Journal of Cancer Therapy
Vol.2 No.5(2011), Article ID:16616,7 pages DOI:10.4236/jct.2011.25093
Different Effects of TERT, TP63, and CYP2A6 Polymorphism on Individual Risk of Tobacco-Related Lung Cancer in MaleJapanese Smokers
![]()
1Showa Pharmaceutical University, Tokyo, Japan; 2Hokkaido University, Sapporo, Japan; 3RIKEN Center for Genomic Medicine, Yokohama, Japan; 4Mitsui Memorial Hospital, Tokyo, Japan.
E-mail: #hyamazak@ac.shoyaku.ac.jp
Received August 26th, 2011; revised September 27th, 2011; accepted October 6th, 2011.
Keywords: TERT, TP63, Cytochrome P450 2A6, lung cancer, tobacco
ABSTRACT
Recent genome-wide association studies have identified lung cancer susceptibility loci, such as chromosome 5p15 (telomerase reverse transcriptase, TERT and cleft lip and palate transmembrane protein 1-like, CLPTM1L), 15q25 (nicotinic cholinergic receptor α, CHRNA3-CHRNA5), and 3q28 (tumor protein p63, TP63). Replication study was performed to confirm the association of the recently-identified susceptible loci (i.e., TERT-CLPTM1L, CHRNA3-CHRNA5, and TP63) in a total of 1460 male Japanese smokers (885 lung cancer cases and 575 healthy control subjects), which were previously studied for a low odds ratio of impaired or deletion polymorphism in cytochrome P450 2A6 (CYP2A6) for lung cancer risk. The minor allele frequency (0.442) of rs2736100 on 5p15 (TERT) was significantly higher in lung cancer cases than that (0.395) of controls, with an odds ratio of 1.27 (95% CI of 1.07 - 1.50, p = 0.00504). A series of subgroup analyses revealed the significant associations of rs4488809 (TP63, odds ratio of 1.21, p = 0.0422) and rs2736100 (TERT, odds ratio of 1.47, p = 6.40 × 10–5) with the risk of lung adenocarcinoma. No significant association of CHRNA3-CHRNA5 and CLPTM1L was found in this population. The present results support replication of the association of TERT and TP63 loci with lung adenocarcinomas and suggest subtype-specific effects of these loci on higher risk of lung cancer in smokers. The CYP2A6 including copy number polymorphism, uninvestigated in large-scale genome-wide association studies, may influence lower risk to heavy tobacco use-related lung cancer.
1. Introduction
Recent association findings for nicotine dependence, smoking behavior, and smoking-related diseases implicate genetically derived individual differences [1-5]. Among several reports from genome-wide association studies (GWAS), nicotinic cholinergic receptor α (CHRNA3- CHRNA5, rs1051730 or rs667282) has been identified as a lung cancer susceptible locus [6-8]. Similarly, recent GWAS with respect to lung cancer risk in different ethnic populations have suggested a variety of susceptible genes, such as telomerase reverse transcriptase (TERT, rs2736100) [7,9,10], cleft lip and palate transmembrane protein 1-like (CLPTM1L, rs401681) [6,11], HLA-B associated transcript-3 (BAT3, rs3117582) [6], and mutS homolog 5 (MSH5, rs3131379) [6]. Tumor protein p63 (TP63, rs4488809, rs9816619, or rs10937405) was newly identified as a susceptible gene for lung adenocarcinomas in the Japanese and Korean from GWAS [10]. In our previous report [12], we demonstrated that genetic polymorphisms in cytochrome P450 2A6 (CYP2A6) are determinants of smoking behavior and tobacco-related lung cancer risk, particularly squamous cell and small cell carcinoma, which are known to be associated with cigarette smoking [4], in heavy smokers of male Japanese. Some of the single nucleotide polymorphisms (SNPs) found by GWAS have been investigated extensively, however, there have been few reports as to newly identified SNPs, including TP63 [10].
In the current study, we performed a replication study of the recently-identified candidate genes (i.e., CHRNA3- CHRNA5, TERT-CLPTM1L, and TP63) in a population of male Japanese smokers. Herein, we report the replication of TP63 and TERT SNPs in one of the subgroups (lung adenocarcinoma). The polymorphisms of CYP2A6, including copy number polymorphism (CNP), which has not been studied yet in the platform of GWAS, may mostly influence susceptibility to heavy tobacco use-related lung cancer risk.
