Advances in Microbiology
Vol. 2 No. 2 (2012) , Article ID: 19691 , 10 pages DOI:10.4236/aim.2012.22022
Glutathione Production by Yarrowia lipolytica Showing Both Methyglyoxal Resistance and a High Activity of Glutathione Synthetase
Graduate School of Human Sciences, Kobe College, Kobe, Japan
Email: *n-shiomi@mail.kobe-c.ac.jp
Received March 16, 2012; revised April 9, 2012; accepted April 25, 2012
Keywords: Glutathione; Yarrowia lipolytica; Glutathione synthetase; GSH2; Methylglyoxal
ABSTRACT
Although Yarrowia lipolytica is an important host strain, there have so far been few studies on the production of glutathione by the strain. We therefore performed a study to obtain an improved strain of Y. lipolytica ATCC20688, which could produce a high yield of glutathione. First, the capability of glutathione production in the ATCC20688 strain was estimated. In comparison with other yeasts, the yield of this strain was higher than those in Pichia strains. Furthermore, this strain could produce glutathione by assimilating sodium oleate. We next performed mutation and gene cloning to improve the yield. After the yield of glutathione was improved in the isolated methylglyoxal-resistant mutant (MGR3), the glutathione synthetase gene was cloned into the MGR3 strain. By using this recombinant strain, we could reach the maximum yield and intracellular content of glutathione of 54 mg/L-medium and 30 mg/g-dry cell weight, respectively.
1. Introduction
Glutathione, which is a tripeptide composed of glutamic acid, cysteine, and glycine, is present in most living organisms, and its biochemical characteristics have been investigated in detail [1,2]. Glutathione plays an important role in the metabolism of toxic compounds, and therefore, it has been used as a drug for hepatic insufficiency in humans, dogs and fish. Increasing attention has also been paid to its use as an antiaging compound [3]. The oxidation of lipids and proteins by active oxygen is a major factor involved in the aging process. Glutathione peroxidase eliminates active oxygen, and thus, glutathione has antiaging activity [4]. Glutathione tablets and dried-yeast powder containing a high concentration of glutathione are widely used as supplements in many countries.
Many studies have reported different processes for glutathione production. The process of glutathione extraction from yeast cells is one of the inexpensive methods. Since the 1960s, glutathione has been industrially produced in Japan with waste water in pulp production by using Candida utilis, and with exhaust molasses by using S. cerevisiae. Several studies on the culture conditions for S. cerevisiae and C. utilis have also been performed to enhance their productivity. Glutathione productivity could be increased by the addition of amino acids, especially cysteine [5]. Another study found that the control of the glucose and ethanol concentrations [6, 7], high density culture [8] and optimal controls of oxygen and pH values [9] resulted in high productivity.
Genetic approaches have been also performed to improve the strains. Glutathione is synthesized by way of a two-step reaction with γ-glutamyl cysteine ligase and glutathione synthetase. The GSH1 or GSH2 genes corresponding to these enzymes were cloned into Escherichia coli, S. cerevisiae [10,11] and Hansenula polymorpha [12], and the productivity of the recombinant strains was investigated. A high expression of GSH1 and GSH2 in recombinant E. coli and S. cerevisiae strains resulted in high glutathione productivity [13].
The utilization of an inexpensive carbon source is important for the inexpensive production of glutathione. The glutathione production using methanol has been studied, as methanol can be used as a carbon source by Pichia pastoris and H. polymorphaare [14]. Both of these strains showed a high level of productivity, although methanol is not inexpensive. On the other hand, Y. lipolytica can consume inexpensive waste food oil and crude oil as a carbon source [15-17], and therefore it is considered to be useful for the production of glutathione from economic and environmental aspects. However, very few studies have utilized Y. lipolytica for glutathione biosynthesis.
Thus, we focused on the glutathione production by Y. lipolytica ATCC20688, and improved the strain by mutation and gene manipuration. The improved strain showed both methylglyoxal resisance and a high activity of glutathione synthetase, and the productivity in this strain was higher than that of glutathione producing yeast. Therefore, this strain may be useful for the production of glutathione.
2. Materials and Methods
2.1. Strains and Media
We used the following yeast strains: Saccharomyces cerevisiae NBRC1661, Saccharomyces cerevisiae FMT1, Pichia pastoris GS115 (mut+, trp1), Pichia stupitis NBRC- 10063, and Yarrowia lipolytica ATCC20460 and Yarrowia lipolytica ATCC20688 (leu2, ura3). These strains were obtained from the American type culture collection and NITE biological resource center, and from Kobe College. YPD medium (20 g/L glucose, 20 g/L bactopeptone, 10 g/L yeast extract), YNB medium (yeast minimal medium: 20 g/L glucose, 7.0 g/L yeast nitrogen base w/o amino acids) and SOL medium (oleate minimal medium: 0.761 g/L sodium oleate, 7.0 g/L yeast nitrogen base w/o amino acids) were used to culture the yeast. The initial concentration of sodium oleate in the SOL medium was determined to be 0.761 g/L (2.5 mM), because the concentration was approximately equal to the saturated solubility of the medium. In the culture of P. pastoris GS115 cells, 2.5 mM of tryptophan was supplied to the medium, and 2.5 mM of uracil and leucine were supplied in the culture of the Y. lipolytica ATCC20688 or its mutant strain, and 2.5 mM of uracil were supplied in the culture of the recombinant strains. The competent E. coli DH5α strain was purchased from Takara Corp. (Shiga, Japan). Modified Luria-Bertani (LB) medium (10 g/L bactopeptone, 10 g/L yeast extract, 5 g/L NaCl, 1 g/L glucose, pH 7.2) was used for the culture of the E. coli DH5α cells.
