Advances in Bioscience and Biotechnology
Vol.3 No.1(2012), Article ID:17202,9 pages DOI:10.4236/abb.2012.31014
Molecular cloning and characterization of two differentially expressed cellulose synthase gene isoforms in Leucaena leucocephala: A pulp yielding tree species
![]()
1Plant Tissue Culture Division, National Chemical Laboratory, Maharashtra, India
2Faculty of Medicine, Adult Cancer Program, Lowy Cancer Research Centre, University of New South Wales, Sydney, Australia
Email: *bm.khan@ncl.res.in
Received 13 October 2011; revised 17 November 2011; accepted 28 December 2011
Keywords: Cellulose Biosynthesis; Cellulose Synthase; Expression Analysis; Leucaena leucocephala; Tree Species
ABSTRACT
Leucaena leucocephala is fast growing leguminous tree species, acclimatized to variety of soil and climatic conditions. It is widely used for pulp production in India. Pulp mainly consists of cellulose, which is a simple polymer of unbranched β-1, 4-linked glucan chains. The polymerization of glucose residues into a β-1, 4-linked backbone is catalysed by the enzyme cellulose synthase (CesA). Here, cDNAs encoding CesA genes from Leucaena were isolated and characterized. The two complete cDNAs of 3.228 kb and 3.222 kb encoding CesA gene from L. leucocephala were designated as Ll-7CesA (FJ871987) and Ll-8CesA (GQ267555) respectively. In-silico studies showed that Ll-7CesA has 95.2% identities and Ll- 8CesA has 95.8% identities with Acacia mangium CesA2. Phylogenetic analysis revealed significant similarity with known dicot CesA genes. The deduced amino acid sequence of both CesA genes contained the conserved D, D, D, QxxRW motif, eight membrane spanning regions and a putative zinc binding domain, which are characteristic of glycosyltransferases. DNA blot analysis suggested, CesA gene to be in multiple copies in Leucaena genome. Semi quantitative and quantitative real-time PCR expression analysis of Ll-7CesA gene showed more expression in stem than leaf and not detected in root where as Ll-8CesA gene was expressed more in stem than leaf and root. Overall Ll-8CesA was expressed in all tested tissues and could be involved in active cellulose biosynthesis.
1. INTRODUCTION
Trees are reservoirs of many economically and biotechnologically significant products. Wood is one such gift of nature, which has diverse applications for mankind. One of the most well known applications of wood is paper and paper products. World paper production was around 330 × 106 tons per annum in the year 2003 and is increasing on yearly basis to a great extent. In India paper and pulp industry uses Bamboo, Eucalyptus and Leucaena as a major source of pulp. Leucaena is fast growing, multipurpose, tropical tree species, acclimatized to variety of soil and climatic conditions and is extensively used in India as a source of pulp [1]. The genus is native of Central America and has a wide range of usage from forage for domestic animals to fuel wood to timber and pulpwood [2].
The major constituents of wood are cellulose, hemicelluloses, and lignin. Pulp is obtained after removal of lignin and hemicelluloses from wood. Pulp consists of cellulose, which contains apparently simple linear chains of β-1, 4-linked glucan residues, but these chains aggregate to form immensely strong microfibrils. The polymerization of glucose residues into a β-1, 4-linked backbone is catalyzed by the enzyme cellulose synthase (CesA), which utilizes UDP-glucose as the substrate [3]. Despite the importance of cellulose, only recently the studies have started to unravel details of its synthesis [4]. Genes encoding plant cellulose synthases were first identified in cellulose-enriched cotton fibres [5]. With the advent of molecular approaches coupled with genome sequence information, 10 CesA genes have been identified in Arabidopsis [6].
The CesA gene has also been isolated from various plant species such as maize [7], rice [8], barley [9], Eucalyptus [10] and aspen [11,12]. Based on mRNA localization, in situ localization and qRT-PCR expression studies CesAs have been classified as primary or secondary wall-related CesAs [10,13,14]. The primary aim of above studies was to characterize the CesA isoform, responsible for cell wall development.
To obtain better quality cellulose, in terms of increased degree of polymerization and crystallinity, with special reference to wood production, it is essential to identify the CesA which is directly involved in cellulose biosynthesis. CesA genes from few tree species have been isolated and characterized [11,12]. Our knowledge about CesA isoforms and its activity in cellulose biosynthesis in tree species is very limited till date. With an aim to meet the increasing demand of high quality wood for paper industry, it was crucial to identify the CesA isoform involved in cellulose biosynthesis from L. leucocephala. Here we report two different cDNA clones for CesA gene from L. leucocephala. Expression pattern of these two genes was also studied which suggests that Ll-8CesA might be more active form of CesA than Ll-7CesA, expressed in all tested tissues.
2. MATERIALS AND METHODS
2.1. Plant Material
Approximately three months old cultured Leucaena plantlets were used for the analysis. To obtain cultured Leucaena plantlet, seeds of L. leucocephala were treated according to the protocol described by Shaik et al. (2009) [15].
2.2. RNA Isolation and cDNA Synthesis
Total RNA was isolated from L. leucocephala tissue samples (Three months old germinated seeds) according to the TRIZOL (Sigma) method. First-strand cDNA synthesis, primed with an oligo (dT)15 primer, was performed with avian myeloblastosis virus reverse transcriptase (AMV RT) according to the manufacturer’s protocol (Promega Corp., Madison, USA). One microgram of total RNA was used to prepare cDNA from stem, leaf and root tissue.