2. Materials and Methods
2.1. Subjects
This study was approved by the ethics committees of Hokkaido University and Showa Pharmaceutical University. The sample population comprised 1460 unrelated male Japanese smokers (885 case and 575 control subjects) out of the 1705 participants of our previous study [12]. The remaining 245 subjects could not participate in this study because of sample limitations; there was apparently no difference after removal of these missing samples. The patient group consisted of 885 men who had received a pathological diagnosis of lung cancer (squamous cell or small cell carcinoma, n = 409 or adenocarcinoma, n = 476) with a mean (± SD) age of 62.6 ± 9.3 years (in the range 22 - 86 years). The control group consisted of 575 male smokers with a mean age of 52.9 ± 11.3 years (range 20 - 92 years) without a history of cancer. The age of the lung cancer patients was defined at the time when pathological diagnosis of lung cancer was first confirmed. Smokers included current and ex-smokers and were defined as individuals who had ever smoked cigarettes with a minimum smoking history of 10 cigarettes per day for at least 1 year. Light and heavy smokers were categorized by the 50th percentile Brinkman index value (daily cigarettes × years) among control subjects and were defined as less than 690 (n = 286) and more than or equal to 690 (n = 289), respectively. Pathological classification of lung cancers was determined by more than three pathologists according to the criteria described in the literature [12].
2.2. Genotyping
Genomic DNA was prepared from peripheral lymphocytes [12]. SNPs were selected from previous GWAS reports [6,8-11] ; rs4488809, rs9816619, rs10937405, and rs4600802 on 3q28; rs2736100, rs401681, rs4634969, rs402710, and rs31489 on 5p15; rs3117582 and rs3131379 on 6p21.33 (containing BAT3-MSH5); and rs1051730, rs667282, rs8034191, rs206534, rs16969968, rs12910984, rs12914385, and rs6495309 on 15q25. By consideration of linkage disequilibrium (LD), SNPs in high LD to another SNP (r2 > 0.8 in HapMap—Japanese in Tokyo (JPT)) were excluded and the most significantly-associated SNPs in each locus were analyzed [13]. The SNPs on 6p21.33 were excluded because this SNP is not polymorphic in HapMap JPT [13]. A total of 7 SNPs were genotyped using commercially available TaqMan predesigned probes and primers (Applied Biosystems, Foster City, CA) on ABI PRISM 7500 (Applied Biosystems). Genotyping for the CYP2A6 was carried out by the methods described previously [12].
2.3. Statistical Analysis
The associations between the genotype distributions and the patients’ status were assessed by odds ratios and 95% confidence intervals (CIs) that were calculated by unconditional logistic regression adjusting for age and cigarette smoking (Brinkman index value), unless otherwise mentioned. A p value less than 0.05 was considered to be statistically significant. Statistical computations were carried out by using the statistical software SAS, version 5.1 (SAS Institute, Inc., Cary, NC) or PLINK 1.07 [14].
3. Results
Table 1 shows the association results of eight genetic polymorphisms analyzed in terms of lung cancer risk in a population of 1460 male Japanese smokers. The minor allele frequencies of three SNPs on 3q28 (TP63) were not significantly different between case and control groups (p ≥ 0.186). The SNP rs2736100 on 5p15 (TERT), but not rs401681 (CLPTM1L), was significantly associated with lung cancer risk (odds ratio of 1.27, 95% CI of 1.07 - 1.50, p = 0.00504). In two SNPs on 15q25 (CHARN3- CHRAN5), there was no significant association. The odds ratio for subjects carrying CYP2A6 variants was calculated to be 0.73 (95% CI of 0.56 - 0.90, p = 0.000230), suggesting a decreased risk of tobacco-related lung cancer in subjects with variants of CYP2A6, which had been previously seen in expected phenotype groups estimated from CYP2A6 gene analyses [12].