2.2. Mutation and Screening Methods for the Isolation of Methylglyoxal-Resistant Mutants
Y. lipolytica ATCC20688 cells were precultured in 10 mL of YPD medium for 3 days at 30˚C. The precultured cells were inoculated at a concentration of 0.1 OD (610 nm) in 50 mL of YPD medium, and cultured at 30˚C with shaking at 120 rpm. After 1 day, the cells were harvested into a 2 mL microtube, washed 3 times with 10 mL of 0.1 M citric buffer (pH 5.5), and suspended in 1 mL of NTG solution (1.0 mg N-nitrosoguanidine dissolved in 1 mL of citric buffer). The cell suspension was incubated for 10 min at 30˚C for mutation, and was washed 3 times with 1.0 mL of 0.1 M citric buffer (pH 5.5). The treated cells were spread on agar plates of YNB medium. Later, all colonies grown on the plates were collected and diluted, and 1000 cells were spread on each agar plate of YNB medium containing 0.20 M methylglyoxal. The 10 plates were incubated at 30˚C for 5 days, and the colonies were isolated as methyl-glyoxal resistant mutants.
2.3. Construction of the Plasmid to Express the GSH2 Gene
The five DNA fragments, including the glutathione synthetase (GSH2) gene of S. cerevisiae [11], URA3 [18], ARS68 [19], pyruvate kinase (PYK1) promoter and the lipase (LIP2) terminator genes in Y. lipolytica [20,21], were cloned using the following method. The sense and antisense primers and strains used as template DNA are listed in Table 1. The chromosomal DNA samples were digested with NotI, and the digested fragments were used as the template DNA. PCR was carried out using a Hot Start PCR kit (Takara Corp.) in the following manner: 50 µL of the reaction mixture containing 0.5 µL of Hot Start DNA polymerase, 3 µL of template DNA, 4 µL of dNTP and 4µ Leach of the 10 µM forward and reverse primers were subjected to 35 cycles of treatment at 95˚C for 30 s, 55˚C for 30 s, and 72˚C for 1 or 3 min. The amplified DNA fragments were separated by gel electrophoresis with 1% low-gelling agarose A-4018 (SigmaAldrich Japan, Tokyo, Japan), and the target DNA fragments were purified from the gel. The purified five DNA fragments, the PYK1 promoter, LIP2 terminator, URA3, ARS68 and GSH2 genes, were respectively inserted into the EcoRV site of the pT7Blue(R) vector with the Ligation Kit 2 (Takara Corp.). The constructed plasmids were respectively named pPK, pTER, pYL-URA, pYL-ARS, and pSC-GSH2.
The plasmid for the expression of GSH2 gene was constructed using the following procedure: After the plasmid pPK was digested with both PshBI and NdeI, the 0.42-kb fragment containing the PYK1 promoter was inserted into the NdeI and NotI sites of the plasmid, pTER. The constructed plasmid was named pPKLIP. The plasmids pYL-URA and pYL-ARS were digested with BamHI and Sse8731I, respectively. The 2.22 kb and 1.38 kb fragments containing the URA3 and ARS68 genes were inserted at the BamHI and Sse8731I sites of the plasmid pPKLIP. The constructed plasmid was named pYL-ARS68. The 1.48 kb DNA fragment containing the GSH2 gene was inserted between the NdeI and NotI sites of the plasmid pYL-ARS68, after the plasmid pSCGSH2 was digested with both NdeI and NotI. Finally, the completed plasmid (8.87 kb) for the expression of GSH2 was named pYL-GSH2 (Figure 1). The transformation

Table 1. List of the primers and template DNA sources.
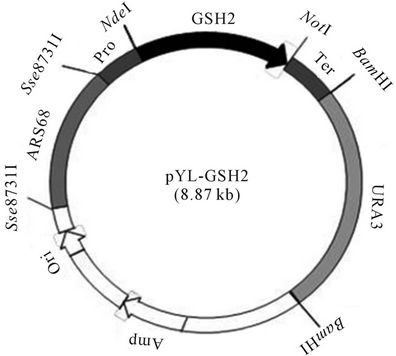
Figure 1. A schematic diagram of the pYL-GSH2 plasmid. Pro, PYK1 promoter; GSH2, glutathione synthetase (GSH2); Ter, Lip2 terminator; URA3, URA3 gene; ori, origin of replication in E. coli; Amp, ampicillin resistance gene; ARS 68, ARS68 gene.
of the pYL-GSH2 plasmid into Y. lipolytica was carried out with the Yeast Maker Kit (CLONTECH Laboratories, Inc., CA, USA).
2.4. Culture Conditions
The yeast were precultured in 20 mL YPD medium at 30°C for three days, and washed once with 20 mL of water, and then were suspended in 1 mL of the water. The pre-cultured cells were inoculated at optical densities (OD) of 0.1 at 610 nm into 100 mL of medium in a 300 mL flask with 4 baffles (Hario glass corp., Tokyo, Japan) or 20 mL of medium in a 100 mL flask, and the cells were cultured for 3 days at 30˚C with periodic shaking at 120 rpm. Four culture tests (three independent experiments in each test) were performed to examine different combinations of yeasts and media.
To test the effect of additives, the ATCC20688 strain was cultured in 100 mL of YNB medium, SOL medium, or YNB medium containing 2.5 mM glutamic acid, cysteine or glycine. To compare the glutathione productivity in various yeast strains, S. cerevisiae NBRC1661, S. cerevisiae FMT1, P. pastoris GS115, P. stupitis NBRC- 10063, Y. lipolytica ATCC20460 and Y. lipolytica ATCC20688 cells were cultured in 100 mL of YNB medium. To examine the growth inhibition of methylglyoxal, the ATCC20688 or the isolated methylglyoxalresistant mutant strain was cultured in 20 mL of YNB media with and without the addition of 0.32 M of methylglyoxal. To examine the yield of glutathione, the Y. lipolytica ATCC20688, methylglyoxal-resistant mutant or recombinant strain was cultured in 100 mL of YNB medium.
2.5. Assays
In the glutathione assay, 20 mL of culture broth (containing cells) were harvested by centrifugation and washed once with water. The cell pellet was suspended in 2 mL of distilled water, and the intracellular glutathione was extracted from cells by incubation for 10 min in boiling water. The concentration of the extracted glutathione was measured by the highly-sensitive method reported by Owens and Belcher [22]. Under this method, the sample (100 µL) was mixed with the reaction mixture [1.25 mL of 0.05 M sodium phosphate buffer pH 7.0, 400 µL of 1 mM EDTA, 15 µL of 10 mM DTNB, 50 µL of 40 mg/L NADPH, 7 µL of glutathione reductase solution (300 unit)], and the increasing rate of absorbance was measured at 412 nm by the formation of 5-mercapto- 2-nitrobenzenzoic acid (Δε412 = 1.55 × 104/mM·cm). The method has 2 merits: the sensitivity is much higher than that of the method without glutathione reductase, and the presence of other thiol compounds do not disturb the reaction. The yield and content of glutathione were defined as mg of intracellular glutathione per 1 L of culture broth (mg/L) and as the mg of intracellular glutathione per 1 g of dry cell weight (mg/g). These two values were utilized as the indicators of productivity.
