Open Journal of Stomatology
Vol. 2 No. 4 (2012) , Article ID: 25464 , 8 pages DOI:10.4236/ojst.2012.24056
Salivary cytokines and levels in denture stomatitis: An exploratory case-control study*
![]()
1Department of Periodontology, School of Dentistry, University of North Carolina, Chapel Hill, USA
2Department of Prosthodontics, School of Dentistry, University of North Carolina, Chapel Hill, USA
3Department of Pharmacology, School of Medicine, University of North Carolina, Chapel Hill, USA
4Department of Pharmaceutical and Biomedical Sciences, Touro College of Pharmacy, New York, USA
5Department of Biochemistry and Molecular Biology, New York Medical College, Valhalla, USA
6GlaxoSmithKline Consumer Healthcare, Parsippany, USA
Email: barross@dentistry.unc.edu
Received 14 August 2012; revised 17 September 2012; accepted 6 October 2012
Keywords: Cytokines; Serum C-Reactive Protein; Denture; C. albicans; Stomatitis; Salivary
ABSTRACT
We aimed to evaluate the microbial and inflammatory characteristics associated with Denture Stomatitis (DS) analyzing: l) Levels of salivary cytokines and cultivable C. albicans; 2) DNA-DNA checkerboard on biofilm associated with mucosal tissue-bearing denture surfaces; 3) Serum C-reactive protein (CRP) levels. Thirty-two subjects were enrolled in the study with control (n = 17) and DS types II and III (n = 15) subjects. Samples were collected from unstimulated whole saliva, serum and swabs from denture surfaces. Salivary levels of inflammatory mediators and CRP were measured by multiplex. Samples from denture and mucosal surfaces were analyzed by DNA-DNA checkerboard. Saliva from DS subjects showed increase in IL-8 (p = 0.04) and IL-1β (p = 0.04) with trend for increase in IL-1β, TNFα and IL-6 levels. C. albicans higher counts in DS saliva (p = 0.03) showed association with elevated levels of IL-8 (p = 0.03) and IL-1α (p = 0.01). CRP levels were not different among groups (p = 0.74). DNA-DNA checkerboard analyses indicated typical periodontal pathogens below the detection threshold of 104 organisms on both denture and inflamed mucosal surfaces. The data suggest that DS is associated with elevation of salivary IL1 and IL-8 together with increased C. albicans. There was no evidence of systemic inflammation as measured by serum C-reactive protein levels.
1. INTRODUCTION
Candida-associated denture stomatitis (CADS) is characterized by inflammation of the palatal mucosa in contact with the maxillary denture and is the most common form of oral candidal infection with Candida albicans being the principal etiological agent [1-3]. CADS has been reported in 11% - 67% of the otherwise healthy denture wearers [4], and has been classified according to Newton’s work in 1962 [5] into 3 clinical types: Type I— a localized simple inflammation or a pinpoint hyperemia; Type II (mild): an erythematous or generalized simple type presenting as more diffuse erythema, and Type III (severe): a granular or papillary type. This condition has been considered to have a multifactorial etiology, which can include factors related to 1) Trauma such as denture wearing; 2) Local trauma resulting from salivary hypofunction [6,7]; 3) Infection with candida species; or 4) Factors related to impaired host defense as in systemic diseases such as HIV infection, leukemia, lymphoma, radiation therapy for head and neck malignancies, chemotherapy, diabetes, hormonal imbalance, anemia, malnutrition, or long-term use of corticosteroid or antibiotics. The multi-factorial etiology for denture stomatitis is reflected in the current treatment regimens which vary from antifungals [8], to denture hygiene measures, antiseptics, and use of denture reline materials containing antifungals [9].
Although some progress has been made in revealing the mechanisms of epithelial and immunoregulatory interactions with Candida, there are major gaps in our knowledge regarding processes that lead to the establishment of candidiasis as an infection and the relationship between mucosal and systemic inflammatory responses. Primary innate defense mechanisms are known to play key roles in preventing yeast colonization of the oral cavity by providing epithelium as a physical barrier as well as lingual antimicrobial peptide, defensins, secretory IgA and salivary factors [10]. The main sources of cytokines generated in the oral mucosa during infection with Candida have been reported to be of oral epithelial cell origin, although infiltrating neutrophils are likely important contributors. Cytokines in saliva are also derived from salivary gland cells (secretory, epithelial cells, fibroblasts, endothelial cells), and leukocyte aggregates within the salivary gland tissues, tonsils, and the oral mucosa, including the periodontium. Therefore, cytokine profiles in whole saliva can depict the collective state of cytokine responses to infectious agents by all tissues in the oral cavity, including accessory salivary glands which are major contributors of secretory IgA [11]. However, the influence of whole saliva composition on Candida adherence and invasion is yet to be fully understood.