The subgroup analyses according to histological types of cancer were investigated by classifying the lung cancer cases into two groups, adenocarcinoma cases and squamous cell or small cell carcinoma cases (Table 2). A significantly higher odds ratio of 1.47 was observed for rs2736100 G allele in TERT (95% CI of 1.22 - 1.77, p = 6.40 × 10–5) in the adenocarcinoma subgroup; however, no significant association was observed in the squamous cell or small cell carcinoma subgroup. The SNP rs4488809 in TP63 showed significant association in the adenocarcinoma cases (odds ratio of 1.21, 95% CI of 1.01 - 1.46, p = 0.0422), suggesting roles of TP63 and TERT as possibly susceptible genes for lung adenocarcinoma. In contrast, significantly lower odds ratios of 0.74 (p = 0.00203) and 0.67 (p = 0.000386) were obtained for CYP2A6 both

Table 1. Association of polymorphisms of TP63, TERT-CLPTM1L, CHRNA3-CHRNA5, and CYP2A6 with lung cancer risk.

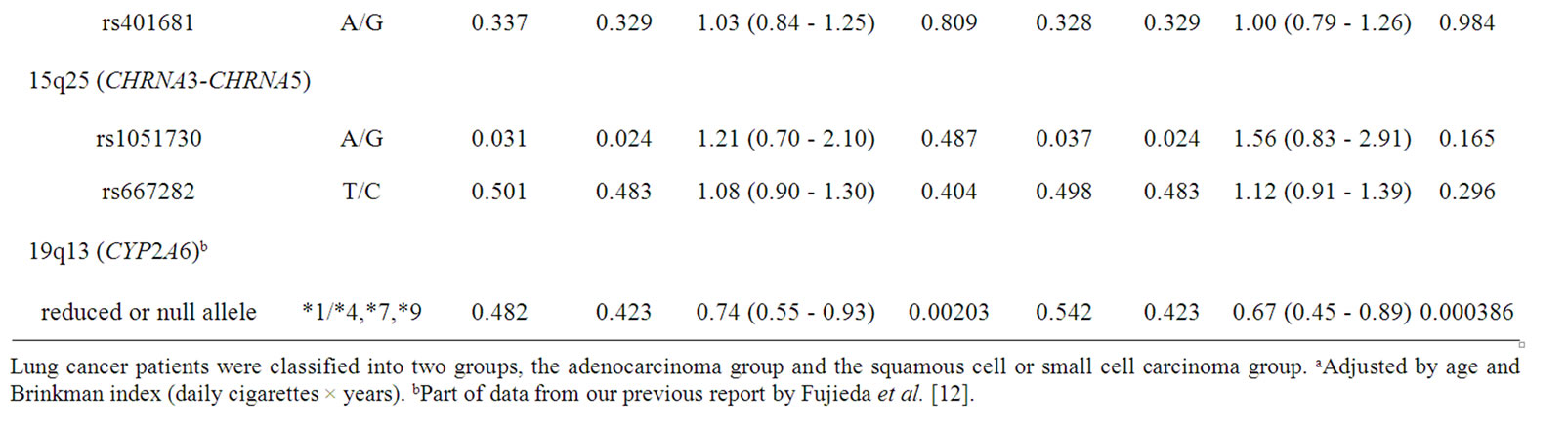
Table 2. Association of polymorphisms of TP63, TERT-CLPTM1L, CHRNA3-CHRNA5, and CYP2A6 with lung cancer risk: subgroup analysis of histological types of lung cancer.
in the adenocarcinoma group and squamous cell and small cell carcinoma group, respectively.
Genetic polymorphisms in the subgroups of smoking status were analyzed (Table 3). In this subgroup analysis, light smokers and heavy smokers were categorized according to the 50th percentile Brinkman index value (690 daily cigarettes × years) among the control subjects. In the light smokers, a significant higher odds ratio of 1.71 was observed for rs2736100 in TERT (95% CI of 1.29 - 2.26, p = 0.000193), whereas no significant association was observed (odds ratio of 1.06, p = 0.558) in the heavy smokers. On the other hand, a significantly lower odds ratio of 0.64 (95% CI of 0.42 - 0.86, p = 6.02 × 10–5) was obtained for CYP2A6 in heavy smokers.
