are options that can avoid high cost and long time needed for marker development [10,11]. The markers are useful for investtigating population structure and phylogeography of the introduced species, and are also useful for the study of comparative studies of different species [10], the process of population divergence and speciation process [12]. No molecular markers have previously been developed for sickleweed that would allow us study the genetic structure of this plant. Here we report on inter-generic transferability of six nuclear microsatellite markers from Daucus carota to F. vulgaris, and two polymorphic chloroplast microsatellite markers.
2. Methods
2.1. Screening of Microsatellite Markers
We reconstructed a phylogeny of the family Apiaceae based on nuclear ribosomal Internal Transcribed Spacer (ITS) DNA sequences available in GenBank and searched for microsatellite markers developed for taxa closely related to Falcaria vulgaris. Based on our phylogenetic analysis, we decided to screen microsatellite markers developed for Daucus carota and Heracleum mantegazzianum. We selected 85 microsatellite markers with di-, triand tetranucleotide repeats developed for Daucus [13,14] and six markers developed for Heracleum [15] for the genetic analysis of sickleweed. Fresh leaf tissues were collected in silica gel from eight populations (Table 1) from Iowa (one population), Nebraska (three populations) and South Dakota (four populations). The voucher specimens except for the populations from Boyd County, Nebraska were deposited at South Dakota State University Herbarium (SDC). The silica gel dried leaf samples were ground to a fine powder and total DNA was extracted using DNeasy Plant Minikit (Qiagen Corp., Valencia, CA).
For screening the microsatellite markers, PCR was carried out in a reaction mixture of 15 μl containing 50 ng genomic DNA, 3 μl of 5 X buffer (Promega), 1.2 μl of 10 mM dNTPs (Promega), 2 μl of 25 mM MgCl2 (Promega), 1 μl each of 10 pM forward and reverse primers and 2 units of Jump Start Taq polymerase (Sigma-Aldrich). The PCR conditions were an initial denaturation of 4 minutes at 94˚C followed by 40 cycles of 1 minute denaturation at 94˚C, 20 seconds of annealing temperature and 1 minute extension at 72˚C, and final extension of 5 minutes at 72˚C. Electrophoresis was carried out in 1.2% agarose gel to evaluate the quality of PCR products and the presence of repeat motif in the amplicons was verified by re-sequencing the PCR products.
For chloroplast microsatellite, we sequenced the trnL intron and trnL-F intergenic spacer (using the primer pair 5’-CGAAATCGGTAGACGCTACG-3’ and 5’-ATTTGAACTGGTGACACGAG-3’) of chloroplast gion and found two mononucleotide repeats with more
Table 1. Collection sites of the sickleweed population.

DNA. We searched for nucleotide repeats within the rethan ten mononucleotide repeats and designed the primers using primer3 software (http://primer3.wi.mit.edu/) to amplify these mono-nucleotide repeats.
2.2. Genotyping, Test for Potential Artifacts and Data Analyses
For genotyping, PCR was carried out in a reaction mixture of 15 μl containing 50 ng genomic DNA, 3 μl of 5 X buffer, 1.2 μl of dNTPs, 2 μl of 25 mM MgCl2, 0.5 μl of 10 pM forward primer, 0.5 μl of 10 pM forward primer tagged with M13 tail, 1 μl of 10 pM reverse primers each and 2 units of Taq polymerase. The PCR conditions were similar to that of primer screening PCR conditions. The PCR products were genotyped using 3730 x1 DNA analyzer (Applied Biosystems) at Iowa State University DNA Facility.
Genemarker V2.4.0 (Softgenetics) was used to visualize the genotyping data and create allele reports. The possible genotyping artifacts such as stuttering, large allele drop-out and presence of null allele were tested using Micro-Checker [16]. The analysis of microsatellite polymorphisms including number of alleles, observed and expected heterozygosity were performed using Arlequin V3.1 [17], the polymorphism information content (PIC) value was computed using the excel microsatellite toolkit [18] and haploid diversity for chloroplast microsatellite was computed using Genalex V. 6.41 [19].