Biofilms are structured microbial communities in which the cells bind tightly to a surface and become embedded in a matrix of extracellular polymeric substances produced by these cells. Indeed components of saliva or serum proteins, such as mucin, fibrinogen and complementary factors specifically bind to Candida blastospores and germ tubes, possibly modifying the biofilm formation [12]. Biofilms of differing compositions colonize oral surfaces including the teeth, the mucosa, the tongue and acrylic denture surfaces. These self-replenishing biofilms gain notoriety from their ability to resist antimicrobials and immune cell challenge [13,14].
The purpose of this study was to identify the candidate inflammatory mediators present within the saliva of subjects with denture stomatitis. We sought to confirm previous reports establishing the relationship between DS and increased salivary levels C. albicans and to determine if the level of C. albicans was associated with the levels of salivary cytokine biomarkers. Finally, since DS is such a common infection among edentate people we accessed whether there was any evidence of systemic inflammatory response as indexed by serum levels of C-reactive protein. Although this was a relatively small exploratory pilot investigation, the data suggest that increased levels of certain salivary cytokines are associated with the presence of DS and related to salivary levels of C. albicans.
2. MATERIAL AND METHODS
An IRB approved case-control study was performed. Thirty-two subjects were enrolled and grouped into two groups: a control group (n = 17) with no signs or symptoms of denture stomatitis (DS), and a diseased group (n = 15) with type II or III DS (Newton’s classification). Inclusion and exclusion criteria, and demographics of this cohort of patients are described in Table 1.

Table 1. Inclusion and exclusion criteria.
The selection of candidate cytokines was based upon the findings of enhanced mRNA expression of specific cytokines within mucosal biopsy samplescollected from these subjects using whole transcriptome gene expression arrays.
2.1. Unstimulated Saliva Collection and Cytokines Analysis
Subjects were instructed to remove their dentures and refrain from eating, drinking, smoking, brushing their teeth or chewing gum for 15 minutes prior to salivary collections. All collections were performed between 9:00 and 11:00 AM. Subjects were instructed to swallow to clear the mouth of any accumulated saliva, and whole unstimulated saliva was allowed to pool in a sterile polypropylene graduated collection vial for 5 minutes. A fraction of the sample was sent immediately to the microbiology lab to prevent overgrowth of the Candida species. Samples were aliquoted into Eppendorf tubes, centrifuged for 10 minutes at 3000 g, and the supernatant stored at −80˚C until analysis. Before cytokines analysis, the samples were thawed and centrifuged for 10 minutes at 3000 g to remove any solids. Specific proteins (IL-1α, IL-1β, IL-6, IL-8, and TNFα) levels in Saliva samples were quantified by ELISA and/or using Fluorokine® MAP cytokine multiplex kits (R&D Systems, Minneapolis, MN) and a Bio-Plex 200® analyzer system (BioRad, Inc., Hercules, CA). Protein levels were log-transformed prior to application of multivariate tests comparing subjects with denture stomatitis to those without denture stomatitis, and comparing diseased and non-diseased sites within subjects with clinical stomatitis.
A fraction of the unstimulated saliva was sampled in Sabouraud Dextrose agar containing quemicetine succinate. Candida colonies were counted after 48 h and the patients were classified according to the number of CFU (Colony-forming units) as follows: negative (CFU/ml = 0), carrier (CFU/ml < 400), and positive (CFU/ml > 400).
The template is used to format your paper and style the text. All margins, column widths, line spaces, and text fonts are prescribed; please do not alter them. You may note peculiarities. For example, the head margin in this template measures proportionately more than is custo-mary. This measurement and others are deliberate, using specifications that anticipate your paper as one part of the entire journals, and not as an independent document. Please do not revise any of the current designations.
2.2. Microbiology
Four bacterial biofilm samples were collected from the tissue and the corresponding surface area of the denture using a plastic curette. The palate was divided into four zones (according to the method described by Barbeau et al. [16] as shown in Figure 1, and one biofilm sample was collected from each zone and recorded on a diagnostic diagram. The samples were pooled (4 mucosal samples and 4 denture samples) and placed into a separate Eppendorf tube containing 150 mL of TE buffer. 150 mL of 0.5 M NaOH were added to each sample and frozen at −80˚C prior to analyses. The pooled biofilm samples were analyzed quantitatively and qualitatively to measure the presence, absence and levels of 18 dental biofilm organisms by DNA-DNA checkerboard tissue, where median cell count levels of 16 organisms was produced for comparison of subjects with clinical stomatitis to those without clinical stomatitis using a nonparametric multivariate test. Mean Candida CFU counts were produced for comparison of the diseased and non-diseased groups. Data has been published elsewhere [15].