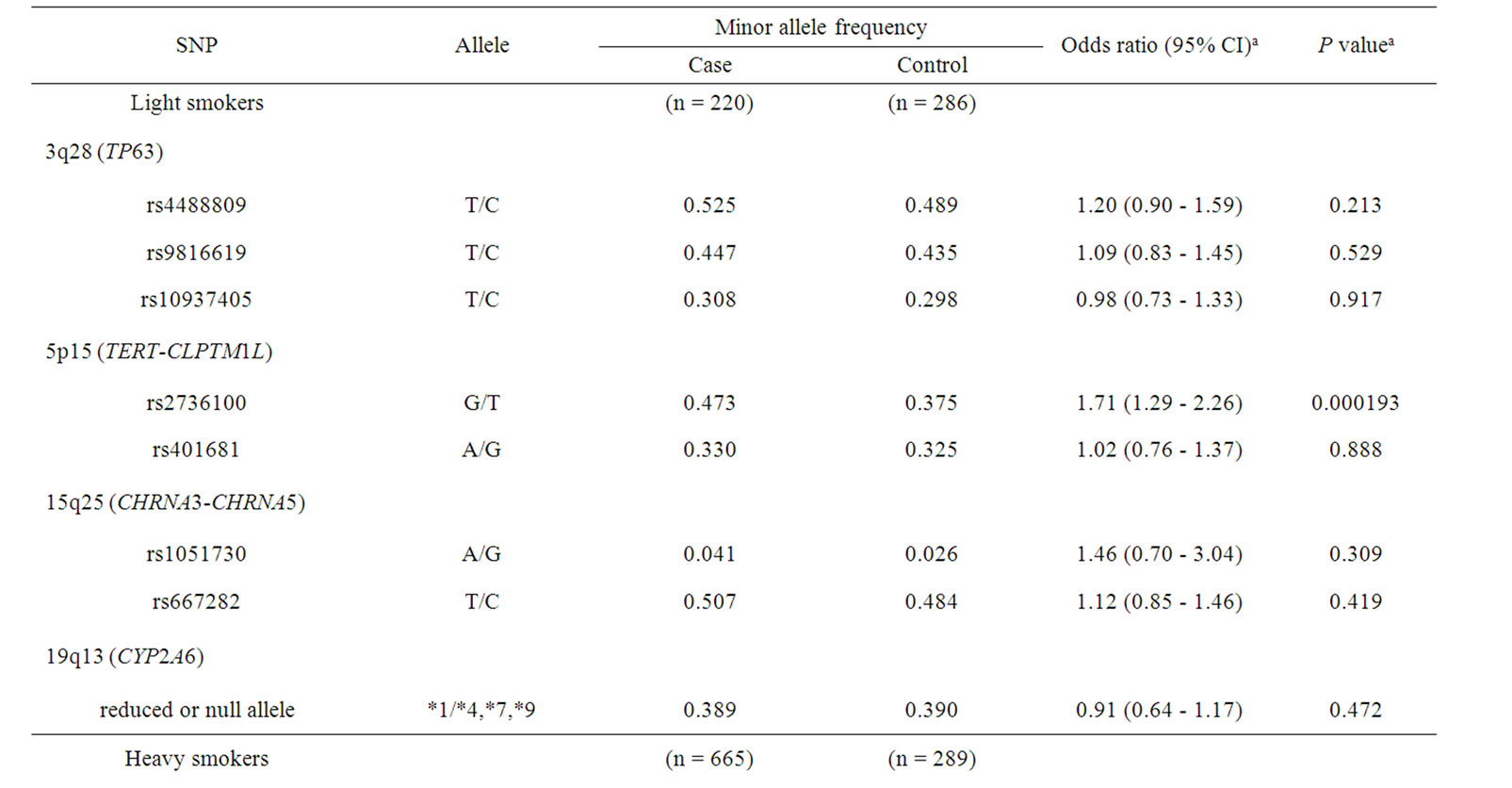

Table 3. Association of polymorphisms of TP63, TERT-CLPTM1L, CHRNA3-CHRNA5, and CYP2A6 with lung cancer risk: subgroup analysis of smoking status.
Consequently, together with the above-mentioned results, the further subgroup analysis was performed (Table 4) to understand different contributions of TERT, TP63, and CYP2A6 polymorphisms to odds ratios in tobacco userelated lung cancer risk. In patients with lung adenocarcinoma, the highest odds ratio of 1.75 (95% CI of 1.29 - 2.38, p = 0.000345) for rs2736100 in TERT was observed in light smokers. The lowest odds ratio of 0.60 (95% CI of 0.34 - 0.86, p = 0.000148) was obtained for CYP2A6 for squamous cell and small cell carcinoma in heavy smokers.