Cell growth was measured by the optical density (OD) of a 1 cm light passing at 610 nm. The dry cell weight (DCW) was calculated by weighing the cell masses after drying at 95˚C for 16 h. One OD corresponded to 412 mg-DCW per 1 L of medium. The concentration of sodium oleate was measured using a free fatty acid measurement kit (NEFC-Test Wako, Wako Pure. Chem. Ind., Osaka, Japan). The average values, standard deviation (SD) and p values (by the t-test) of the cell growth and glutathione productivity were calculated from the results of three independent culture tests.
2.6. Enzymatic Activities
The enzyme solution was prepared as follows: the four kinds of cells (Y. lipolytica ATCC20688, its mutant and those transformants carrying pYL-GSH2) cultured in 200 mL of YNB medium for 2 days were harvested by centrifugation and washed twice with water. The cells were suspended in 1 mL of 10 mM TE buffer (pH 7.5) and transferred to 2 mL microtubes containing 1mL of glass beads (0.25 - 0.3 mm in diameter). After the cells were disrupted with a cell disrupter (MINI-BEADBEATER, WAKENYAKU Co. Ltd., Kyoto, Japan) for 3 min, the whole solution was centrifuged at 12,000 × g for 15 min. Each supernatant was respectively put in seamless cellulose tubing (Wako Chem. Ind., Osaka, Japan) and dialyzed with 3 L of 10 mM TE buffer (pH 7.5) for 1 day. The resulting enzyme solution was used for the following enzyme assay. The protein concentration in the solution was measured with a protein assay kit (Bio-Rad Laboratories, Inc. Japan, Tokyo, Japan).
The glyoxalase I activity was measured according to the method described by Inoue et al. [23]. The reaction mixture (1 mL) containing 5.0 mM methylglyoxal, 5.0 mM glutathione, 100 mM potassium phosphate buffer (pH 7.5), and enzyme solution was incubated at 30˚C. The initial formation rate of S-lactoylglutathione (Δε240 = 3.37/mM·cm) was measured at 240 nm. The activity of glyoxalase II was measured according to the method reported by Allen et al. [24]. The reaction mixture contained 50 mM Tris/HCl buffer (pH 7.4) and 300 µM S-D-lactoylglutathione. The initial rate of the hydrolysis of S-lactoylglutathione (Δε240 = −3.1/mM·cm) was measured at 240 nm.
The activity of glutamate cysteine ligase was measured according to the method developed by White et al. [25]. The reaction mixture contained 40 mM ATP, 2 mM L-cysteine, 20 mM L-glutamic acid, 2 mM EDTA, 20 mM sodium borate, 2 mM serine, and 40 mM MgCl2. The activity of glutathione synthetase was measured by evaluating the glutathione levels. The reaction mixture (100 µL) containing 100 mM Tris/HCl buffer (pH 7.5), 10 mM MgCl2, 20 mM KCl, 1 µM γ-glytamyl-cysteine, 1 µM glycine, and 2 µM ATP was incubated for 5 min and heated for 10 min at 95˚C to inactivate the enzymes. The amount of glutathione produced was determined by the high-sensitive method (described previously). The specific activities of these enzymes, other than glutamate cysteine ligase, were expressed as µmol/min·mg-protein. The activity of glutamate cysteine ligase was expressed as the fluorescence density/min·mg-protein. The enzymatic activities were measured twice, and the mean value was presented.
2.7. Stability and Copy Number of pYL-GSH2
The stability of the plasmid was examined according to the previously described method [26]: The recombinant cells (the ATCC20688 strain having the pYL-GSH2 plasmid) were cultured in YNB medium containing leucine (without the supplementation of uracil) for 52 h. They were diluted with water and then spread on agar plates of YNB medium containing leucine and uracil. The auxotrophy of uracil for 300 colonies was evaluated using two kinds of agar plates (one contained both uracil and leucine and the other contained just leucine).
The length and copy number of the plasmid present in the recombinant strain were estimated by the following manner: The recombinant cells cultured in 50 mL of YNB medium were collected in a 1.5 mL microtube, and 200 µL of TE buffer (pH 7.8) containing 1% SDS, 200 µL phenol-chloroform and 0.25 - 0.3 mm of glass beads were added to the cell pellet. The cells were disrupted by a cell disrupter (MINI-BEADBEATER) for 5 min. The all-aqueous phase was taken out, and the DNA was purified.
After purified DNA solution was transformed into the E. coli DH5α cells for amplification, the presence of the plasmid in E. coli DH5α was confirmed by electrophoresis on 0.8% agarose gels. The rest of the purified DNA was digested with NotI, and the copy number was determined by a real-time PCR method. The primers for the PYK promoter sequence (target) which was present in both pYL-GSH2 and chromosomal DNA, and the LIP2 promoter sequence (control) which were only present in chromosomal DNAs, were used. Real-time PCR was performed by using a Rotor-GeneTM device (Qiagen K. K., Tokyo, Japan). The reaction mixture was prepared with Roter-Gene SYBR Green PCR Kit (Qiagen K. K.), and performed for 70 cycles of treatment at 95˚C for 10 s and 65˚C for 30 s. The threshold line and Ct values were determined by using the Roter-Gene series software program, and the relative amount of DNA in the PYK promoter in comparison with the LIP2 terminator was determined by the ΔΔCt method. Three independent experiments were performed, and the average and SD were calculated.