2.3. DNA-DNA Checkerboard
2.3.1. Sample Deposition on Nylon Membranes
The samples were boiled in a water bath for 5 minutes and neutralized using 0.8 ml 5 M ammonium acetate. The released DNA was placed into the slots of a Minislot device (Immunetics, Cambridge MA) and thereby deposited as “lanes” onto a (formerly Boehringer Mannheim, now Roche Diagnostics nylon membrane). The DNA was then permanently fixed to the membrane by exposure to ultraviolet light, followed by baking at 120˚C, for 20 min. The Minislot device permits the deposition of up to 30 different bacterial samples in individual “lanes” on a single 15 × 15 cm nylon membrane. In order to create reference signals on each membrane, two of the ‘lanes’ were used to deposit pooled DNA of known bacterial numbers (105 and 106) for each one of the microorganisms included in the analysis.
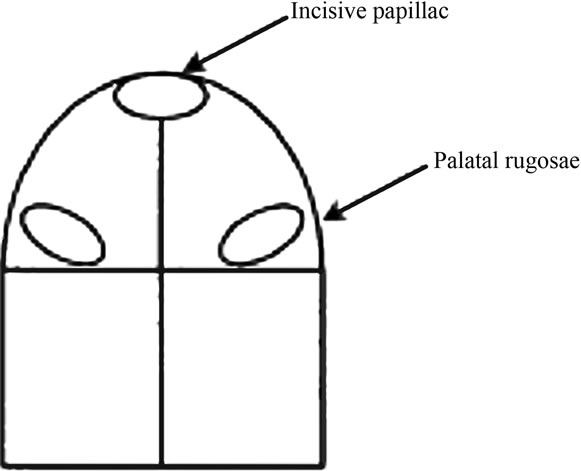
Figure 1. A diagram showing the division of the palate area into quadrants for bacterial biofilm collection from each of the 4 zones.
2.3.2. DNA-DNA Hybridization
The membranes were prehybridized in “prehybridization” buffer [50% formamide, 5 × SSC (1 × SSC = 150 mM NaCl, 15 mM Na citrate, pH 7.0), 1% casein (Sigma), 5 × Denhardt’s reagent, 25 mM sodium phosphate (pH 6.5) and 0.5 mg/ml yeast RNA (Boehringer-Mannheim)], at 42˚C, for 1 hour and then placed back into a “Miniblotter 45”, turned 90˚ to the original orientation. A 30 × 45 “checkerboard” pattern was produced. Digoxigenin-labeled DNA probes dissolved in “hybridization” buffer [45% formamide, 5 × SSC, 1 × Denhardt’s reagent, 20 mM Na phosphate (pH 6.5), 0.2 mg/ml yeast RNA, 20 ng/ml of labeled probe, 10% dextran sulfate and 1% casein], were laid in individual lanes of the Miniblotter and the whole apparatus was placed in a sealed plastic bag. Hybridization was allowed to occur overnight, in a shaking water bath, at 42˚C. Membranes were then washed once at low stringency to remove loosely bound probe and then twice at high stringency (68˚C, 0.1 × SSC, 0.1% SDS, 20 min), in a Disk Wisk apparatus (Schleicher and Schuell).
2.4. Blood Collection (Serum CRP)
Venous blood (5 - 7 ml) was collected under a sterile technique in a purple top vaccutainer. Blood was processed into serum within 2 hours after collection: the whole blood was kept at room temperature for 30 - 45 minutes to allow a clot to form, and then centrifuged for 12 minutes to separate the serum from the clot. Serum was then aliquoted into barcode labeled microfuge tubes and quickly frozen at −80˚C and stored until analysis using ELISA method. Mean serum CRP levels (mg/ml) were compared in diseased and non-diseased groups.
3. STATISCAL ANALYSIS
Salivary cytokine levels were not normally distributed and between group comparisons were made using the non-parametric, non-paired Wilcoxon test. Associations between salivary C. albicans CFU/ml and cytokine levels were compared by one-way ANOVA and regression analyses. The association between the presence or absence of C. albicans and the presence (pooling Type II and Type III) or absence of DS was determined by Chi Square test. A p value < 0.05 was considered significant, however no adjustments were made for multiple comparisons and the results should be interpreted with caution.
4. RESULTS
4.1. Salivary Markers Comparing Control and Denture Stomatitis
We measured salivary levels of IL-1α, IL-1β, IL-6, IL-8 and TNFα. The data from the DS and control subjects appear in Table 2. There was a statistically significant increase in salivary levels of IL-8 (p = 0.04) and IL-1β (p = 0.04) in DS as compared to health. In DS the apparent increase in salivary levels of IL-6, TNFα and IL-1α were not statistically significant. These increases in protein expression were consistent with the increased tissue mRNA expression (data not shown).