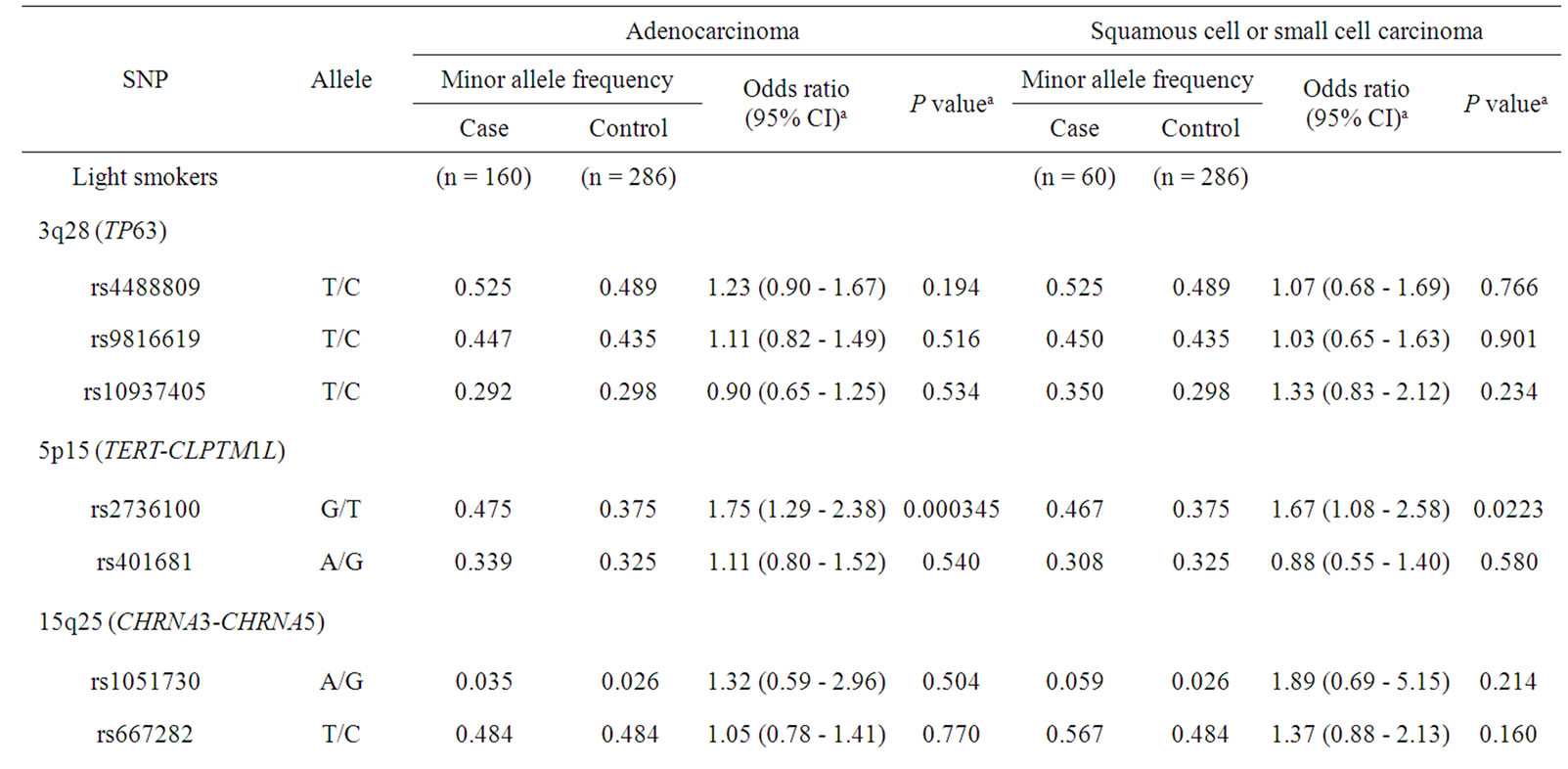

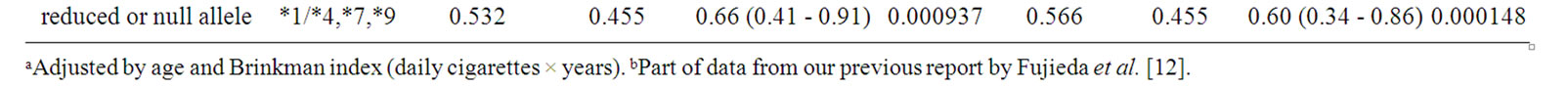
Table 4. Association of polymorphisms of TP63, TERT-CLPTM1L, CHRNA3-CHRNA5, and CYP2A6 with lung cancer risk: subgroup analysis of both histological types of lung cancer and smoking status.