3. Results and Discussions
The probability of microsatellite marker transferability reduces with increased phylogenetic relationship [20]. There are several evidences of microsatellite marker transferability within genus but few evidences at higher taxonomic level than genus. For example, microsatellite transferability success rate within the genera in eudicots are approximately 60% compared to 10% across genera [10]. Therefore, it is always beneficial to identify phylogenetically close relatives before screening the microsatellite markers for cross species transferability when microsatellites markers are not available for sister species within the genus. In our phylogenetic analysis, we found Apium graveolens, D. carota, Eryngium alpinum and H. mantegazzianum were the only taxa with microsatellite markers developed within the clade and they were distantly related to F. vulgaris. Falcaria is a monotypic genus. Previous studies have shown transferability of Daucus and Heracleum microsatellite markers to other species [13,15]; therefore, we chose microsatellite markers developed for these two species to test their transferability to sickleweed.
None of the six H. mantegazzianum microsatellites markers amplified the F. vulgaris DNA; however, all six markers did amplify the DNA of H. maximum which was used as a positive control in this test. The transferability of these microsatellite markers (see [15] for primer sequences) from H. mantegazzianum to H. maximum has not been previously reported. With Daucus microsatellite primers, 26% of the primers amplified Falcaria DNA. Based on the quality of electrophoretic bands, Daucus primers tested for the amplification of Falcaria were classified: six primers producing a clean band with nearly expected amplicon size (7%), 16 primers producing multiple bands (19%) and 63 primers did not produce any band (74%). The six markers that produced clean bands are presented in Table 2. The identified six nuclear microsatellites and two chloroplast microsatellite primers were used for genotyping sickleweed samples collected from eight populations.
The signals of the genotyping results were clean and did not show stuttering bands. Micro-Checker showed no evidence of scoring error due to stuttering and large allele drop-out. The program did not identify any genotypeing error due to null alleles either. Our test of genotyping artifacts suggest that any deviation from HardyWeinberg equilibrium in our analysis is the result of change in allele frequency but not due to genotyping artifacts.
All six nuclear microsatellite loci were polymorphic with mean number of allele per locus of 8.8 (range 3 to 19) and most of these loci except ESSR80 have high polymorphism content value (Figure 1). Three out of six nuclear microsatellite loci were monomorphic for population from Iowa. This could be because of small sample size of the population. Two, three and five loci showed significant deviation from Hardy-Weinberg equilibrium for the populations from Iowa, Nebraska and South Dakota, respectively (Table 3).
Among six nuclear microsatellite loci, three loci (ESSR9, GSSR24 and GSSR25) produced three to four alleles for some samples (Figure 2). These multiple alleles per sample indicates that sickleweed in the novel range might have undergone gene duplications perhaps through polyploidization [21].
Chloroplast DNA markers are often used for the study at higher taxonomic level as they have very slow evolutionary rate and do not reveal much variation within species [22]. However, chloroplast microsatellites have been effectively used for the study of intra-specific variation of the plant [11]. The two chloroplast microsatellites markers used in our study were also polymorphic and detected two (CSSR1) and three alleles (CSSR2). The average mean haploid diversity for these two loci was 0.23 and these two loci detected three chloroplast haplotypes. The number of alleles and the haploid diversity of these two loci in different populations are presented in Table 4.
These polymorphic microsatellite markers can be used for the study of population structure and gene flow in the introduced as well as native range and will ultimately be useful in the identification of source population(s). Use of non-recombinant chloroplast microsatellite markers and nuclear microsatellite markers will exactly determine if the gene flow is the result of seed or pollen flow [11]. The nuclear and chloroplast microsatellite markers together can provide important insights about the genetic
Table 2. Characteristics of microsatellite markers for F. vulgaris.
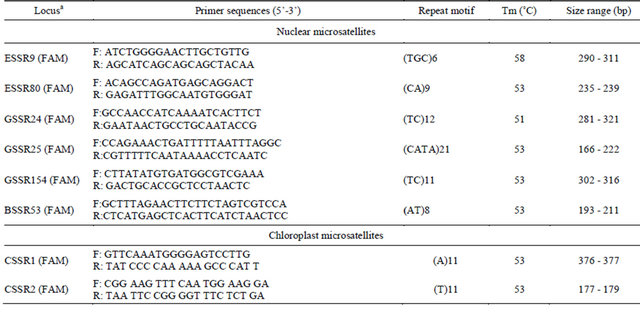
aThe fluorescent dye used to label forward primer is given in parentheses.