A statistically significant association between salivary counts of C. albicans and salivary levels of IL-8 (r2 = 0.16, P = 0.03), and IL-1α (r2 = 0.20, p = 0.01) was seen as illustrated in Figure 2.
C. albicans was detected in saliva of 80% (12/15) of the DS subjects, and in 35.3% (6/17) of the Healthy subjects, (p = 0.03), using the positive cut-off value of (CFU/ml > 400).
4.2. Bacterial Composition of Mucosal and Denture Surface Biofilm Determined by DNA-DNA Checkerboard
Tissue and denture surfaces were sampled using a plastic curette to measure the presence, absence and levels of 16 dental biofilm organisms by DNA-DNA checkerboard. Biofilm samples from the palatal tissue surfaces did not show any detectible organisms, suggesting a level below 104 counts. Organisms were detected in the biofilm samples collected from dentures with (3/17) control subjects, (1/ 8) DS Type II, and (1/7) DS Type III. The data for the 3 healthy are compared to the Type II and Type III DS in Table 3. There were no significant differences in the counts comparing Health to Diseased samples.
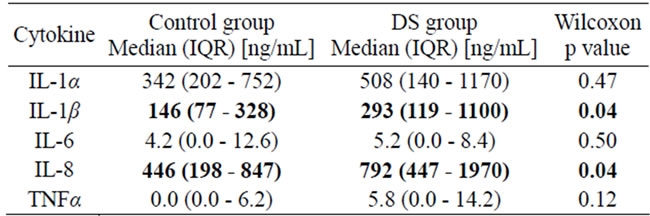
Table 2. Salivary Cytokines levels in control and DS groups.
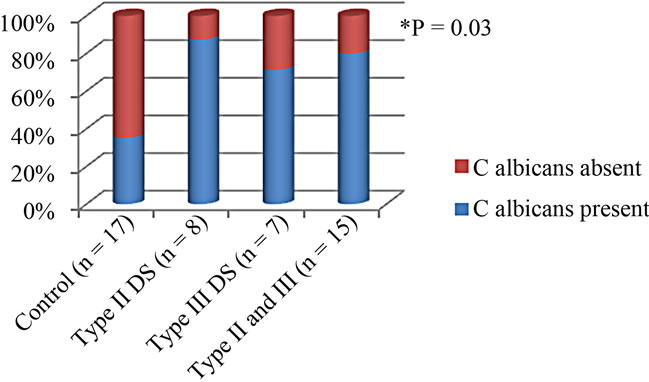
Figure 2. Salivary C. albicans prevelance according to DS severity.
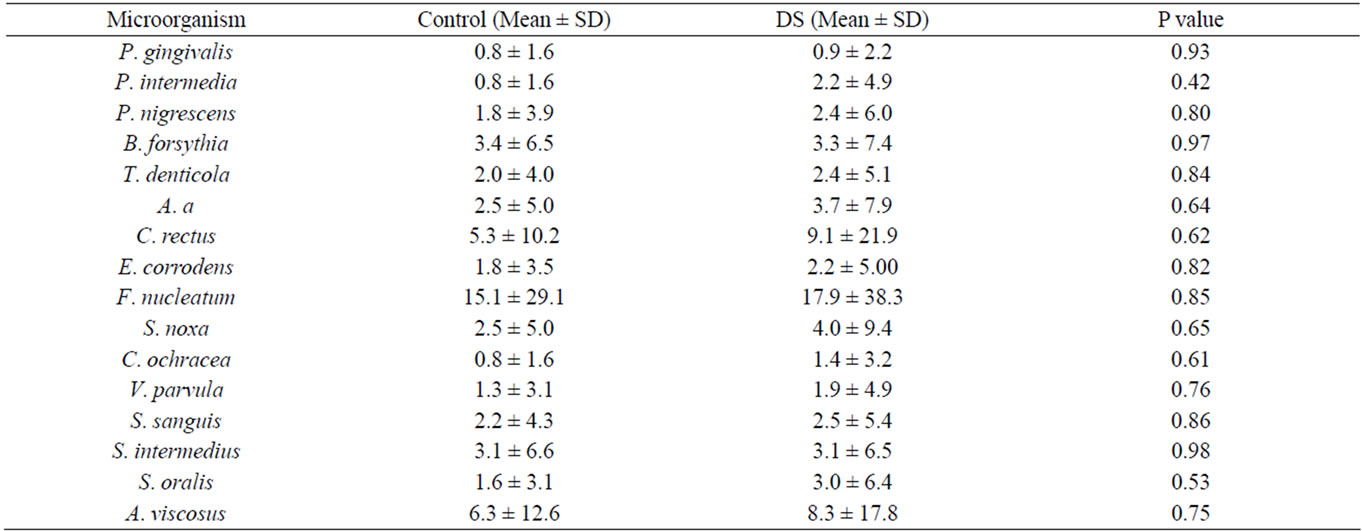
Table 3. Organisms detected with DNA-DNA checkerboard of biofilm from denture surfaces. Numbers reflect counts × 106 CFU.