3. Results
3.1. The Glutathione Production Capacity of the Y. lipolytica ATCC20688 Strain
The Y. lipolytica ATCC20688 strain is one of the most useful host strains for gene cloning. Thus, we examined the glutathione production using the Y. lipolytica ATCC- 20688 strain. Figure 2 shows the cell growth and glutathione productivity when the ATCC20688 cells were cultured in YNB medium. Glutathione was gradually produced in cells until 52 h (late-exponential phase), but the content at 28 h was higher than that at 52 h. Generally, glutathione is gradually secreted into the medium before and after the stationary phases after the decrease of the pH value in the broth, and it is not secreted during the exponential growth phase. We therefore compared the values of the yields at 28 h or 52 h and the content at 28 h to determine the native capability for glutathione production.
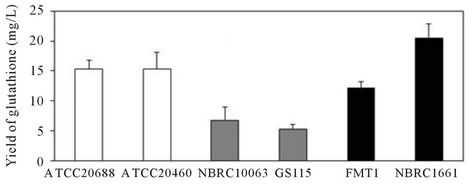
The amount of glutathione in the Y. lipolytica ATCC- 20688 strain was compared to that of Y. lipolytica ATCC- 20460, Saccharomyces cerevisiae NBRC1661, FMT1 and Pichia pastoris GS115 and NBRC10063. Figure 3 shows the glutathione productivity when yeast cells were cultured in YNB medium for 28 h. The content in the ATCC20688 cells was 2.1 times higher than that in the Y. lipolytica ATCC20460 cells, but the overall yield was similar. Furthermore, the average yield in the ATCC20688 cells was 1.3, 2.9 and 2.3 times higher than those in the S. cerevisiae FMT1, P. pastoris GS115 and NBRC10063 strains (p < 0.05), although the value was 25% lower than that in the Saccharomyces strain NBRC- 1661 (p = 0.14). In addition, the average content in the ATCC20688 cells was 1.3 5.4 and 6.3 times higher than those in the S. cerevisiae FMT1, P. pastoris GS115 and NBRC10063 strains (all p < 0.05), and the value was similar to that of the Saccharomyces strain NBRC1661.
Figure 2 shows the cell growth and glutathione production when the ATCC20688 cells were cultured in SOL medium (oleate was the sole carbon source). The ATCC20688 strain could efficiently assimilate sodium oleate as a carbon source, and the yield at 52 h and content of glutathione at 28 h were 11.2 mg/L and 7.6 mg/g. The concentration of sodium oleate at 52 h was decreased below 5 mg/L. Therefore, the ATCC20688 strain could produce glutathione by assimilating sodium oleate as a carbon source. Further, the effects of glutamic acid, cysteine and glycine (the components of glutathione) in the culture with the YNB medium were examined. As shown in Figure 2, the cell growth of the ATCC20688 strain was inhibited in case of the supplementation with cysteine, and the content of glutathione at 28 h was lower than that of the control (without supplement). The supplementation of glutamic acid or glycine caused no
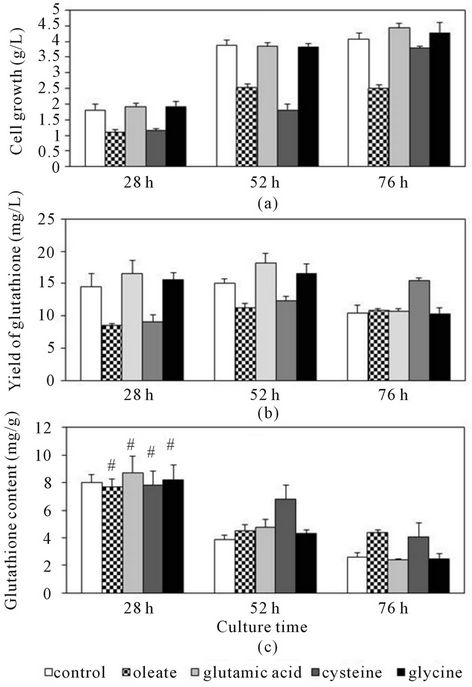
Figure 2. The glutathione productivity of the Y. lipolytica ATCC20688 strain. The ATCC20688 cells were grown in YNB medium without any supplement (control), SOL medium (oleate), or in YNB media with 2.5 mM of amino acid (glutamic acid, cysteine or glycine). (a) Growth; (b) Yield of glutathione; (c) Content of glutathione. Bars: means ± SD of 3 independent experiments. **p < 0.01, *p < 0.05 and #P > 0.05 vs. control. The p-values were determined for the results of the content at 28 h.
significant difference in the production compared with the control.
3.2. Enhancement of Glutathione Production in a Methylglyoxal-Resistant Mutant
Breeding by mutation was performed to enhance the glutathione biosynthesis in the yeast. Methylglyoxal was utilized as an indicator for isolation. The ATCC20688 cells were mutated with NTG solution, spread on agar plates of YNB medium containing 0.2 M methylglyoxal, and methylglyoxal-resistant mutants were isolated by selecting large colonies. Ten mutants were finally isolated, and 4 of these mutants showed higher glutathione productivity than the ATCC20688 parent strain. One mutant used in further investigation was named MGR3. Figure 4 shows the typical cell growth curves (A) and the growth at 70 h (B) of the MGR3 mutant and the
 (a)
(a)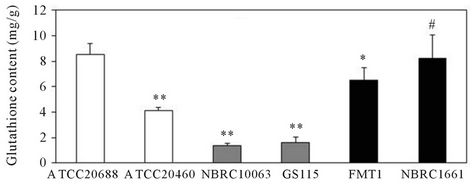 (b)
(b)
Figure 3. A comparison of the glutathione production in the Yarrowia, Saccharomyces and Pichiastrains. Yeast were grown in YNB medium for 28 h, and the yield and content of glutathione was determined to compare the glutathione production capability among the Yarrowia, Saccharomyces and Pichia strains. (a) Yield of glutathione; (b) Content of glutathione. Bars: means ± SD of 3 independent experiments. **p < 0.01, *p < 0.05 and #p > 0.05 vs. control (ATCC20688). The p-values were determined for the results of the content.
