Journal of Environmental Protection
Vol. 4 No. 8 (2013) , Article ID: 35697 , 12 pages DOI:10.4236/jep.2013.48100
Aβ-Like Peptide Displayed on Bacteriophage T7 Catalyzes Chromate and Uranyl Reduction
![]()
1Fundamental & Computational Sciences Directorate, Pacific Northwest National Laboratory, Richland, USA; 2Department of Radiological Science, Washington University School of Medicine, St. Louis, USA; 3Xinhua Hospital Affiliated to Shanghai Jiaotong University, Shanghai, China; 4Earth Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, USA.
Email: *hongjunj@mir.wustl.edu
Copyright © 2013 Hongjun Jin et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received November 18th, 2012; revised February 7th, 2013; accepted April 5th, 2013
Keywords: chromate; uranyl; bioremediation; bacteriophage
ABSTRACT
In order to discover genes capable of catalyzing the reductive immobilization of toxic chromate and uranyl ions, we have created a T7 bacteriophage library containing cDNA from environmental microbes (i.e., Geobacter sulfurreducens and Shewanella oneidensis MR-1) that are known to mediate the reduction of chromate and uranyl ions. After three rounds of screening, ten bacteriophage mutants were found to mediate the NADH-dependent reduction of chromate and uranyl ions whose cDNA encodes polypeptide chains ranging 14 to 73 amino acids in length. All identified sequences contain disordered structural motifs similar to the β-amyloid peptide (Aβ) known to promote aggregation and formation of high-affinity metal binding sites. Confirmation of this structural similarity involved phage display of the 42 amino-acid Aβ-peptides that have been found to catalyze the NADH-dependent reduction of both chromate and uranyl ions. Transmission electron microscopy (TEM) and X-ray absorption near edge structure (XANES) measurements confirm that reduced uranium is present on the surface of bacteriophage expressing the Aβ-peptide. The surface-displayed Aβ-like peptide on bacteriophage has the potential to couple naturally occurring electron transfer shuttles present in soils to promote economically viable remediation of contaminated sites containing toxic chromate and uranyl ions.
1. Introduction
Bioremediation of chromate and uranyl toxic metal ions in contaminated soils remains a major technical challenge. Current solutions involve a range of biological approaches that include the targeted feeding of specific anaerobic microbes (e.g. Geobacter sulfurreducens) whose specialized metabolism permits the biosorption, biosequestration, and reductive immobilization of extracellular chromate and uranyl ions that can serve as terminal electron acceptors of cellular respiration [1-7]. Likewise, Shewanella oneidensis MR-1 contains specialized metal reductases located on the outer membrane that selectively associate with metal oxides to mediate their reduction [4,5,8]. The recent structural determination of these metal reductases and the understanding that cellular machinery regulates the targeted assembly of a metal reductase complex on the outer membrane both suggest the possibility of utilizing synthetic biology approaches to enhance these pathways and promote more effective bioremediation at contaminated sites [9,10]. Likewise, the identification of soluble enzymes that are capable of the reductive immobilization of chromate and uranyl ions under both anaerobic and aerobic conditions open a new approach by reengineering of microbes to enhance bioremediation [11,12]. That may be said in that bacteriophages present at natural sites offer a potential means to serve as gene delivery vectors for these synthetic biology applications [13-15]. Additional mechanisms of bioremediation may take advantage of the ability to display catalytic protein moieties on the surface of bacteriophages where the reducing potential of available humic compounds acts as electron shuttles that allow transfer of reducing equivalents to extracellular catalysts [16-19].
Critical to such effective bioremediation is the identification of additional protein catalysts that have optimal function and stability and that remain functional when displayed on bacteriophages. This would enable engineering and applications at contaminated field sites. Accordingly, we have analyzed the genomes of the environmentally relevant microbes Geobacter sulfurreducens and Shewanella oneidensis MR-1 for additional catalytic functionalities that remain functional following their display on T7 bacteriophage. These measurements involved the creation of cDNA genomic libraries that were introduced into the T7 bacteriophage to allow for highthroughput functional screening (Figure S1). Bacteriophage capable of reducing chromate were isolated and sequenced, permitting the identification of ten sequences ranging 14 to 73 amino acids in length that possess a structural similarity to the 42 amino-acid Aβ-peptide, which has been found to catalyze chromate reduction. As these structural motifs can be expressed at high-abundance on bacteriophage, these peptide catalysts have the ability to couple naturally occurring electron shuttles present in soils to promote economic remediation of contaminated sites.
2. Materials and Methods
2.1. Chemicals and Reagents
Agar, agarose, CsCl, K2CrO4, UO2(CH3COOH)2, NADH, 1,5-diphenylcarbazide(DPC), 2-(2-thiazo-lyazo-p-cresol), N-acetyl-N,N,N-trimethyammonium bromide, PEG 8000, and triethanolamine were purchased from Sigma (St. Louis, MO). Restriction enzymes EcoRI and HindII were purchased from Novagen EMD Biosciences (San Diego, CA). LB medium, and LB-M9 medium were purchased from Thermo Fisher Scientific (Cole-Parmer Instrument Company, Vernon Hills, IL). The copper grid (FCF100-cu) was purchased from Electron Microscope Sciences (Hatfield, PA) and negative staining regent Nano-W (Cat# 2018) was purchased from Nanoprobes, Inc. (Yaphank, NY). EpoThin epoxy for section of XANES samples, and other supplies were purchased from Buehler Inc (Lake Bluff, IL).