4.3. Serum CRP
There was no significant difference in the serum CRP level comparing health to DS individuals (p = 0.74). We also examined levels of clinically elevated CRP values using CRP < 3 mg/ml as a threshold, and no statistical differences between the groups could be detected (Table 4).
5. DICUSSION
We investigated levels of the selected markers: IL-1α, IL-1b, IL-6, IL-8, and TNFα in saliva and found a statistically significant two fold increase in median salivary levels of IL-8 (p = 0.04) and a 1.8-fold increase in IL-1β. These increases are in accordance with other studies that looked at these pro-inflammatory cytokines in oral epithelial cells infected with C. albicans [17-19]. Increased levels of GM-CSF, IFN-γ, TNF-α IL-8, IL-18 and IL-1α, IL-1β, IL-6, have been identified in the oral mucosa epithelial cell infection models and saliva of subjects with oral candidiasis and in experimental models of oral candidiasis [11,18,19]. Recognition of C. albicans by oral epithelial cells through specific patternrecognition receptors (PRRs) leads to activation of intracellular signaling pathways that modulate inflammatory responses including release of cytokines such as IL-1α, IL-6, IL-8, IL-15, IL-18, TNFα as well as upregulation of synthesis of specific antimicrobials such as defensins and nitrous oxide (NO) [19]. These mediators could also exhibit a regulatory effect on the secretion of each other, for example TNF-α, which showed a, increase, albeit no significant, in the DS group is known to activate secretion of other cytokines, such as IFN-γ, IL-1, IL-6, and IL-12 which further amplifies the protective immune response to C. albicans infection [19,20]. Furthermore, IL-1β which was significantly increased in the DS group,
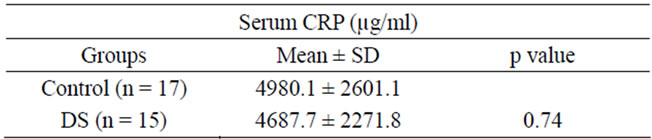
Table 4. CRP levels in control and DS groups.
is known to play a crucial role in activating production of IL-1inducible cytokines, which attract and activate cells of the immune and inflammatory systems, e.g., IL-6, IL-8, granulocyte-macrophage (GM)-CSF, and G-CSF [21]. We found a significant association between the salivary counts of C. albicans and the levels of IL-8 and IL-1α (Figure 2). We have recently found regarding C. albicans colonization in denture stomatitis patients, that the denture appears to be the primary source of salivary C. albicans rather than the inflamed mucosa. C. albicans was detected in saliva of 80% (12/15) of the DS subjects, but only 41.2% (7/17) of the Healthy subjects, (p = 0.03). Thus, C. albicans was twice as likely to be present in saliva if mild or severe DS was present. Therefore, the high levels in saliva likely represent the organisms released from denture surfaces with minor contribution from the mucosal surfaces. IL8 levels in saliva ranged from 447 - 1970 ng/ml in the DS group, this cytokine acts as a potent chemokine for neutrophils at nanomolar concentrations, thus this increase in IL-8 levels would lead to neutrophil recruitment within the inflamed mucosa and passage into the saliva via transepithelial migration. Such an increase in neutrophils within the saliva has previously been reported in mucositis and in DS subjects [11,18,22].
It is well understood that adequate attachment of candidal organisms to denture surface is a crucial step in the pathogenesis of denture stomatitis. The formation of biofilms on the surface of the denture protects these organisms from detachment and removal by salivary flow and physical forces [4,14]. It’s also worth noticing that quorum sensing (QS) occurs both in single species bacterial communities and in complex mixed bacterialyeast communities [23,24]. However, the role played by saliva or serum pellicles during the colonization process and subsequent multilayer biofilm formation is poorly understood [12,24-26].
Measuring the levels of 16 dental biofilm organisms by DNA-DNA checkerboard in the mucosa and dentures, only samples collected from dentures were within the detectable range of 1 × 104 counts, however, organisms could be detected in the biofilm samples collected from dentures in three control cases, 1-DS Type 2, and 1-DS Type 3 patients and there were no significant differences in the counts comparing Health to Diseased samples. Those findings suggest that a more sensitive method, like 16 s RNA, should be utilized to better assess changes in the bacterial mucosal bacterial flora when comparing DS and control samples [27-33].