Table 3. Number of samples (n), Number of alleles (A), observed heterozygosity (Ho) and expected heterozygosity (He) of different nuclear microsatellite markers for the samples from Iowa, Nebraska and South Dakota.
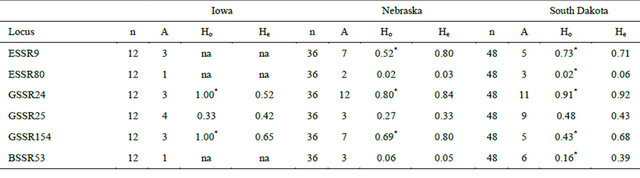
*Departure from Hardy-Weinberg equilibrium at p < 0.05; na—Locus is monomorphic and test for H-W equilibrium was not done.
Table 4. Number of alleles and haploid diversity of two chlo- roplast microsatellite markers.
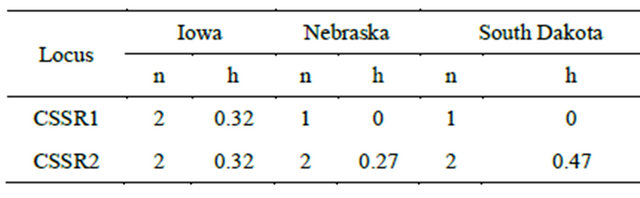

Figure 1. Polymorphism information content values displayed by the microsatellite loci.
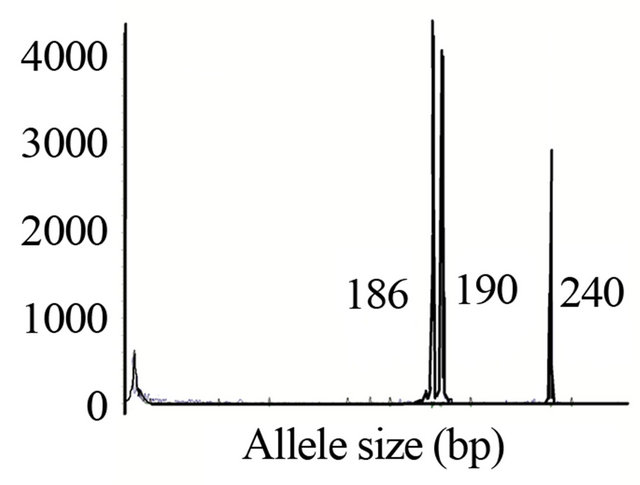
Figure 2. Electropherograms of a sample at GSSR 25 locus. Similar multiple peaks were present at GSSR24 and ESSR9 loci. Multiple peaks displayed by several loci suggest that sickleweed in the United States might have undergone polyploidization.
structure of populations. Because nuclear DNA markers are bi-parental while the chloroplast DNA markers are maternally inherited, they differ in their utilities to reveal real-time population processes operating in the populations. Therefore, analyses of sickleweed populations from native and introduced range using these markers provide insights into sickleweed evolution [7] and invasion pathways [23] and may contribute in the risk assessment and effective management of this species in the United States.
Screening of more microsatellite markers particularly markers based on Expressed Sequence Tags (ESTs) from other species of Apiaceae family may reveal other microsatellites markers that may be useful for sickleweed because studies have shown that ESTs based microsatellite markers show greater transferability than anonymous microsatellites as the genes are highly conserved across different genera [24,25]. Similarly, there are several other universal chloroplast primers (see [11]) that can be sequenced to identify chloroplast microsatellite markers. These microsatellite markers are useful not only for the population genetic study of sickleweed but may be useful for the comparative study of species across Apiaceae family.