2.2. T7 Bacteriophage Display
A T7 Select phage display system was purchased from Novagen EMD Biosciences (San Diego, CA) using the BLT5403 E. coli strain, which carries the T7 gene 10 under the T7 ϕ10 promoter. Randomly primed cDNA was synthesized from 4 μg of bacterial mRNA and directionally cloned into the T7 Select 10 - 3 vector arms between EcoRI and Hind III sites using the T7 Select 10 - 3 Orient Express cDNA cloning system (EMD Biosciences). The cloned cDNA was packaged into phage particles, and a serial dilution was made of a small proportion (5%) of the packaged phage to determine the primary size of the library. To make an amplified phage lysate for screening, the remainder (95%) of the phage was used to infect and lyse E. coli strain BLT5403. All the packaging, titration, and preparation of phage lysates were performed according to protocols provided with the T7 Select 10 - 3 cloning system.
2.3. cDNA T7 Phage Library and Engineering Polypeptides on T7 Phage
For the construction of the T7 Selection cDNA display library, mRNA was extracted from the total RNA of Geobacter sulfurreducens (strain ATCC 51573) and Shewanella oneidenesis (MR-1, ATCC 70050) using the RNA extraction RNeasy Mini Kit (Valencia, CA). cDNA was synthesized with the OrientExpress cDNA synthesis kit (Novagen EMD Biosciences) using random hexanucleotide primers and overhang PCR as previously reported [20]. The cDNA fragments between 500 and 2000 bp length were ligated in the T7 Select 10 - 3 vector arms (Novagen EMD Biosciences) between EcoRI and HindIII. For amyloid beta (1-42aa) (Aβ) peptide construction, human amyloid beta (1-42aa), DNA (gi:157878441) was synthesized from DNA 2.0 Inc. (San Diego, CA), and digested with EcoRI and HindIII, and then ligated into the T7 Select 10 - 3 vector arms (Novagen EMD Biosciences) between EcoRI and HindIII.
2.4. T7 Phage Amplification and Purification
Phage titers were determined by mixing serial dilutions of the produced phages with an overnight culture of bacteria containing the original E. coli strain BLT5403. The bacteria and phages were mixed with warm top agar (44˚C), plated on LB agar plates, and incubated at 37˚C). The phages needed for purification were amplified in a 300 ml culture of E. coli BLT5403 by infecting with 4 × 1010 p.f.u. of T7 phage stock. The culture was incubated at 37˚C). The culture was not shaken during the first hour of incubation, and was shaken (200 rpm) for the next 2 - 3 hours of incubation. After incubation period, the cloudy culture became clear, indicating lysis of the bacteria. 5 M NaCl was added to a final concentration of 500 mM and the cell lysate containing the phage particles was separated from the cell debris by centrifugation for 10 min at 8000 g at 4˚C) in a Sorval RC 6 plus Centrifuge (Thermo Scientific). Then 20% (g/ml) final PEG 8000 was added to the supernatant. The lysate-PEG mixture was stored overnight at 4˚C). The next day the phages were precipitated by centrifugation at 8000 g at 4˚C). The pellet was resuspended in 5 mL of 1 M NaCl, 10 mM Tris-HCl, pH 8.0. The resuspended phages were loaded on a centrifuge tube with CsCl gradient (20% - 62% w/v) and purified in a Beckman SW41 rotor centrifuged at 35,000 g for 1 h at 20˚C). The phages layer from the CsCl gradient was collected and dialyzed against PBS and stored at 4˚C) for later use.
2.5. High-Throughput Phage Chromate Reductase Screening
Three rounds of bio-panning (screening) towards efficient chromate reduction were performed. The amplified crude T7 phage lysate bearing the cDNA library was achieved by infection of BLT-5403 with the phage library without PEG precipitation as described above. After titration (see above), 10 µL of the initial pool of mixed T7 phage mutants (2 × 1012 pfu) were serially diluted with M9-LB medium to 2 × 107 pfu/ml. This stock was used as the starting lysate for the first-round screening.
2.6. The First Round of Screening
5 × 104 pfu of the diluted lysate (pool of the mutation library) were dispensed to 96-deep-well microplates (1.5 ml). Equal volumes of freshly cultured BLT-5403 (OD 0.75 - 0.8) were then added to each well and mixed by pipetting the contents up and down several times. The plates were sealed with silicone lids and incubated at 37˚C) overnight while being shaken at 250 rpm. 5 M NaCl was added to the well to give a final concentration of 500 mM. Cell debris in wells was removed by centrifugation at 4000 g in Sorval RC 6 plus Centrifuge (Thermo Scientific) for 40 minutes. In order to screen chromate-reduction responsive wells in plates, an aliquot (80 µl) of the clear lysate from each well was transferred to a new 96-well microplate. The well-position for each lysate in the micro-plates was assigned during transfer. The residual Cr (VI) in the chromate-responsive wells was then detected with the plate reader by measuring the absorbance at 370 nm. M9-LB with and without NADH were used as controls in the place of lysate. The wells that showed significantly low absorbance at 370 nm (<60% of the control) after a reaction period of 16 hours were selected as the chromate-responsive wells.