Sachdeo et al. [34] have used a similar methodology to the one performed in our study to characterize the microbiota of edentulous oral cavities using checkerboard DNA-DNA hybridization, analyzing samples from mucosal and denture surfaces, and saliva from edentulous patients for 41 bacterial species where they found that A. actinomycetemcomitans and P. gingivalis, which were thought to be eliminated with the extraction of all natural teeth, were seen in significant numbers in the edentulous subjects. These pathogens were detected in our study as well. Also, when those authors compared the mean total DNA probe counts among the dorsal surfaces of the tongue, denture palates, and the subjects’ palates, the highest mean counts were found on the tongue dorsum, followed by the polished (exterior) surface of the denture palate, and were lowest on the hard palate. This finding is also consistent with our study where no organisms were detected from the mucosal samples. However, no samples were taken from the polished surfaces of the denture in our samples, as we collected the tissue-bearing surface.
Other studies have shown that the predominant cultivable flora of denture plaque was Gram-positive cocci (Streptococci) and rod-shaped bacteria [35-37]. Results from such studies suggest similarity between denture plaque bacterial composition and the periodontal niche in dentate patients. Therefore, consistent with the association of periodontal disease and specific periodontal pathogens on conditions such as cardiovascular disease and poor glycemic control among diabetics it has been suggested that denture cleaning is important to oral and systemic health and that the denture-wearing population requires care and follow-up similar to their non-denturewearing counterparts [34].
Analyzing levels of CRP in those patients, we found no significant difference in the levels of CRP between the two groups of patients. The findings are consistent with those of Kostial el. al who reported that CRP values in patients with acute fungal stomatitis in non-immunocompromised patients were within normal ranges [38]. In contrast, another study, Ajwani et al. reported elevated levels of CRP and microbial counts in edentulous individuals with signs of oral candidiasis or denture stomatatis [39]. It has been reported that CRP levels are higher in patients with periodontitis [40-44], and our data showed there is similarity between the microbial content of denture plaque and that associated with periodontitis. Thus, it is plausible that fungal infections, which activate a different microbial pathogen recognition system than bacteria, may specifically trigger the release of hepatic acute phase reactants, other than CRP, such as alpha antitrypsin, transferrin or orosomucoid synthesis.
6. CONCLUSION
Elevated levels of salivary IL-8 and IL-1β are associated with DS, and IL-8 is positively associated with increased levels of C. albicans in saliva. Our findings suggest that the overgrowth and colonization of C. albicans on acrylic denture surfaces may act as an early colonizer in the succession of biofilm formation (replacing many streptococci) and may serve to facilitate the emergence of anaerobic microorganisms. These data suggest that an unclean denture can serve as the primary source of C. albicans that enhances mucosal inflammation, pointing to the importance of denture hygiene in reducing denture stomatitis.
REFERENCES
- Webb, B.C., Thomas, C.J., Willcox, M.D., et al. (1998a) Candida-associated denture stomatitis. Aetiology and management: A review. Part 1. Factors influencing distribution of Candida species in the oral cavity. Australian Dental Journal, 1, 45-50. doi:10.1111/j.1834-7819.1998.tb00152.x
- Budtz-Jorgensen, E. (1990) Etiology, pathogenesis, therapy, and prophylaxis of oral yeast infections. Acta Odontologica Scandinavica, 1, 61-69. doi:10.3109/00016359009012735
- Chandra, J., Mukherjee, P.K., Leidich, S.D., et al. (2001) Antifungal resistance of candidal biofilms formed on denture acrylic in vitro. Journal of Dental Research, 3, 903-908. doi:10.1177/00220345010800031101
- Ramage, G., Tomsett, K., Wickes, B.L., et al. (2004) Denture stomatitis: A role for Candida biofilms. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontics, 1, 53-59. doi:10.1016/j.tripleo.2003.04.002
- Newton, A.V. (1962) Denture sore mouth. A possible etiology. British Dental Journal, 112, 357-360.