2.3. Amplification of Internal Region and RACE PCR for CesA Gene
A PCR based approach was followed to isolate the internal regions of CesA gene. Known CesA sequences at GenBank database were aligned using Clustal X software and primers were designed from the conserved regions. Four primer sets were designed, CesAF1, CesAF2, CesAR1, CesAR2 (Table 1). These primers were used to amplify internal regions of CesA gene. BLAST result of these amplicons showed a significant similarity to Populus, Betula and Acacia CesA genes. CesA genes from these three species were aligned and two different degenerate primer sets CesFullF1 and CesA3kR (Table 1) were designed. The primers were designed from the start codon and near to stop codon (3’end) of CesA gene. The expected size fragment was sub cloned and sequenced.

Table 1. List of primers.
RACE PCR was attempted to isolate the full length CesA gene. Gene racer RACE kit (Invitrogen) was used according to the manufacturer’s instruction. The information obtained from the amplification of internal regions of CesA gene was used to perform the RACE PCR reaction. In brief, total RNA was treated according to manufacturer’s instruction and cDNA was prepared for 3’ RACE reaction. 3’ RACE PCR fragment was obtained after two step PCR reaction using RaceCesF and RaceCesNF primers (Table 1). The amplified fragment was purified and cloned in pGEM-T Easy cloning vector (Promega, USA). Plasmids were isolated from 10 clones and sequenced.
Sequence analysis of 3’ RACE PCR clones, allowed us to design primers for full length CesA gene. The following primers were designed: CesAFullF2, CesAFullR (Table 1). PCR amplification was performed using above primers and Leucaena cDNA as a template. HiFi taq DNA polymerase (Stratagene, USA) was used for the PCR reaction. The amplified fragment was cloned in pGEM-T Easy cloning vector (Promega, USA) and sequenced.
2.4. Phylogenetic Analysis
CesA deduced amino acid sequences from GenBank database were used to construct a phylogenetic tree. The nucleotide sequences were conceptually translated into amino acid sequence (www.expasy.ch) and evolutionary history was inferred using the Neighbor-Joining method [16]. CesA sequence from prokaryotic system Cronobacter sakazakii CesA (CrsaCes) and Mesotaenium caldariorum (MecaCesA) were chosen as an out-group. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches [17]. The evolutionary distances were computed using the Poisson correction method [18] and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). Phylogenetic analyses were conducted in MEGA4 [19].
2.5. In-Silico Analysis of Two CesA Sequences
Ll-CesA gene sequences were characterized with the Genscan software and homology was verified by database searching at the National Center for Biotechnology Information server using BLAST algorithm (http://www. ncbi.nlm.nih.gov). The deduction of the amino acid sequences, calculation of the theoretical molecular mass and pI, was performed with ExPASy Proteomic tools provided at http://www.expasy.ch/tools/. Global alignment of two nucleotide or amino acid sequences and percentages of identity were calculated using the EMBOSS Pairwise Alignment Algorithms (http://www.ebi. ac.uk/emboss/). Multiple alignments of the amino acid sequences were carried out with the Clustal W1.8 program (http://www.ebi.ac.uk/clustalw/). Conserved domains in amino acid sequence were defined by searching the Pfam protein families database (http://pfam.janelia. org/) and with RPS-BLAST (Search the conserved domain database) on NCBI server. Transmembrane regions were identified using following programme TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
2.6. Semi Quantitative and Quantitative (Real-Time) PCR
Total RNA was extracted individually from shoot, leaf and root of three month old cultured L. leucocephala on MS media. Total RNA (1 μg) was used for making cDNA. Brilliant SYBRGreen QPCR kit (Stratagene, USA) and Stratagene Mx3000P Real-Time PCR machine were used for all reactions. The primer sequences that were designed for Ll-CesAs and 5.8S rRNA is given in Table 1. Semi quantitative PCR conditions are as follows: 5 min at 94˚C, followed by 30 sec at 94˚C, 30 sec at 55˚C and 1 min at 72˚C (25 cycles for 5.8S rRNA and for Ll-CesAs 30, 31 and 32 cycles). Last step was for 5min at 72˚C. qRT-PCR reactions were performed under the following conditions: 2 min at 50˚C, followed by 10 min at 94˚C, 40 cycles of 30 sec at 94˚C, 30 sec at 55˚C, 30 sec at 72˚C. Prior to start of qRT-PCR and semi quantitative PCR reaction, equal quantity of cDNA was normalised for each reaction. Optimal numbers of PCR cycles for visible amplification for each gene were determined in preliminary experiments. Fold of expression was calculated by ΔΔCt method (2–ΔΔCt) [20,21].
2.7. Genomic DNA Isolation, Restriction Enzyme Digestion and DNA Blotting
Good quality genomic DNA was isolated using modified protocol as described by Lodhi et al. (1994) [22]. Four different restriction enzymes namely EcoRI, BamHI, DraI and XhoI were used for identifying as the multigene family of CesA gene. Standard protocol as described by Sambrook et al. (1989) [23], was followed for the experiment. In brief, Digested DNA was electrophoretically separated on 0.8% agarose gel and transfered to a Nylon Hybond-N+ membrane (Amersham, USA). Membrane was prehybridized for 8 h at 62˚C in hybridization buffer (1% BSA; 0.5 M Na2HPO4×2H2O, pH-7.2; 7% SDS and 1 mM EDTA, pH-8.0) (Sambrook et al., 1989). Hybridization was performed at 62˚C for 18 h in same buffer containing radiolabeled probe (ά-P32). Common region of ~800 bp fragment was used to prepare radiolabeled probe using Amersham’s random labelling kit. The blot was washed with moderate wash buffer (2X SSC and 0.1% SDS) before exposing to Storage Phosphor Screen (GE, Healthcare) for 4 h. The Phosphor Screen was scanned at 200 micron resolution in Typhoon Trio+ (GE).