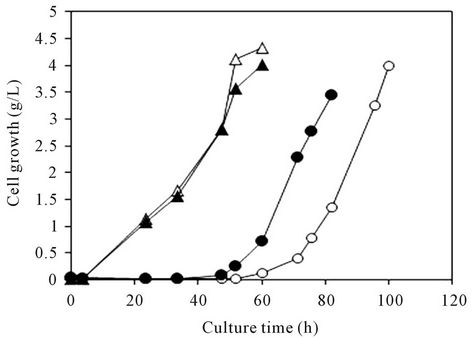
ATCC20688 parent strains, when these cells were cultured in YNB medium and YNB medium containing 0.32 M of methylglyoxal. Both of the cell growth curves in the yeasts cultured with methylglyoxal supplementation were shifted to the right (Figure 4(a)) because of the toxicity of methylglyoxal. However, the increase of growth in the MGR3 mutant was earlier than that of the ATCC20688 parent strain in these curves. The average cell growth of the MGR3 strain at 70 h of culture was 6.0 times higher, therefore, the ability of the MGR3 mutant strain to degrade methylglyoxal was much higher than that of the ATCC20688 strain.
Figure 5 shows the growth and glutathione productivity of the MGR3 mutant and the ATCC20688 parent strain when the cells were cultured in YNB medium. The growth of Y. lipolytica MGR3 and ATCC20688 cells were almost the same, but the average yields of glutathione in the MGR3 strains was 1.7 times higher than that of the ATCC20688 parent strain after 52 h of culture.The content at 28 h was also 2.0 times higher (p < 0.05). The enzymatic activities in the MGR3 mutant and ATCC20688 parent strains were also determined (Table 2). The activities of glyoxalase I and II in the MGR3 mutant were 2.0 and 2.4 times higher than those in the ATCC20688 parent strain. On the other hand, the activity of glutathione synthetase was 1.23 times higher, and the
 (a)
(a) (b)
(b)
Figure 4. Methylglyoxal resistance in the Y. lipolytica ATCC- 20688 and MGR3 strains. The MGR3 mutant and ATCC- 20688 parental strains were grown in YNB medium and YNB medium containing 0.32 M methylglyoxal. (a) Typical growth curves; (b) Growth at 70 h. Symbols: white and black triangles, ATCC20688 and MGR3 strains without supplementation; white and black circles, ATCC20688 and MGR3 strains cultured with 0.32 M methylglyoxal. Bars: means ± SD of 3 independent experiments. **p < 0.01 vs. control (ATCC20688).
increase of glutamate-cysteine ligase was a little (1.07 times higher).
3.3. Improvement in the Production of Glutathione by the Recombinant Strain Expressing the GSH2 Gene
We newly constructed a plasmid (pYL-GSH2; Figure 1) that expressed the GSH2 gene of Saccharomyces cerevisiae in Y. lipolytica ATCC20688. The pYL-GSH2 plasmid was transformed into the Y. lipolytica ATCC20688 parent and MGR3 mutant strains, and were named Y. lipolytica ATCC20688/pYL-GSH2 and MGR3/pYLGSH2, respectively. Approximately 1000 colonies were obtained by the transformation operation with 1 µg of the plasmid. The transformation efficiency was approximately 1000 times higher than that in the plasmid without

Figure 5. The growth and glutathione production in the methylglyoxal-resistant mutant and recombinant strains. Yeast were grown in YNB medium. (a) Growth; (b) Yield of glutathione; (c) Content of glutathione. Bars: means ± SD of 3 independent experiments. **p < 0.01, *p < 0.05 and #p > 0.05 vs. control. The p-values were determined for the results of the content at 28 h.
ARS68 (when the plasmid without ARS68 was transformed, approximately one colony per 1 µg of plasmid appeared).
We then examined the effects of the pYL-GSH2 plasmid on the enzymatic activities. The activities of glutathione synthetase and glutathione cysteine ligase in these strains are shown in Table 2. The activity of glutathione synthetase in the ATCC20688/pYL-GSH2 recombinant strain was 1.46 times higher than that of the ATCC20688 host strain, but was almost the same as that of the MGR3/pYL-GSH2 recombinant strain. However, the activity of glutathione cysteine ligase of the MGR3/ pYL-GSH2 recombinant strain was 1.11 times higher than that of the ATCC20688/pYL-GSH2 recombinant strain.
Figure 5 shows the cell growth and glutathione production when the recombinant ATCC20688/pYL-GSH2 and MGR3/pYL-GSH2 cells were cultured in YNB medium. The cell growth of the recombinant strains were not significantly different compared to the host strains (all p > 0.1). The yield and content of the ATCC- 20688/pYL-GSH2 recombinant strain at 52 h and 28 h were 3.5 and 3.2 times higher than those of the ATCC- 20688 host strain, and those of the MGR3/pYL-GSH2 strain were a little higher than those of the ATCC- 20688/pYL-GSH2 strains. The maximum yield of glutathione in the ATCC20688/pYL-GSH2 and MGR3/ pYL-GSH2 strains at 52 h reached 51 and 54 mg/L, and the maximum contents of these strains at 28 h were 29 mg/g and 30 mg/g, respectively.
3.4. Stability and Copy Number of the pYL-GSH2 Plasmid
To examine the stability of the plasmid, the auxotrophy of uracil was checked using 300 recombinantcells cultured in selective medium (YNB medium with leucine but without uracil). Only 4 colonies showed auxotrophy of uracil. Therefore, 1.3% of the cells lost the plasmid in every segmentation. Further, after the plasmid was extracted from the ATCC20688/pYL-GSH2 cells and amplified in the E. coli cells, the length of the plasmid was confirmed by agarose gel electrophoresis. The length of the extract plasmid was the same as the plasmid pYLGSH2. These results suggest that the plasmid was stably amplified in the cytoplasm without insertion into chromosomal DNA. Further, the copy number of the plasmid was estimated by a real-time PCR method. The promoter sequence of the pyruvate kinase gene, which was present in both the chromosome and plasmid, was amplified. The estimated amount of DNA in the ATCC-20688/pYLGSH2 strain was 2.8 ± 0.5 times higher than that in the ATCC20688 strain, indicating that the average copy number is 1.8 ± 0.5.
4. Discussion
Glutathione is widely used as a drug and a supplement for health, and therefore, many studies have reported processes to increase glutathione production using yeast. However, the production of glutathione by Y. lipolytica has not been investigated, although Y. lipolytica, which can utilize inexpensive waste food oil or alkanes as a carbon source, is considered to be an important yeast strain for the production in terms of both economic and environmental aspects. We focused on the production of glutathione by the Y. lipolytica ATCC20688 strain, and improved the strain in three steps.