- Lyon, J.P., da Costa, S.C., Totti, V.M., et al. (2006) Predisposing conditions for Candida spp. carriage in the oral cavity of denture wearers and individuals with natural teeth. Canadian Journal of Microbiology, 5, 462-467. doi:10.1139/w05-148
- Abraham, C.M., Al-Hashimi, I. and Haghighat, N. (1998) Evaluation of the levels of oral Candida in patients with Sjogren’s syndrome. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontics, 1, 65-68. doi:10.1016/S1079-2104(98)90151-2
- Webb, B.C., Thomas, C.J., Willcox, M.D., et al. (1998b) Candida-associated denture stomatitis. Aetiology and management: A review. Part 3. Treatment of oral candidosis. Australian Dental Journal, 43, 244-249. doi:10.1111/j.1834-7819.1998.tb00172.x
- Ellepola, A.N. and Samaranayake, L.P. (2000) Oral candidal infections and antimycotics. Critical Reviews in Oral Biology and Medicine, 11, 172-198. doi:10.1177/10454411000110020301
- Cannon, R.D., Holmes, A.R., Mason, A.B., et al. (1995) Oral candida: Clearance, colonization, or candidiasis? Journal of Dental Research, 74, 1152-1161. doi:10.1177/00220345950740050301
- Dongari-Bagtzoglou, A. and Fidel Jr., P.L. (2005) The host cytokine responses and protective immunity in oropharyngeal candidiasis. Journal of Dental Research, 84, 966-977. doi:10.1177/154405910508401101
- Nikawa, H., Nishimura, H., Makihira, S., et al. (2000) Effect of serum concentration on Candida biofilm formation on acrylic surfaces. Mycoses, 43, 139-143. doi:10.1046/j.1439-0507.2000.00564.x
- Nett, J. and Andes, D. (2006) Candida albicans biofilm development, modeling a host-pathogen interaction. Current Opinion in Microbiology, 9, 340-345. doi:10.1016/j.mib.2006.06.007
- Ramage, G., Vandewalle, K., Wickes, B.L., et al. (2001) Characteristics of biofilm formation by Candida albicans. Revista Iberoamericana de Micología, 4, 163-170.
- AlTarawneh, S.K., Bencharit, S., Mendoza, L., et al. (2011) Role of C. albicans in saliva and on mucosal and denture surfaces in denture stomatitis: An exploratory casecontrol study. Journal of Prosthodontics, accepted for publication.
- Barbeau, J., Seguin, J., Goulet, J.P., et al. (2003) Reassessing the presence of Candida albicans in denture-related stomatitis. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology and Endodontics, 95, 51-59. doi:10.1067/moe.2003.44
- Mostefaoui, Y., Claveau, I. and Rouabhia, M. (2004) In vitro analyses of tissue structure and interleukin-1beta expression and production by human oral mucosa in response to Candida albicans infections. Cytokine, 25, 162-171. doi:10.1016/j.cyto.2003.11.015
- Dongari-Bagtzoglou, A. and Kashleva, H. (2003) Candida albicans triggers interleukin-8 secretion by oral epithelial cells. Microbial Pathogenesis, 34, 169-177. doi:10.1016/S0882-4010(03)00004-4
- Villar, C.C. and Dongari-Bagtzoglou, A. (2008) Immune defence mechanisms and immunoenhancement strategies in oropharyngeal candidiasis. Expert Reviews in Molecular Medicine, 10, e29. doi:10.1017/S1462399408000835
- Kennedy, A.D., Willment, J.A., Dorward, D.W., et al. (2007) Dectin-1 promotes fungicidal activity of human neutrophils. European Journal of Immunology, 37, 467-478. doi:10.1002/eji.200636653
- Mizutani, H., Black, R. and Kupper, T.S. (1991) Human keratinocytes produce but do not process pro-interleukin-1 (IL-1) beta. Different strategies of IL-1 production and processing in monocytes and keratinocytes. The Journal of Clinical Investigation, 87, 1066-1071. doi:10.1172/JCI115067
- Ali, A., Rautemaa, R., Hietanen, J., et al. (2006) Expression of interleukin-8 and its receptor IL-8RA in chronic hyperplastic candidosis. Oral Microbiology and Immunology, 21, 223-230. doi:10.1111/j.1399-302X.2006.00280.x
- Blankenship, J.R. and Mitchell, A.P. (2006) How to build a biofilm: A fungal perspective. Current Opinion in Microbiology, 9, 588-594. doi:10.1016/j.mib.2006.10.003
- Pereira-Cenci, T., Del Bel Cury, A.A., Crielaard, W., et al. (2008) Development of Candida-associated denture stomatitis: New insights. Journal of Applied Oral Science, 16, 86-94. doi:10.1590/S1678-77572008000200002
- Nikawa, H., Mikihira, S., Egusa, H., et al. (2005) Candida adherence and biofilm formation on oral surfaces. Japanese Journal of Medical Mycology, 46, 233-242. doi:10.3314/jjmm.46.233