3. RESULTS
3.1. Isolation and Cloning of Two CesA Genes from L. leucocephala
Amplification of internal regions of CesA gene using primer combination CesAF1-CesAR1 and CesAF1-CesAR2 gave amplification of approximately 600 and 800 bp respectively (Table 1, Figure 1). In silico study of 800 bp clones showed 88% identity with Acacia mangium and Betuala platyphylla, and 86% with Populus trichocarpa nucleotide sequences. Above three sequences were aligned and degenerate primers were designed from start codon (CesAFullF1) and near 3’ end (CesA3kR) of the CesA sequence (Table 1, Figure 1). Approximately 3.0 kb amplicon was amplified and cloned in pGEM-T Easy vector and sequenced. Sequence analysis of the clones led us to conclude the possibility of two different CesA genes. Now as 5’ end was known, 3’ RACE was performed to reach the 3’ end of the CesA coding sequence. Sequencing of 10 3’ RACE clones again suggested presence of two CesA isoform in Leucaena and allowed us to design primers (CesAFull F2 and CesAFull R) for amplification of full length CesA coding region (Table 1, Figure 1). An approximately 3.2 kb fragment was amplified, cloned and sequenced. Two different types of cDNA sequences were designated as Ll-7CesA and Ll-8CesA (GenBank Accession No. FJ871987 and GQ267555) respectively. Identities and similarities between both Ll-CesAs were 96.3% and 97.6% respectively.
3.2. Molecular Characterization of Ll-7CesA and Ll-8CesA Gene from L. leucocephala
Like other plant cellulose synthase both Ll7-CesA and Ll-8CesA proteins contains the D, D, D, QxxRW (QVLRW) motifs respectively (Figure 2(a)), which is a characteristics of beta-glycosyltransferases [11,24]. The regions surrounding the D, D, D, QxxRW residues shows similarity between other cellulose synthases and is known as U1, U2, U3 and U4 domains [24,25]. There are eight highly conserved cysteine residues in four pairs of CxxC in N-terminal region of both Ll-CesA proteins (CxxC motif underlined) were observed. These residues form the putative LIM like zinc-binding domain [26]. Like other cellulose synthases these two CesAs from Leucaena also have six transmembrane domains in carboxy terminal region of the protein and two transmembrane domains (T1-T8) in the amino terminal region. The amino terminal region of Ll-CesAs protein also contains SPXX motifs (Figures 2(a) and (b)) that are characteristics of nucleic acid binding proteins [27].
3.3. Ll-CesA: A Multigene Family
To investigate the multigene nature of CesA genes in L. leucocephala, DNA blot analysis was performed using genomic DNA. Good quality genomic DNA was isolated and digested with following restriction enzymes: EcoRI, DraI, XhoI and BamHI. Out of these restriction enzymes Bam HI and DraI cuts inside the Ll-CesA gene. Radioactive probe of approximately 800 bp was prepared from a common region of both Ll-CesAs cDNA sequence. Multiple bands were seen on blot in all four lanes; which clearly suggest that existence of multiple copies of CesA gene (Figure 3). The presence of multiple copies of CesA in Leucaena was in coherence with other known plant species.
3.4. Ll-CesA Genes: Its Similarity to CesAs Involved in Cellulose Biosynthesis
An unrooted phylogenetic tree was drawn to investigate the evolutionary and similarity index of Ll-CesAs with homologous CesA sequences by using deduced amino acid sequences. Ll-7CesA and Ll-8CesA from Leucaena share a 95.8% and 95.2% identity and 97.8% and 96.9% similarity with Acacia; 90.8% and 89.7% identity and 95.3% and 94.2% similarity with Populus and 89.4% and 88.4% identity and 93.3% and 92.4% similarity with Eucalyptus respectively. The tree was divided into four clusters. Cluster I subdivided into two groups. The group I comprises monocotyledonous plant species and group II comprises dicotyledonous plant species. Both Ll-7CesA and Ll-8CesA were grouped with CesAs of dicotyledonous

Figure 1. Schematic representation of designing of primers and expected amplicon sizes: Start sites of primers used for isolation of Ll-CesA genes.