As a first step, we estimated the possibility of utilizing the Y. lipolytica ATCC20688 strain for the glutathione production. In batch culture without pH control, the content of glutathione present in cells is almost constant
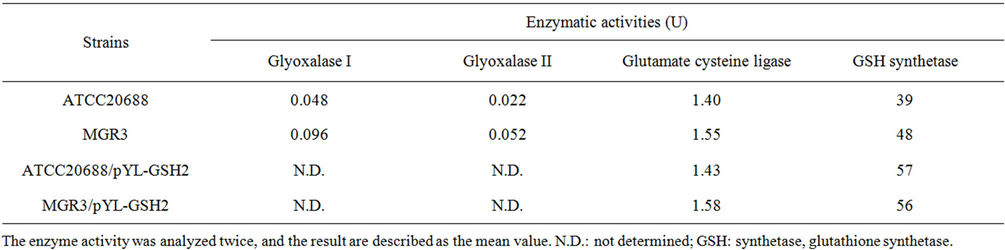
Table 2. The enzymatic activities of Y. lipolytica ATCC20688, MGR3, and their transformants.
during the exponential phase, but glutathione is gradually secreted into the broth before and after the stationary phases due to the decrease of the pH value in the broth [9]. The content of glutathione in the ATCC20688 strain also decreased similarly during the stationary phases (52 h and 76 h). Thus, we utilized the values of the yield at 28 h or 52 h and the content at 28 h to represent the native capability. The yield and content of glutathione in the ATCC20688 strain cultured in the YNB medium at 28 h were compared with those insome strains of S. cerevisiae and P. pastoris, because S. cerevisiae and P. pastoris are also important strains that are frequently used for gene manipulation. As shown in Figure 3, the yield of glutathione in the Y. lipolytica strain was higher than the P. pastoris strains, although the values were a little lower than those of the S. cesrevisiae NBRC1661 strain. However, the content in the ATCC20688 strain was the highest (or similar) among these strains. Based on these results, we concluded that the ATCC20688 strain has sufficient potential for the production of glutathione.
Furthermore, the effects of additions to the media were examined to clarify the characteristics of the ATCC- 20688 strain. The utilization of waste food oil for glutathione production is important to ensure inexpensive production. We used sodium oleate as a model compound of waste food oil. As shown in Figure 2, the ATCC20688 strain could efficiently produce glutathione by assimilating sodium oleate, but the content of the glutathione was a little lower than that of the cells cultured with glucose. Moreover, the effect of the supplementation of glycine, glutamine or cysteine on the productivity of glutathione was examined. Glutathione was formed by yeast cultured with these 3 amino acids, and it was reported that the productivity was enhanced by the addition of cysteine to S. cerevisiae [5]. However, the supplementation of cysteine caused the inhibition of cell growth and no significant enhancement of the yield of glutathione in the present study (Figure 2). The increase of the yield and content were not significant even when glycine and glutamic acid were added as supplements. Therefore, these amino acids might already be present at sufficient quantities in the ATCC20688 strain. These results suggest that enhancement of the activities directly related to the glutathione synthesis is required to enhance the production.
As a second step, genetic improvements by mutation were performed to enhance the productivity of the yeast. For the improvement by mutation, we utilized methylglyoxal resistance to select cells. Methylglyoxal is degraded to lactic acid by glyoxalase I and II with the help of glutathione at a low concentration [27]. However, it works as an inhibitor of cell growth at high concentrations. Interestingly, some mutants showing a marked ability to degrade methylglyoxal (a characteristic known as methylglyoxal resistance) can lead to high production of glutathione in S. cerevisiae (unpublished result). The isolated mutant in the present study, MGR3, showed higher methylglyoxal resistance (Figure 4) and glutathione productivity (Figure 5) than the ATCC20688 strain. To confirm the reason for this, the enzymatic activities were examined (Table 2). The activities of glyoxalase I and II in the MGR3 mutant were much higher (2.0 and 2.4 times higher) than those in the ATCC20688 parent strain, but the activities of glutathione synthetase and glutamate-cysteine ligase were just 1.23 and 1.07 times higher. These results suggest that the mutation is related to a release of the repression of the glyoxalase I and II genes (GLO1 and GLO2), resulting in an increase in the degradation of methylglyoxal. As a result, these changes might induce the high glutathione synthesis.
As a third step, we performed experiments to increase the glutathione synthetase activity by cloning the GSH2 gene of S. cerevisiae into the Y. lipolytica strain, because a small increase in the glutathione synthetase activity in the MGR3 strain was effective for increasing the glutathione biosynthesis. We made two additional changes to the newly-constructed pYL-GSH2 plasmid (Figure 1): the GSH2 gene was inserted downstream of the pyruvate kinase (PYK1) promoter to obtain high expression, and the autonomously replicating sequence, ARS68, was inserted to induce high stability and a high copy number.
After the ATCC20688/pYL-GSH2 and MGR3/pYLGSH2 recombinant strains were obtained, the capability of the plasmid was examined. We found that the pYLGSH2 plasmid was stably amplified in the cytoplasm, and that the average copy number of the plasmid was 1.8. As shown in Table 2, the glutathione synthetase activity of the ATCC20688/pYL-GSH2 recombinant strain was 1.46 times higher than that of the ATCC20688 host strain, but the activity was almost the same as the MGR3/ pYL-GSH2 transformant. The copy number and the activity were lower than we expected, although we designed the plasmid to be highly expressed. Some studies have reported that a plasmid containing an ARS region in Y. lipolytica [28] generated 2 - 3 copies per generation and had high stability (97%), although it had low stability in S. cerevisiae [29]. Our results are similar to those of other plasmids in Y. lipolytica, and therefore, the ARS68 gene may not lead to an increase in the copy number. Moreover, the enhancement of the activity of glutathione synthetase was not high (1.46 times), and the activities of the ATCC20688/pYL-GSH2 and MGR3/pYLGSH2 strains were similar. Although the reason is not clear and further studies are required, these results may suggest that something (glutathione or an inhibitory gene) inhibits the expression, and that this leads to an upper limit for the production of glutathione.