4. Conclusion
Our results demonstrated successful inter-generic transferability of microsatellite markers from Daucus carota to Falcaria vulgaris. Since these two species belong to two distantly related genera, transferability of microsatellite markers between these species indicates that these microsatellite markers may work for other genera within the Apiaceae family. We are also reporting two chloroplast microsatellite markers for sickleweed. Sequencing other chloroplast region using universal chloroplast markers may reveal more chloroplast microsatellite markers. These nuclear and chloroplast microsatellite markers are polymorphic and are useful for population genetics and phylogeographic study of sickleweed.
5. Acknowledgements
The authors thank Carol Erickson, Teresa Y. Harris and Ryan Frickel who helped with sampling of sickleweed specimens. Drs. Gary Larson and Jack Butler provided valuable comments/suggestions on the manuscript. This study was supported by faculty startup fund to MPN and in part by the U.S. Forest Service, Rocky Mountain Research Station.
REFERENCES
- S. Y. Larina, “Falcaria vulgaris Bernh. Interactive Agricultural Ecological Atlas of Russia and Neighboring Countries,” 2009. http://www.agroatlas.ru/en/content/weeds/Falcaria_vulgaris
- E. M. Gress, “Falcaria rivini, a Plant New to the United States,” Rhodora, Vol. 25, 1923, pp. 13-14.
- S. Piya, M. P. Nepal, A. Neupane, G. E. Larson and J. L. Butler, “Inferring Introduction History and Spread of Falcaria vulgaris Bernh. (Apiaceae) in the United States Based on Herbarium Records,” Proceedings of the South Dakota Academy of Sciences, Vol. 91, 2013, pp. 113-129.
- B. L. Korman, “Biology and Ecology of Sickleweed (Falcaria vulgaris) in the Fort Pierre National Grassland of South Dakota,” M.Sc. Thesis, South Dakota State University, Brookings, 2011.
- Nebraska Invasive Species Council, “Invasive Plants of Nebraska,” 2011. http://snr5.unl.edu/invasives/pdfs/Invasive%20Plant%20Lists/NE%20Invasive%20Plants%20List%20Only%204-14-11.pdf
- A. K. Sakai, F. W. Allendorf, J. S. Holt, D. M. Lodge, J. Molofsky, K. A. With, et al., “The Population Biology of Invasive Species,” Annual Review of Ecology and Systematics, Vol. 32, 2001, pp. 305-332. doi:10.1146/annurev.ecolsys.32.081501.114037
- P. Sunnucks, “Efficient Genetic Markers for Population Biology,” Trends in Ecology and Evolution, Vol. 15, No. 5, 2000, pp. 199-203. doi:10.1016/S0169-5347(00)01825-5
- K. A. Selkoe and R. J. Toonen, “Microsatellites for Ecologists: A Practical Guide to Using and Evaluating Microsatellite Markers,” Ecology Letters, Vol. 9, No. 5, 2006, pp. 615-629. doi:10.1111/j.1461-0248.2006.00889.x
- J. Squirrell, P. M. Hollingsworth, M. Woodhead, J. Russell, A. J. Lowe and M. Gibby, “How Much Effort Is Required to Isolate Nuclear Microsatellites from Plants?” Molecular Ecology, Vol. 12, No. 6, 2003, pp. 1339-1348. doi:10.1046/j.1365-294X.2003.01825.x
- T. Barbara, C. Palma-Silva, G. M. Paggi, F. Bered, M. F. Fay and C. Lexer, “Cross-Species Transfer of Nuclear Microsatellite Markers: Potential and Limitations,” Molecular Ecology, Vol. 16, No. 18, 2007, pp. 3759-3767. doi:10.1111/j.1365-294X.2007.03439.x
- J. Provan, W. Powell and P. M. Hollingsworth, “Chloroplast Microsatellites: New Tools for Studies in Plant Ecology and Evolution,” Trends in Ecology and Evolution, Vol. 16, No. 3, 2001, pp. 142-147. doi:10.1016/S0169-5347(00)02097-8