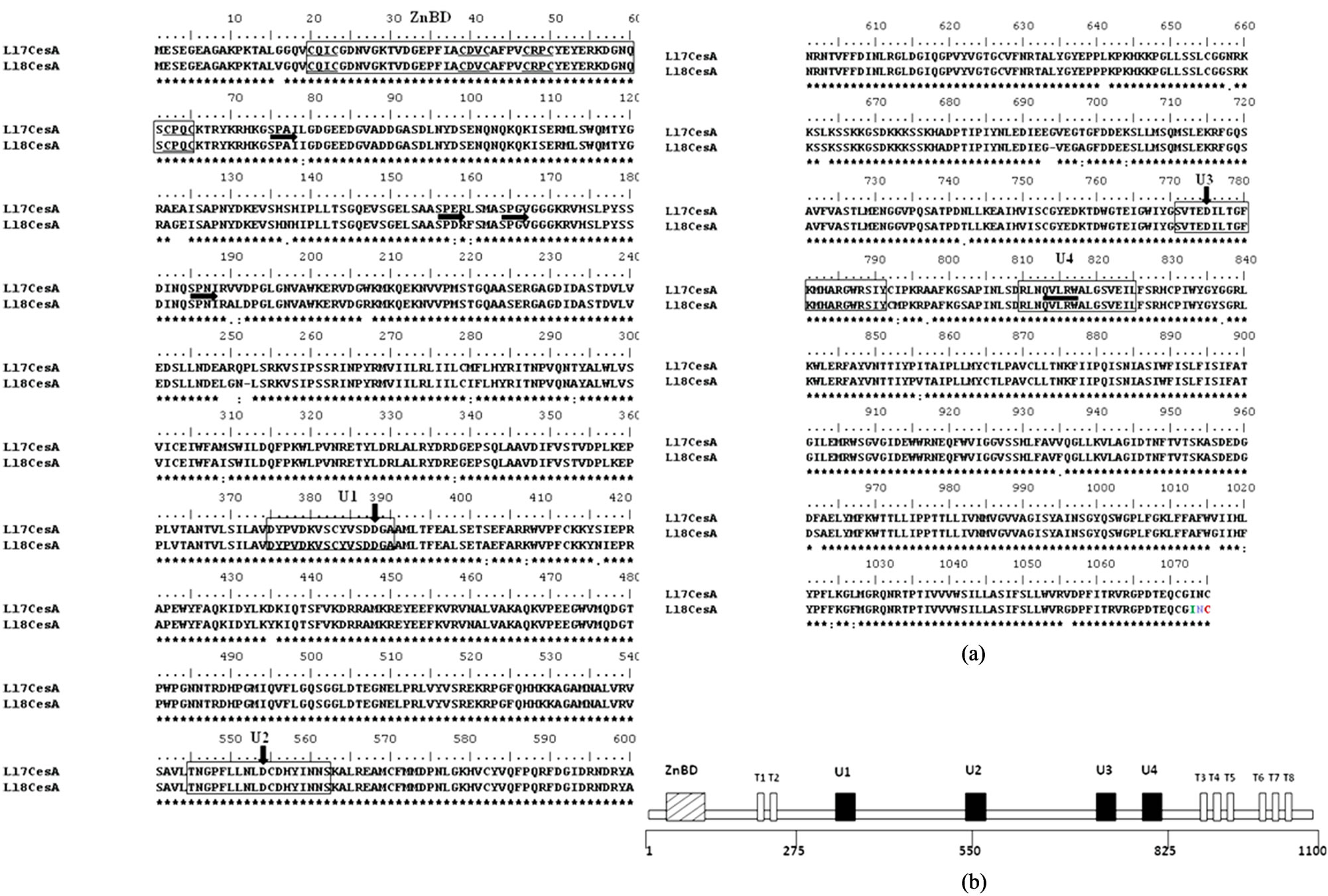
Figure 2. (a) Alignment of deduced amino acid sequences of Ll-7CesA and Ll-8CesA: Alignment was performed by Clustal analysis. Conserved residues are boxed. ZnBD is LIM-like zinc binding domain containing eight highly conserved cysteine residues in four pairs, underlined with thin horizontal lines. SPXX, motifs characteristic of Nucleic Acid Binding Proteins are highlighted with thick horizontal arrows. Ubiquitous (U) sequences U1, U2, U3 and U4 are boxed in thin rectangle. The positions of three aspartic acid (D) and QxxRW motifs are indicated by thick vertical arrows and horizontal line respectively; (b) Schematic representation of protein model and conserved motifs for cellulose synthase (CesA): Eight predicted transmembrane regions are indicated as T1-T8. Conserved ‘U’ motifs originally identified in bacterial cellulose synthase, also present in higher plants cellulose synthase [24].

Figure 3. Genomic DNA blot analysis for Ll-CesA gene in L. leucocephala: Leucaena genomic DNA (20 µg) was digested with restriction enzymes EcoRI (lane 1), DraI (lane 2), XhoI (lane 3) and BamHI (lane 4). Blot was hybridized with approximately 800 bp randomly 32P-labeled probe. Dark arrows represents band in the lanes.
species Eucalyptus, Populus and Acacia in group II of cluster I (Figure 4).