Finally, we estimated the capability of the MGR3/ pYL-GSH2 recombinant strain to produce glutathione. As shown in Figure 5, the yield and content of glutathione in the ATCC20688/pYL-GSH2 recombinant strain were much higher than those in the ATCC20688 host strain, because of the higher glutathione synthetase activity in the recombinant strain. In addition, the values in the MGR3/pYL-GSH2 recombinant strain were higher than those in the ATCC20688/pYL-GSH2 recombinant strain, because of the higher activity of glutamate cysteine ligase in the MGR3/pYL-GSH2 recombinant strain. Consequently, the maximum values of the glutathione yield and content in the MGR3/pYL-GSH2 recombinant strain were 54 mg/L and 30 mg/g. The glutathione producing yeasts, C. utilis WSH-02-08 and S. cerevisiae FF- 8 strains, were widely utilized in recent studies [7,9], and the contents of those strains cultured in batch process without pH control were below 30 mg/g, as calculated from the results in the reports. Therefore, the Y. lipolytica MGR3/pYL-GSH2 strain may be useful for the production of glutathione.
The glutathione production of the WSH-02-08 and FF-8 strains reached a much higher level under optimal controls of oxygen and pH value [7,9]. Therefore, further improvements in our strain are required before it can be used on an industrial scale. Higher productivity can be achieved by the additional expression of the glutamatecysteine ligase gene, GSH1, and by the enhancement of the copy number by insertion of the ribosomal RNA gene [30,31], under optimal high density culture. Further studies regarding these possibilities are now underway, and will be described in a subsequent manuscript.
5. Conclusion
The yield of glutathione in the Y. lipolytica ATCC20688 strain was considered to be sufficiently high in comparison to the other yeasts, and it could produce glutathione by assimilating sodium oleate. This strain was improved by both mutation and gene manipulation. In the methylglyoxal-resistant mutant (MGR3), the activity of glutathione synthetase and the glutathione contents were 1.23 and 1.7 times higher than those in the parent strain. Additionally, the glutathione synthetase gene of S. cerevisiae was cloned into the mutant strain by using the newly constructed vector pYL-GSH2, and the activity of glutathione synthetase in the recombinant strain (MGR3/ pYL-GSH2) could thus be enhanced (1.4 times higher). Moreover, the maximum yield and intracellular content of glutathione in the recombinant strain were 54 mg/Lmedium and 30 mg/g-dry cell weight, respectively. From these results, we concluded that the recombinant strain is therefore useful for the production of glutathione, although further improvements are required before it can be used on an industrial scale.
6. Acknowledgements
Our group is grateful thank Ms. Takayo Adachi, Kana Ishizuka, Rina Okada, Noriko Ashida, Konomi Ishizuka, Ayami Kadoma, Midori Kuriyama, and Aya Nomura for their assistance with our research. This research was supported by grants-in-aids from the Central Laboratory and the School of Human Sciences at Kobe College.
REFERENCES
- M. E. Anderson, “Glutathione: An Overview of Biosynthesis and Modulation,” Chemico-Bioligal Interactions, Vol. 111-112, 1998, pp. 1-14. doi:10.1016/S0009-2797(97)00146-4
- Y. Li, G. Wei and J. Chen, “Glutathione: A Review on Biotechnological Production,” Applied Microbiology and Biotechnology, Vol. 66, No. 3, 2004, pp. 233-242. doi:10.1007/s00253-004-1751-y
- E. Beutler, “Nutritional and Metabolic Aspects of Glutathione,” Annual Review of Nutrition, Vol. 9, 1989, pp. 287-302. doi:10.1146/annurev.nu.09.070189.001443
- B. M. Lomaestro and M. Malone, “Glutathione in Health and Disease: Pharmacotherapeutic Issues,” The Annals of Pharmacotherapy, Vol. 29, No. 12, 1995, pp. 1263-1273.
- C. Alfafara, K. Miura, H. Shimizu, S. Shioya and K. Suga, “Cysteine Addition Strategy for Maximum Glutathione Production in Fed-Batch Culture of Saccharomyces cerevisiae,” Applied Microbiology and Biotechnology, Vol. 37, No. 2, 1992, pp. 141-146. doi:10.1007/BF00178160
- K. O. Udeh and B. Achremowicz, “High-Glutathione Containing Yeast Sacchromyces cerevisiae: Optimization and Production,” Acta Microbiologica Polonica, Vol. 46, No. 1, 1997, pp. 105-114.
- J. Y. Cha, J. C. Park, B. S. Jeon, Y. C. Lee and Y. S. Cho, “Optimal Fermentation Conditions for Enhanced Glutathione Production by Saccharomyces cerevisiae FF-8,” Journal of Microbiology, Vol. 42, No. 1, 2004, pp. 51-55.
- K. Sakato and H. Tanaka, “Advanced Control of Glutathione Fermentation Process,” Biotechnology and Bioengineering, Vol. 40, No. 8, 1992, pp. 904-912. doi:10.1002/bit.260400806
- W. Nie, G. Wei, G. Du, Y. Li and J. Chen, “Enhanced Intracellular Glutathione Synthesis and Excretion Capability of Candida utilis by Using a Low pH-Stress Strategy,” Letters in Applied Microbiology, Vol. 40, No. 5, 2005, pp. 378-384. doi:10.1111/j.1472-765X.2005.01687.x
- Y. Ohtake and S. Yabuuchi, “Molecular Cloning of the Gamma-Glytamylcysteine Synthetase Gene of Sacchromyces cerevisiae,” Yeast, Vol. 7, No. 9, 1991, pp. 953- 961. doi:10.1002/yea.320070907
- Y. Inoue, K. Sugiyama, S. Izawa and A. Kimura, “Molecular Identification of Glutathione Synthetase (GSH2) Gene from Saccharomyces cerevisiae,” Biochimica Biophysoca Acta, Vol. 1395, No. 3, 1998, pp. 315-320.