- Nikawa, H., Egusa, H., Yamashiro, H., et al. (2006) The effect of saliva or serum on bacterial and Candida albicans colonization on type I collagen. Journal of Oral Rehabilitation, 33, 767-774. doi:10.1046/j.0305-182X.2003.01149.x
- Riep, B., Edesi-Neuss, L., Claessen, F., et al. (2009) Are putative periodontal pathogens reliable diagnostic markers? Journal of Clinical Microbiology, 47, 1705-1711. doi:10.1128/JCM.01387-08
- Lazarevic, V., Whiteson, K., Hernandez, D., et al. (2010) Study of interand intra-individual variations in the salivary microbiota. BMC Genomics, 11, 523. doi:10.1186/1471-2164-11-523
- Milward, M.R., Chapple, I.L., Wright, H.J., et al. (2007) Differential activation of NF-kappaB and gene expression in oral epithelial cells by periodontal pathogens. Clinical and Experimental Immunology, 148, 307-324. doi:10.1111/j.1365-2249.2007.03342.x
- Kirakodu, S.S., Govindaswami, M., Novak, M.J., et al. (2008) Optimizing qPCR for the quantification of periodontal pathogens in a complex plaque biofilm. The Open Dentistry Journal, 2, 49-55. doi:10.2174/1874210600802010049
- Gomes, S.C., Nonnenmacher, C., Susin, C., et al. (2008) The effect of a supragingival plaque-control regimen on the subgingival microbiota in smokers and never-smokers: Evaluation by real-time polymerase chain reaction. Journal of Periodontology, 79, 2297-2304. doi:10.1902/jop.2008.070558
- Casarin, R.C., Ribeiro, E.P., Mariano, F.S., et al. (2010) Levels of Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, inflammatory cytokines and species-specific immunoglobulin G in generalized aggressive and chronic periodontitis. Journal of Periodontal Research, 45, 635-642. doi:10.1111/j.1600-0765.2010.01278.x
- Colombo, A.P., Boches, S.K., Cotton, S.L., et al. (2009) Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. Journal of Periodontology, 80, 1421-1432. doi:10.1902/jop.2009.090185
- Sachdeo, A., Haffajee, A.D. and Socransky, S.S. (2008) Biofilms in the edentulous oral cavity. Journal of Prosthodontics, 17, 348-356. doi:10.1111/j.1532-849X.2008.00301.x
- Theilade, E. and Budtz-Jorgensen, E. (1988) Predominant cultivable microflora of plaque on removable dentures in patients with denture-induced stomatitis. Oral Microbiology and Immunology, 3, 8-13. doi:10.1111/j.1399-302X.1988.tb00597.x
- Theilade, E., Budtz-Jorgensen, E. and Theilade, J. (1983) Predominant cultivable microflora of plaque on removable dentures in patients with healthy oral mucosa. Archives of Oral Biology, 28, 675-680. doi:10.1016/0003-9969(83)90101-2
- Radford, D.R., Challacombe, S.J. and Walter, J.D. (1999) Denture plaque and adherence of Candida albicans to denture-base materials in vivo and in vitro. Critical Reviews in Oral Biology and Medicine, 10, 99-116. doi:10.1177/10454411990100010501
- Kostiala, I. (1984) C-reactive protein response induced by fungal infections. The Journal of Infection, 8, 212-220. doi:10.1016/S0163-4453(84)93883-0
- Ajwani, S., Mattila, K.J., Narhi, T.O., et al. (2003) Oral health status, C-reactive protein and mortality—A 10- year follow-up study. Gerodontology, 20, 32-40. doi:10.1111/j.1741-2358.2003.00032.x
- Beck, J.D. and Offenbacher, S. (2002) Relationships among clinical measures of periodontal disease and their associations with systemic markers. Annals of Periodontology, 7, 79-89. doi:10.1902/annals.2002.7.1.79
- Craig, R.G., Yip, J.K., So, M.K., et al. (2003) Relationship of destructive periodontal disease to the acute-phase response. Journal of Periodontology, 74, 1007-1016. doi:10.1902/jop.2003.74.7.1007
- D’Aiuto, F., Parkar, M. and Tonetti, M.S. (2005) Periodontal therapy: A novel acute inflammatory model. Inflammation Research, 54, 412-414. doi:10.1007/s00011-005-1375-4
- Ebersole, J.L., Cappelli, D., Mathys, E.C., et al. (2002) Periodontitis in humans and non-human primates: Oralsystemic linkage inducing acute phase proteins. Annals of Periodontology, 7, 102-111. doi:10.1902/annals.2002.7.1.102
- Slade, G.D., Offenbacher, S., Beck, J.D., et al. (2000) Acute-phase inflammatory response to periodontal disease in the US population. Journal of Dental Research, 79, 49-57. doi:10.1177/00220345000790010701
NOTES
*This work was supported by NIH research grant UL-1-RR025746 and GSK Consumer Health Care.