3.5. Tissue Specific Differential Expression of Two Ll-CesA Genes
The expressions of both Ll-CesAs were determined using semi quantitative PCR and quantitative real-time PCR (qRT-PCR). The expression patterns of Ll-CesAs genes were examined in shoots leaves and roots. Semi quantitative PCR suggested that both the genes are differentially expressed. Equal quantity of cDNA was determined by 5.8S rRNA primers (Figure 5, lane A). The expression of Ll-7CesA was observed in stem and leaf only. However expression of Ll-7CesA in shoots was higher than leaves (Figure 5, lane B). In case of Ll- 8CesA, expression level was observed highest in shoot than leaves and roots (Figure 5, lane C). Moreover the expression level of Ll-8CesA was comparatively higher than Ll-7CesA. qRT-PCR was used for determining fold

Figure 4. Evolutionary Relationship of Ll-CesAs among various CesA proteins: Unrooted Neighbor-joining Phylogenetic tree of Ll-CesAs: The phylogenetic tree was constructed on the basis of ClutalX2 multiple sequence alignment of deduced amino acids of Ll-CesAs with 42 CesA protein sequences. For development of tree 500 bootstrap replicates and only branches with 50% or greater support were considered. Species were abbreviated as follows and accession numbers as well as type of CesAs are given in brackets. Arabidopsis thaliana (AtCesA1-NP_194967; AtCesA2-NP_195645; AtCesA4-NP_199216; AtCesA5-NP_196549; AtCesA6-NP_201279; AtCesA9-NP_179768; AtCesA10-NP_180124); Zea mays (ZmCesA1-NP_001104954; ZmCesA2- NP_001105574; ZmCesA4-NP_001105621; ZmCesA5-NP_001104955; ZmCesA6-NP_00110- 4956; ZmCesA7-NP_001104957; ZmCesA8-NP_001104958; ZmCesA9-NP_001104959; ZmCesA10- NP_001105672; ZmCesA11-NP_001105236); Oryza sativa (OsCesA1-NP_001051830; OsCesA2- NP_001051648; OsCesA4-NP_001059162; OsCesA8-NP_001059303); Populus tremula (PtCesA1- AAT09894; PtCesA2-AAT09895; PtCesA3-AAT09897; PtCesA4-AAT09898; PtCesA5-AAL- 23710; PtCesA6-AAP40636); Eucalyptus grandis (EugCesA1-ABY25277; EugCesA2-ABY25278; EugCesA3-ABY25279; EugCesA4-AAY60846; EugCesA5-AAY60847); Triticum aestivum (TaCesA-BAD06322); Mesotaenium caldariorum (MecaCesA-AAT48369); Nicotiana alata (NaCesA-AAK49454); Ze-Zinnia elegans (ZeCesA-BAG06272); Gossypium hirsutum (GhCesA4- AAL37718); Leucaena leucocephala (Ll-7CesA-ACU87559; Ll-8CesA-ACU80553); Acacia mangium (AmCesA2-AAT66941); Pinus radiata (PrCesA2-AAQ63936); Bambusa oldhamii (BoCesA2-AAY43218); Physcomitrella patens (PhpCesA5-ABI78958); Cronobacter sakazakii (CrsaCes-YP_001440220).
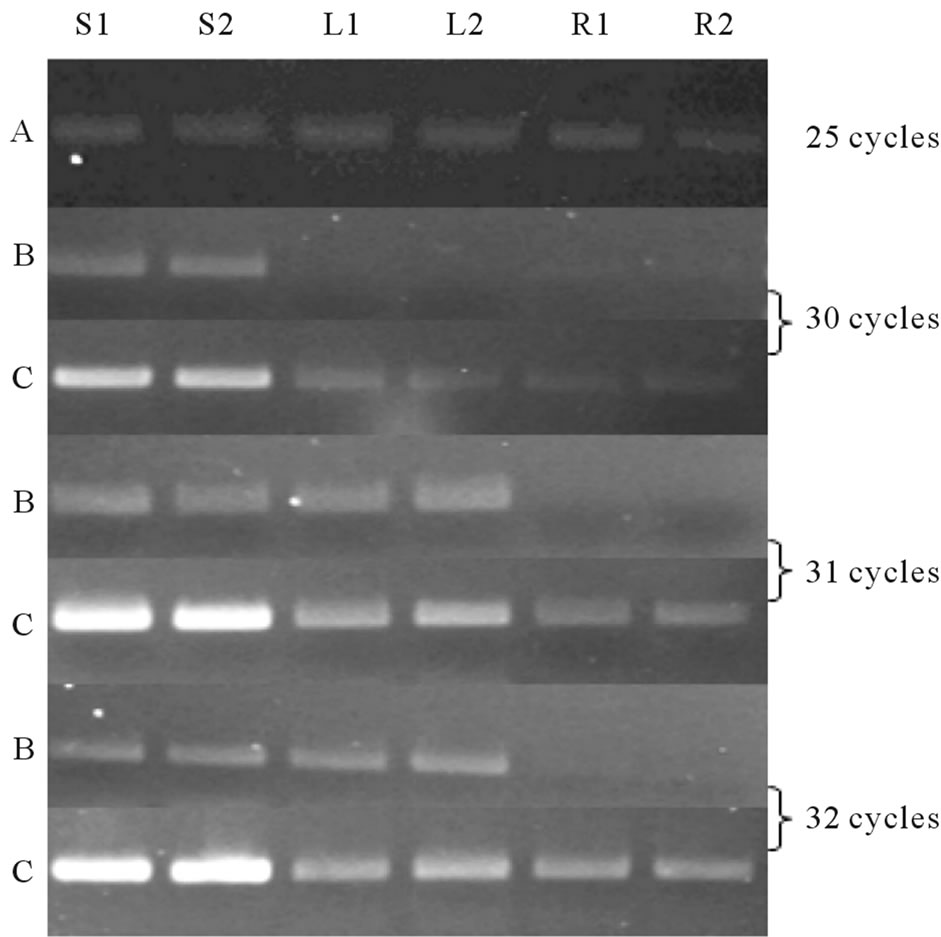
Figure 5. Semi quantitative PCR analysis: PCR was done at three different cycle numbers (30, 31 and 32 cycles) with cDNA from shoot (In duplicate, S1 and S2), Leaf (L1 and L2) and Root (R1 and R2); observed on 2% agarose gel. Lane B and Lane C: Ll-7CesA and Ll-8CesA expression respectively. Lane A: 5.8S rRNA (25 Cycles).
expression of both the Ll-CesAs genes in different tissues. No Ct value was recorded for Ll-7CesA in case of root. When compared to leaves, shoots have 5.35 fold more expression than leaves (Figure 6(a)). However In case of Ll-8CesA, least expression was recorded in roots. The expression was 15.03 fold higher in shoots and 1.51 fold more in leaves when compare to roots (Figure 6(a)). Moreover when expression of individual genes were compared in individual tissues then it was found that Ll-8CesA was 10 fold higher in case of shoot and 5.3 fold higher in case of leave tissue when compared to Ll-7CesA expression (Figure 6(b)).