- V. M. Ubiyvovk, T. Y. Nazarko, O. G. Stasyk, M. J. Sohn, H. A. Kang and A. A. Sibirny, “GSH2, a Gene Encoding Gamma-Glutamylcysteine Synthetase in the Methyltrophic Yeast,” FEMS Yeast Research, Vol. 2, No. 3, 2002, pp. 327-332.
- L. Yin, J. Chen and L. Shiyi,“Production of Glutathione by High Cell Density Culture of Recombinant Escherichia coli,” Chinese Journal of Pharmaceticls, Vol. 30, No. 1, 1999, pp. 1-4.
- M. Rollini, H. Pagani, S. Riboldi and M. Manzoni, “Influence of Carbon Source on Glutathione Accumulation in Methyltrophic Yeasts,” Annals of Microbiology, Vol. 55, No. 3, 2005, pp. 199-203.
- H. P. Fickers, P. H. Benetti, Y. Waché, A. Marty, S. Mauersberger, M. S. Smit and J. M. Nicaud,“ Hydrophobic Substrate Utilization by the Yeast Yarrowia lipolytica, and Its Potential Applications,” FEMS Yeast Research, Vol. 5, No. 607, 2005, pp. 527-543. doi:10.1016/j.femsyr.2004.09.004
- N. Shiomi, Y. Inoue, K. Tomioka and T. Yasuda, “Immobilization of Candida Cells Showing Mycelium-Like Shapes in Porous Polyvinyl Formal Resin and Their Applications,” Journal of Chemical Engineering of Japan, Vol. 36, No. 2, 2003, pp. 161-165. doi:10.1252/jcej.36.161
- T. H. Kim, J. H. Lee, Y. S. Bae and S. J. Kim, “Identification and Characterization of an Oil-Degrading Yeast, Yarrowia lipolytica 180,” Journal of Microbiology, Vol. 37, No. 3, 1999, pp. 128-135.
- S. Mauersberger, H. J. Wang, C. Gaillardin, G. Barth and J. M. Nicaud, “Insertional Mutagenesis in then-AlkaneAssimilating Yeast Yarrowia lipolytica: Generation of Tagged Mutations in Genes Involved in Hydrophobic Substrate Utilization,” Journal of Bacteriology, Vol. 183, No. 17, 2001, pp. 5102-5109. doi:10.1128/JB.183.17.5102-5109.2001
- P. Founier, A. Abbas, M. Chasles, B. Kudla, D. M. Ogrydziak, D. Yaver, J. W. Xuan, A. Peito, A. M. Ribet, C. Feyrol, F. He and C. Gillardin, “Colocalization of Centromeric and Replicative Functions on Autonomously Replicating Sequences Isolated from the Yeast Yarrowia lipolytica,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 90, No. 11, 1993, pp. 4912-4916. doi:10.1073/pnas.90.11.4912
- C. A. Strick, L. C. James, M. M. O’Donnell, M. G. Gollaher and A. E. Franke, “The Isolation and Characterization of the Pyruvate Kinase-Encoding Gene from the Yeast Yarrowia lipolytica,” Gene, Vol. 118, No. 1, 1992, pp. 65-72. doi:10.1016/0378-1119(92)90249-O
- G. Pignede, H. Wang, F. Fudalej, C. Gaillardin, M. Seman and J. M. Nicaud, “Characterization of an Extracellular Lipase Encoded by LIP2 in Yarrowia lipolytica,” Journal of Bacteriology, Vol. 182, No. 10, 2000, pp. 2802-2810. doi:10.1128/JB.182.10.2802-2810.2000
- C. W. Owens and R. V. Belcher, “A Colorimetric MicroMethod for the Determination of Glutathione,” The Biochemical Journal, Vol. 94, No. 3, 1965, pp. 705-711.
- Y. Inoue, H. Rhee, K. Watanabe, K. Murata and A. Kimura, “Metabolism of 2-Ketoaldehydes in Mold: Purification and Characterization of Glyoxalase I from Aspergillusniger,” The Journal of Biochemistry, Vol. 102, No. 3, 1987, pp. 583-589.
- R. E. Allen, T. W. C. Lo and P. J. Thornalley, “Purification and Characterization of Glyoxalase II from Human Red Blood Cells,” European Journal of Biochemistry, Vol. 213, No. 3, 1993, pp. 1261-1267. doi:10.1111/j.1432-1033.1993.tb17877.x
- C. C. White, H. Viernes, C. M. Krejsa, D. Botta and T. J. Kavanagh, “Fluorescence-Based Microtiter Plate Assay for Glutamate-Cysteine Ligase Activity,” Analytical Biochemistry, Vol. 318, No. 2, 2003, pp. 175-180. doi:10.1016/S0003-2697(03)00143-X
- N. Shiomi, H. Fukuda, F. Yasuki, K. Murata and A. Kimura, “Expression and Stability of the Plasmid Carring Ethionine-Resistant Gene in Saccharomyces cerevisiae”, Kagakukougakuronbunsyu, Vol. 17, No. 3, 1991, pp. 674-678.
- Y. Inoue and A. Kimura, “Methylglyoxal and Regulation of Its Metabolism in Microorganisms,” Advances in Microbial Physiology, Vol. 37, 1995, pp. 177-227. doi:10.1016/S0065-2911(08)60146-0
- C. S. Newlon, “Yeast Chromosome Replication and Segregation,” Microbiological Reviews, Vol. 52, No. 4, 1988, pp. 568-601.
- A. B. Fucher and B. S. Cox, “Copy Number and the Stability of 2-Micron Circle-Based Artificial Plasmids of Saccharomyces cerevisiae,” Journal of Bacteriology, Vol. 157, No. 1, 1984, pp. 283-290.
- T. Juretzek, M. T. Le Dall, S. Mauersberger and C. Gaillardin, “Vectors for Gene Expression and Amplification in the Yeast Yarrowia lipolytica,” Yeast, Vol. 18, No. 2, 2001, pp. 97-113. doi:10.1002/1097-0061(20010130)18:2<97::AID-YEA652>3.0.CO;2-U
- M. T. Le Dall and J. M. Nicaud, “Multiple-Copy Integration in the Yeast Yarrowia lipolytica,” Current Genetics, Vol. 26, No. 1, 1994, pp. 38-44. doi:10.1007/BF00326302
NOTES
*Corresponding author.