4. Discussion
Recent GWAS have identified lung cancer susceptibility genes such as TERT [7,9], CLPTM1L, CHRNA3-CHRNA5, and TP63 [10]. We performed a replication study of these associations in a population of male Japanese smokers (a total of 1,460 subjects), which were previously studied for lung cancer risk [12]. Miki et al. [10] have identified that TP63 is a significant candidate gene determining individual risk of lung adenocarcinoma in the Japanese and Korean populations [10]. In that report [10], rs4488809 in TP63 showed the strongest association with lung adenocarcinoma risk. In this context, the present study replicated this association of rs4488809 with p = 0.0422 in TP63 in lung adenocarcinoma cases (Table 2). The SNP rs4488809 was associated with only adenocarcinoma risk, but not squamous cell and small cell carcinoma in the present study. Recent report for association results of TP63 in Caucasian populations showed significant association of SNPs in TP63 with squamous cell carcinoma susceptibility [15]. None of three SNPs tested in this study showed significant association for squamous cell carcinoma (p ≥ 0.765, n = 278; data not shown), although the sample size was not large. Further analysis will be interested to clarify the cancer type-specific effects of TP63 polymorphism.
An important contribution of TERT on 5p15 variants to lung cancer has been suggested by multiple research groups [7,10,16]. The rs2736100 in TERT were significantly associated with lung cancer risk (p = 0.00504), as shown in Table 1. In subgroup analysis shown in Table 4, association of this SNP was detected in both adenocarcinoma group and squamous cell or small cell carcinoma group in light smokers, with similar odds ratios of 1.75 (p = 0.000345) and 1.67 (p = 0.0223), respectively. These results suggest that TERT polymorphisms would be associated with non-smoking-related lung cancer outside the context of histological types of cancer. Data reported by Wang et al. [16], demonstrating that TERTCLPTM1L variants showed the strongest association for adenocarcinoma and weak but similar trend for squamous cell carcinoma in never-smokers, supported our preset results. On the other hand, CHARN3-CHRAN5 also located on 15q25 has been reported one of the most import factors identified in GWAS in Caucasian populations [7,9]; however, the association of these variants were not replicated in this study (Table 1). The cause of these differences was probably because of their extremely low frequency in the Asian population, as has been reported by other Japanese group [17]. Wu et al. [18] have reported that different SNPs on CHARN3-CHRAN5 locus, rs2036534C > T, rs667282C > T, rs12910984G > A, and rs6495309T > C, would be associated with increased lung cancer risk in Chinese populations. However, these associations were not replicated in this study (p ≥ 0.196, data not shown). Contribution of SNPs on CHARN3- CHRAN5 to susceptibility of lung cancer may be concluded to be small in the Asian populations.
In an analysis of all the subjects tested, CYP2A6 variant was significantly associated with lung cancer risk, with the lowest p-value of 0.000230 among polymorphisms tested in this study (Table 1). As we previously reported [12], CYP2A6 was suggested to be associated with smoking-dependent lung cancer risk outside the context of histological types of cancer (Table 4). However, in recent reports on GWAS, CYP2A6 polymorphism was not identified as a risk factor of lung cancer. This is partly because the SNPs analyzed in this study (rs5031016 for CYP2A6*7 and rs28399433 for CYP2A6*9) were not included in the platforms of the previous GWAS. Most of GWAS did not detect CNP, although whole-gene deletion type polymorphism of the CYP2A6 (CYP2A6*4) was observed at relatively high frequency in the Asian populations [12]. The ethnic difference in the CYP2A6 allele frequency between Caucasians and Japanese should be also considered in terms of lower frequencies of CYP2A6*9 and rare CYP2A6*4 and never CYP2A6*7 detected in Caucasians than those highly found in the Asians (~15% - 20%) [12]. From these ethnic differences in polymorphic frequencies, the contribution of CYP2A6 polymorphisms in Caucasians might be smaller. Further analysis is needed to evaluate the contribution of CYP2A6 polymorphisms in the studies of large sample size.