4. DISCUSSION
It has been established in Arabidopsis, Zea mays and Oryza sativa that CesA is a multigene family [6,8,14,28]. Reports have also described that these genes perform individual function such as development of primary and secondary cell wall. On the contrary, presence of more than two sequences in the genome of Eucalyptus, Populus, Arabidopsis, maize and barley for CesA suggests that CesA genes might be involved in performing redundant functions inside the cell [7,26,29-31]. Compared to primary walls, secondary walls contain higher amounts of cellulose with a higher degree of polymerization and crystallinity [25,32]. Therefore, characterization of CesA gene, primarily responsible for development of secondary cell wall is of paramount importance in gener-
 (a)
(a)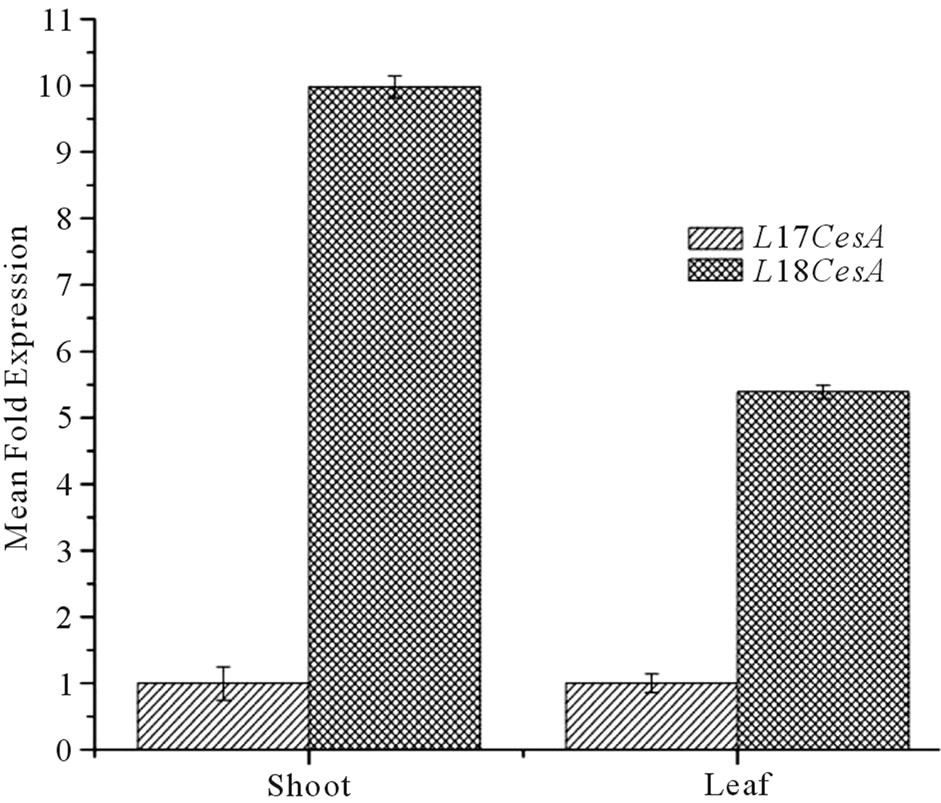 (b)
(b)
Figure 6. (a) Tissue specific normalized expression of Ll-7CesA and Ll-8CesA using qRT-PCR: Light grey shaded histogram represents mean fold expression of Ll-7CesA and dark grey histogram represents mean fold expression of Ll-8CesA; (b) Comparison of mean fold expression level of Ll-7CesA and Ll-8CesA transcripts in individual tissues: Light grey histogram: Ll-7CesA, Dark grey histogram: Ll-7CesA. Error bars are showing standard deviations (SD) for triplicate analysis.
ating elite tree species for paper industry.
With an aim to characterize CesA gene responsible for development of secondary wall, two full length CesA genes of approximately 3.2 kb were isolated from L. leucocephala. They possess 96.3% identity and 97.6% similarity with each other. Size of the cDNA was comparable to known sequence of CesA from other plant species. Primary in-silico analysis showed that both the cDNA sequences are similar to other known homologous CesA sequences. DNA binding, transmembrane, D, D, D, QxxRW, SPXX, CxxC motifs were identified in both the cDNA sequences. All these data classifies them to be member of family glycosyltransferase (Figure 2). Phylogenetic analysis also revealed that both the cDNAs share a significant similarity with CesA sequence from other dicot tree species. Ll-7CesA and Ll-8CesA were grouped with Eucalyptus and Populus which have dual nature and are involved in primary as well as secondary cell wall biosynthesis which suggest that these sequences might be involved in such functions in L. leucocephala (Figure 4).