- M. A. F. Noor and J. L. Feder, “Speciation Genetics: Evolving Approaches,” Nature Reviews Genetics, Vol. 7, No. 11, 2006, pp. 851-861. doi:10.1038/nrg1968
- P. F. Cavagnaro, S. M. Chung, S. Manin, M. Yildiz, A. Ali, M. S. Alessandro, et al., “Microsatellite Isolation and Marker Development in Carrot—Genomic Distribution, Linkage Mapping, Genetic Diversity Analysis and Marker Transferability Across Apiaceae,” BMC Genomics, Vol. 12, 2011, p. 386. doi:10.1186/1471-2164-12-386
- M. Iorizzo, D. A. Senalik, D. Gizebelus, M. Bowman, P. F. Cavagnaro, M. Mativienko, et al., “De-Novo Assembly and Characterization of the Carrot Transcriptome Reveals Novel Genes, New Markers, and Genetic Diversity,” BMC Genomics, Vol. 12, 2011, p. 389. doi:10.1186/1471-2164-12-389
- P. Henry, J. Provan, J. Goudet, A. Guisan, S. Jahodova and G. Besnard, “A Set of Primers for Plastid Indels and Nuclear Microsatellites in the Invasive Plant Heracleum mantegazzianum (Apiaceae) and Their Transferability to Heracleum sphondylium,” Molecular Ecology Resources, Vol. 8, No. 1, 2008, pp. 161-163. doi:10.1111/j.1471-8286.2007.01911.x
- C. Van Oosterhout, W. F. Hutchinson, D. P. M. Wills and P. Shipley, “Micro-Checker: Software for Identifying and Correcting Genotyping Errors in Microsatellite Data,” Molecular Ecology Notes, Vol. 4, No. 3, 2004, pp. 535-538. doi:10.1111/j.1471-8286.2004.00684.x
- L. Excoffier, L. G. Laval and S. Schneider, “Arlequin ver. 3.0: An Integrated Software Package for Population Genetics Data Analysis,” Evolutionary Bioinformatics, Vol. 1, 2005, pp. 47-50.
- S. D. E. Park, “Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection,” Ph.D Thesis, University of Dublin, Dublin, 2001.
- R. Peakall and P. E. Smouse, “GenAlEx 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research,” Molecular Ecology Notes, Vol. 6, No. 1, 2006, pp. 288-295. doi:10.1111/j.1471-8286.2005.01155.x
- R. K. Varshney, A. Garner and M. E. Sorrells, “Genic Microsatellite Markers in Plants; Features and Applications,” Trends in Biotechnology, Vol. 23, No. 1, 2005, pp. 48-55. doi:10.1016/j.tibtech.2004.11.005
- J. A. Coyer, G. Hoarau, G. A. Pearson, E. A. Serrao, W. T. Stam and J. L. Olsen, “Convergent Adaptation to a Marginal Habitat by Homoploid Hybrids and Polyploid Ecads in the Seaweed Genus Fucus,” Biology Letters, Vol. 2, No. 3, 2006, pp. 405-408. doi:10.1098/rsbl.2006.0489
- P. Taberlet, L. Gielly, G. Pautou and J. Bouvet, “Universal Primers for Amplification of Three Non-Coding Regions of Chloroplast DNA,” Plant Molecular Biology, Vol. 17, No. 5, 1991, pp. 1105-1110. doi:10.1007/BF00037152
- A. Estoup and T. Guillemaud, “Reconstructing Routes of Invasion Using Genetic Data: Why, How and So What?” Molecular Ecology, Vol. 19, No. 19, 2010, pp. 4113-4130. doi:10.1111/j.1365-294X.2010.04773.x
- P. C. H. Pashley, J. R. Ellis, D. E. McCauley and J. M. Burke, “EST Databases as a Source for Molecular Markers: Lessons from Helianthus,” Journal of Heredity, Vol. 97, No. 4, 2006, pp. 381-388. doi:10.1093/jhered/esl013
- S. B. Savadi, B. Fakrudin, H. L. Nadaf and M. V. C. Gowda, “Transferability of Sorghum Genic Microsatellite Markers to Peanut,” American Journal of Plant Sciences, Vol. 3, No. 9, 2012, pp. 1169-1180. doi:10.4236/ajps.2012.39142
NOTES
*Corresponding author.