Overall, the different roles of TERT, TP63, and CYP2A6 variants were shown in subgroups indicating smoking status and lung cancer types in male Japanese smokers. Taken together the candidate genes identified in the recent GWAS, lung cancer risk in smokers may be accounted for by individual smoking status and multiple gene variations such as TERT, TP63, and CYP2A6 polymorphism. In conclusion, we firstly clarified that the association of TP63 was replicated in the lung adenocarcinoma group of an independent Japanese population. Genetic variation in CYP2A6 may mostly influence susceptibility to heavy tobacco use-related lung cancer risk in male Japanese smokers.
5. Competing Interests
There are non-financial competing interests in this study.
6. Acknowledgements
This work was supported in part by the Ministry of Education, Culture, Sports, Science and Technology of Japan and an SRF Grant for Biomedical Research in Japan. The authors thank Drs. Masaki Fujieda, Hirotoshi DosakaAkita, Yuichi Sawamura, Jun Yokota, Fumihiko Matsumoto, and Norie Murayama for their support.
REFERENCES
- The Tobacco and Genetic Consortium, “Genome-Wide Metaanalyses Identify Multiple Loci Associated with Smoking Behavior,” Nature Genetics, Vol. 42, No. 5, 2010, pp. 441-447. doi:10.1038/ng.571
- J. Z. Liu, F. Tozzi, D. M. Waterworth, S. G. Pillai, P. Muglia, L. Middleton, et al., “Meta-Analysis and Imputation Refines the Association of 15q25 with Smoking Quantity,” Nature Genetics, Vol. 42, No. 5, 2010, pp. 436-440. doi:10.1038/ng.572
- N. L. Saccone, R. C. Culverhouse, T. H. Schwantes-An, D. S. Cannon, X. Chen, S. Cichon, et al., “Multiple Independent Loci at Chromosome 15q25.1 Affect Smoking Quantity: A Meta-Analysis and Comparison with Lung Cancer and COPD,” PLoS Genetics, Vol. 6, No. 8, 2010, p. e1001053. doi:10.1371/journal.pgen.1001053
- E. D. Pleasance, P. J. Stephens, S. O’Meara, D. J. McBride, A. Meynert, D. Jones, et al., “A Small-Cell Lung Cancer Genome with Complex Signatures of Tobacco Exposure,” Nature, Vol. 463, No. 7278, 2010, pp. 184- 190. doi:10.1038/nature08629
- T. Truong, R. J. Hung, C. I. Amos, X. Wu, H. Bickeboller, A. Rosenberger, et al., “Replication of Lung Cancer Susceptibility Loci at Chromosomes 15q25, 5p15, and 6p21: a Pooled Analysis from the International Lung Cancer Consortium,” Journal of the National Cancer Institute, Vol. 102, No. 13, 2010, pp. 959-971. doi:10.1093/jnci/djq178
- Y. Wang, P. Broderick, E. Webb, X. Wu, J. Vijayakrishnan, A. Matakidou, et al., “Common 5p15.33 and 6p21.33 Variants Influence Lung Cancer Risk,” Nature Genetics, Vol. 40, No. 12, 2008, pp. 1407-1409. doi:10.1038/ng.273
- M. T. Landi, N. Chatterjee, K. Yu, L. R. Goldin, A. M. Goldstein, M. Rotunno, et al., “A Genome-Wide Association Study of Lung Cancer Identifies a Region of Chromosome 5p15 Associated with Risk for Adenocarcinoma,” American Journal of Human Genetics, Vol. 85, No. 5, 2009, pp. 679-691. doi:10.1016/j.ajhg.2009.09.012