Multigene nature of CesAs has been well established in various research articles, Our DNA blot results also suggest that CesAs are present as a multigene family in Leucaena. Presence of multiple bands in the blot suggests that the CesA is present in multiple copies in Leucaena genome (Figure 3). Similar results have also been cited in model plants such as Arabidopsis, rice and poplar [8,33-38].
Now the question was where these transcripts are being expressed. We performed a semi quantitative PCR paralleled with qRT-PCR to investigate the presence of Ll-7CesA and Ll-8CesA transcript in various tissues. Root, stem and leaves were investigated for this experiment. The expression level of Ll-8CesA was higher than Ll-7CesA in all tissues tested. However the overall expression of individual transcript was highest in stem than leaves and roots in both the case. Differential expression pattern of CesAs in Leucaena corroborated with the expression pattern of other homologous CesA transcript. When compared to CesAs of Eucalyptus, Arabidopsis and Populus, it was found that the expression pattern of both Ll-CesAs is similar to primary/secondary cell wall synthesizing CesAs. This suggests that they might have dual role and involved in redundant functions in cell wall biosynthesis in Leucaena. As the semi quantitative and qRT-PCR data suggests that the expression of Ll-8CesA was higher than Ll-7CesA in case of all tissues tested, it can be assumed that this isoform might be involved in active cellulose biosynthesis.
5. ACKNOWLEDGEMENTS
This work was supported by grants from Council of Scientific and Industrial Research-New Millennium Initiative for Technology Leadership (CSIR-NMITLI), India. We wish to thank Director, National Chemical Laboratory and Head Plant Tissue Culture Division. RKV and S. Singh thank CSIR-NMITLI for their fellowship grant and SS thanks CSIR-UGC for his fellowship grant.
REFERENCES
- Prasad, J.V.N.S., Korwar, G.R., Rao, K.V., Mandal, U.K., Rao, G.R., Srinivas, I., Venkateswarlu, B., Rao, S.N. and Kulkarni H.D. (2010) Optimum stand density of Leucaena leucocephala for wood production in Andhra Pradesh, Southern India. Biomass and Bioenergy, 35, 1-9.
- Shelton, H.M. and Jones, R.J. (1995) Opportunities and limitations in Leucaena. Proceedings of a workshop held in Bangor, Indonesia. ACIAR Proceedings, Canberra, 16- 23.
- Saxena, I.M. and Malcolm B.J.R. (2005) Cellulose Biosynthesis: Current views and evolving concepts. Annals of Botany, 96, 9-21. doi:10.1093/aob/mci155
- Taylor, N.G. (2008) Cellulose biosynthesis and deposition in higher plants. New Phytologist, 178, 239-252. doi:10.1111/j.1469-8137.2008.02385.x
- Pear, J.R., Kawagoe, Y., Schreckengost, W.E., Delmer, D.P. and Stalker, D.M. (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proceedings of the National Academy of Sciences, 93, 12637-12642. doi:10.1073/pnas.93.22.12637
- Richmond, T.A. and Somerville, C.R. (2000) The cellulose synthase superfamily. Plant Physiology, 124, 495- 498. doi:10.1104/pp.124.2.495
- Appenzeller, L., Doblin, M., Barreiro, R., Wang, H.Y., Niu, X.M., Kollipara, K., Carrigan, L., Tomes, D., Chapman, M. and Dhugga, K.S. (2004) Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose, 11, 287-299. doi:10.1023/B:CELL.0000046417.84715.27
- Tanaka, K., Murata, K., Yamazaki, M., Onosato, K., Miyao, A. and Hirochika, H. (2003) Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiology, 133, 73-83. doi:10.1104/pp.103.022442
- Trethewey, J.A.K. and Harris, P.J. (2002) Location of (1→3), (1→4)-β-D-glucans in vegetative cell walls of barley (Hordeum vulgare) using immunogold labelling. New Phytologist, 154, 347-358. doi:10.1046/j.1469-8137.2002.00383.x
- Lu, S., Li, L., Yi, X., Joshi, C.P. and Chang, V.L. (2008) Differential Expression of Three Eucalyptus secondary cell wall-related cellulose Synthase genes in response to tension stress. Journal of Experimental Botany, 59, 681- 695. doi:10.1093/jxb/erm350
- Joshi, C.P., Bhandari, S., Ranjan, P., Kalluri, U., Liang, S., Fujino, T. and Samuga, A. (2004) Genomics of Cellulose biosynthesis in poplars. New Phytologist, 164, 53-61. doi:10.1111/j.1469-8137.2004.01155.x
- Kalluri, U., and Joshi, C.P. (2003) Isolation and characterization of a new, fulllength cellulose synthase cDNA from developing xylem of aspen trees. Journal of Experimental Botany, 54, 2187-2188. doi:10.1093/jxb/erg232
- Dhugga, K.S. (2001) Building the wall: Genes and enzyme complexes for polysaccharide synthesis. Current Opinion in Plant Biology, 4, 488-493. doi:10.1016/S1369-5266(00)00205-3
- Holland, D., Helentjaris, T., Dhugga, K., XoconostleCazares, B. and Delmer, D.P. (2000) A comparative analysis of the cellulose synthase (CesA) gene family in plants. Plant Physiology, 123, 1313-1323. doi:10.1104/pp.123.4.1313
- Shaik, N.M., Arha, M., Nookaraju, A., Gupta, S.K., Srivastava, S., Yadav, A.K., Kulkarni, P.S., Abhilash, O.U., Vishwakarma, R.K., Singh, S., Tatkare, R., Chinnathambi, K., Rawal, S.K. and Khan B.M. (2009) Improved method of in vitro regeneration in Leucaena leucocephala—A leguminous pulpwood tree species. Physiology and Molecular Biology of Plants, 15, 311-318. doi:10.1007/s12298-009-0035-5
- Saitou, N. and Nei, M. (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406-425.
- Felsenstein, J. (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution, 39, 783-791. doi:10.2307/2408678
- Zuckerkandl, E. and Pauling, L. (1965) Evolutionary divergence and convergence in proteins. In: Bryson, V. and Vogel, H.J., Eds., Evolving Genes and Proteins, Academic Press, New York, 97-166.
- Tamura, K., Dudley, J., Nei, M. and Kumar, S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596-1599. doi:10.1093/molbev/msm092
- Freeman, W.M., Walker, S.J. and Vrana, K.E. (1999) Quantitative RT-PCR: Pitfalls and potential. Biotechniques, 26, 112-125.
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification Real-time RT-PCR. Nucleic Acid Research, 29, 2002-2007. doi:10.1093/nar/29.9.e45
- Lodhi, M.A., Ye, G.N., Weeden, N.F. and Reisch, B.I. (1994) A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Molecular Biology Reporter, 12, 6-13. doi:10.1007/BF02668658
- Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989) Molecular cloning: A laboratory manual. 2nd Edition, Cold Spring Harbor, New York.
- Saxena, I.M., Brown, R.M., Fevre, M., Geremia, R.A. and Henrissat, B. (1995) Multidomain architecture of h-glycosyltransferases: Implications for mechanism of action. Journal of Bacteriology, 177, 1419-1424.
- Delmer, D.P. (1999) Cellulose biosynthesis: Exciting times for a difficult field of study. Annual Review of Plant Physiology and Plant Molecular Biology, 50, 245- 276. doi:10.1146/annurev.arplant.50.1.245
- Arioli, T., Peng, L., Betzner, A.S., Burn, J., Wittke, W., Herth, W., Camilleri, C., Hofte, H., Plazinski, J. and Birch, R. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science, 279, 717-720. doi:10.1126/science.279.5351.717
- Suzuki, M. (1989) SPXX, a frequent sequence motif in gene regulatory proteins. Journal of Molecular Biology, 207, 61-84. doi:10.1016/0022-2836(89)90441-5
- Hazen, S.P., Scott-Craig, J.S. and Walton, J.D. (2002) Cellulose synthase-like genes of rice. Plant Physiology, 128, 336-340. doi:10.1104/pp.010875
- Ranik, M. and Myburg, A.A. (2006) Six new cellulose synthase genes from Eucalyptus are associated with primary and secondary cell wall biosynthesis. Tree Physiology, 26, 545-556. doi:10.1093/treephys/26.5.545
- Samuga, A. and Joshi, C.P. (2002) A new cellulose synthase gene (PtrCesA2) from aspen xylem is orthologous to Arabidopsis AtCesA7 (irx3) gene associated with secondary cell wall synthesis. Gene, 296, 37-44. doi:10.1016/S0378-1119(02)00864-8
- Samuga, A. and Joshi, C.P. (2004) Differential expression patterns of two new primary cell wall-related cellulose synthase cDNAs, PtrCesA6 and PtrCesA7 from aspen trees. Gene, 334, 73-82. doi:10.1016/j.gene.2004.02.057
- Mellerowicz, E.J., Baucher, M., Sundberg, B. and Boerjan, W. (2001) Unravelling cell wall formation in the woody dicot stem. Plant Molecular Biology, 47, 239-274. doi:10.1023/A:1010699919325
- Taylor, N.G., Scheible, W., Cutler, S., Somerville, C.R. and Turner, S.R. (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary wall synthesis. Plant Cell, 11, 769-779.
- Taylor, N.G., Laurie, S. and Turner, S.R. (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell, 12, 2529- 2540.
- Taylor, N.G., Howells, R.M., Hutty, A.K., Vickers, K. and Turner, S.R. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proceedings of the National Academy of Sciences, 100, 1450- 1455. doi:10.1073/pnas.0337628100
- Joshi, C.P. (2003) Molecular biology of cellulose biosynthesis in plants. In: Pandalai, S., Ed., Recent Research Developments in Plant Molecular Biology, Research Signpost Press, Kerala, 19-38.
- Joshi, C.P. (2003) Xylem-specific and tension stress responsive expression of cellulose synthase genes from aspen trees. Applied Biochemistry and Biotechnology, 105, 17-26. doi:10.1385/ABAB:105:1-3:17
- Gardiner, J.C., Taylor, N.G. and Turner, S.R. (2003) Control of cellulose synthase complex localization in developing xylem. Plant Cell, 15, 1740-1748. doi:10.1105/tpc.012815
Abbreviations
AMV-RT: Avian Myeloblastosis Virus Reverse Transcriptase.
cDNA: Complementary DNA.
CesA: Cellulose Synthase A.
Ll-CesA: Leucaena leucocephala Cellulose Synthase A.
RACE: Rapid Amplification of cDNA Ends.
qRT-PCR: Quantitative real-time PCR.
NOTES
*Corresponding author.
#Rishi K. Vishwakarma and Sameer Srivastava contributed equally to the research article.