- C. I. Amos, X. Wu, P. Broderick, I. P. Gorlov, J. Gu, T. Eisen, et al., “Genome-Wide Association Scan of Tag SNPs Identifies a Susceptibility Locus for Lung Cancer at 15q25.1,” Nature Genetics, Vol. 40, No. 6, 2008, pp. 616- 622. doi:10.1038/ng.109
- J. D. McKay, R. J. Hung, V. Gaborieau, P. Boffetta, A. Chabrier, G. Byrnes, et al., “Lung Cancer Susceptibility Locus at 5p15.33,” Nature Genetics, Vol. 40, No. 12, 2008, pp. 1404-1406. doi:10.1038/ng.254
- D. Miki, M. Kubo, A. Takahashi, K. A. Yoon, J. Kim, G. K. Lee, et al., “Variation in TP63 is Associated with Lung Adenocarcinoma Susceptibility in Japanese and Korean populations,” Nature Genetics, 42, No. 10, 2010, pp. 893-896. doi:10.1038/ng.667
- T. Rafnar, P. Sulem, S. N. Stacey, F. Geller, J. Gudmundsson, A. Sigurdsson, et al., “Sequence Variants at the TERT-CLPTM1L Locus Associate with Many Cancer Types,” Nature Genetics, Vol. 41, No. 2, 2009, pp. 221- 227. doi:10.1038/ng.296
- M. Fujieda, H. Yamazaki, T. Saito, K. Kiyotani, M. A. Gyamfi, M. Sakurai, et al., “Evaluation of CYP2A6 Denetic Polymorphisms as Determinants of Smoking Behavior and Tobacco-Related Lung Cancer Risk in Male Japanese Smokers,” Carcinogenesis, Vol. 25, No. 12, 2004, pp. 2451-2458. doi:10.1093/carcin/bgh258
- International HapMap Consortium, “The International HapMap Project,” Nature, Vol. 426, No. 6968, 2003, pp. 789-796. doi:10.1038/nature02168
- J. C. Barrett, B. Fry, J. Maller and M. J. Daly, “Haploview: Analysis and Visualization of LD and Haplotype Maps,” Bioinformatics, Vol. 21, No. 2, 2005, pp. 263-265. doi:10.1093/bioinformatics/bth457
- Y. Wang, P. Broderick, A. Matakidou, J. Vijayakrishnan, T. Eisen and R. S. Houlston, “Variation in TP63 is Associated with Lung Adenocarcinoma in the UK Population,” Cancer Epidemiology, Biomarkers and Prevention, Vol. 20, No. 7, 2011, pp. 1453-1462. doi:10.1158/1055-9965.EPI-11-0042
- Y. Wang, P. Broderick, A. Matakidou, T. Eisen and R. S. Houlston, “Role of 5p15.33 (TERT-CLPTM1L), 6p21.33 and 15q25.1 (CHRNA5-CHRNA3) Variation and Lung Cancer Risk in Never Smokers,” Carcinogenesis, Vol. 31, No. 2, 2010, pp. 234-238. doi:10.1093/carcin/bgp287
- K. Shiraishi, T. Kohno, H. Kunitoh, S. Watanabe, K. Goto, Y. Nishiwaki, et al., “Contribution of Nicotine Acetylcholine Receptor Polymorphisms to Lung Cancer Risk in a Smoking-Independent Manner in the Japanese,” Carcinogenesis, Vol. 30, No. 1, 2009, pp. 65-70. doi:10.1093/carcin/bgn257
- C. Wu, Z. Hu, D. Yu, L. Huang, G. Jin, J. Liang, et al., “Genetic Variants on Chromosome 15q25 Associated with Lung Cancer Risk in Chinese Populations,” Cancer Research, Vol. 69, No. 12, 2009, pp. 5065-5072. doi:10.1158/0008-5472.CAN-09-0081
Abbreviations
CI: confidence interval; CLPTM1L: cleft lip and palate transmembrane protein 1-like; CNP: copy number polymorphism; CYP2A6: cytochrome P450 2A6; GWAS: genome-wide association studies; CHRNA: nicotinic cholinergic receptor α; LD: linkage disequilibrium; SNP: single nucleotide polymorphism; TERT: telomerase reverse transcriptase; and TP63: tumor protein p63.
NOTES
*These should be considered co-first authors.