2.7. The Second Round of Screening
50 µl of lysates from the chromate-responsive wells were dispensed to newly designated wells in a different 96 deep-well microplate (~100 pfu/well) in which 1.5 ml of BLT 5403 cells (OD 0.75) were preloaded. Following incubation at 37˚C) with shaking at 250 rpm, cell lysis was performed as described above. After an addition of 500 mM NaCl and centrifugation at 4000 g for 40 min, the clear phage lysate was transferred to a clean plate and kept at 4˚C until the next experiment. To measure specific chromate reduction activity, kinetic measurements of chromate consumption were carried out by measuring the spectra of absorbance at 340 nm. 50 µl of chromate solution (20 µM) in TBS-buffer (100 mM Tris-HCl, 250 mM NaCl, pH 7.5) were preloaded in assigned wells in a flat-bottom microplate. 30 µl of lysates were then added to the wells. As controls, 0.1% BSA, with TBS-buffer were used in place of lysate. The plates were vortex-mixed briefly and then pre-warmed to 37˚C. 20 µl of 0.5 mM NADH were then added to all wells as a reducing regent. Decreases in the absorption peak for chromate at 340 nm in all wells were measured simultaneously at intervals of 30 seconds over a period of 60 minutes. The negative slope resulting from 5 minutes and 10 minutes was used for the selection of chromate-responsive wells.
2.8. The Third Round of Screening
A single T7 phage plaque was picked from a chromate responsive well of mutations from the second round of screening on a top agar plate. The single plaque was amplified and phage lysates were collected with the 96-well microplate (~1 pfu/well) protocol. Putative positive plaques were further run through the phage amplification and virus particle assembling. These were then retested using the microplate chromate reduction assay and compared to phage particles produced from vector without any insertion. using a colorimetric reaction with 1,5-diphenylcabazide (DPC), reading the absorption at 540 nm in the presence of sulfuric acid as previously reported [11,12], and allowing the plaques to stand at room temperature for 15 minutes. Only those phage mutations that proved responsive to chromate reduction after three rounds of selection were sent for DNA sequencing. To confirm the mutations in the frame of fusion 10b protein of T7 phage, 0.1 µg/ml T7 Select-Up primer from the Novagen T7 Select phage display kit (EMD Biosciences) was used for the sequencing reaction.
2.9. Chromate Detection
Chromate [Cr(VI)] solution has a distinct maximum absorbance at 370 nm (A370, e = 4700 M−1·cm−1) [21]. A standard curve for chromate [Cr(VI)] was established by its absorbance at 370 nm using serially diluted chromate solution. The chromate reduction assay was performed by measuring the steady state of absorbance at 370 nm with a fixed concentration of NADH. The chromate reduction was also confirmed by endpoint method where the colorimetric dye of 1,5-diphenyl carbazide was used as previously reported [12,22]. 50 µL 1 mM chromate solution, 40 µL 1 × 109 pfu/mL T7 phage, and 10 µL of 1 mM NADH were mixed to start the reaction, and after three days of incubation at room temperature (22 ± 1˚C), the reaction solution was mixed with 100 µL of a colorimetric reaction reagent so as to stop the reaction. After 15 min of color development, the A540 of samples was determined using a microplate reader (Molecular Device LLC, Sunnyvale, CA). As previously reported, the Cr(III) enzymatic reduced product produces a specific absorbance at 580 nm [21]. The reduction product Cr(III) in our kinetics studies was measured using the absorbance at 580 nm.
2.10. U(VI) Determination
U(VI) was quantified as described [12] with a little modification for 96-well plate protocol. Samples were collected after incubation experiments and then mixed with colorimetric reaction reagents. The colorimetric reagent used as an 8 mL mixture containing a complex solution consisting of 5 mL of 1 mM 2-(2-thiazolyazo-pcresol), 1 mL of Triton X-100 (0.1%), 1 mL of 1 mM, N-acetyl-N, N, N-trimethyammonium bromide, and 1 mL of 0.1 M triethanolamine buffer (pH 6.5). This method depends on the binding of 2-(2-thiazolyazo-pcresol) to U(VI), a process aided by Triton X-100 and N-acetyl-N, N, N-trimethy-ammonium bromide. 50 µL 1 mM uranium (VI) acetate, 40 µL 1 × 109 pfu /mL T7 phage (A260 = 1.0), and 10 µL of 1 mM NADH were mixed to start the reaction. After three days of incubation at room temperature (22 ± 1˚C), the reaction solution was mixed with 100 µL of a colorimetric reaction reagent to stop the reaction. After 15 min of color development, the A625 of the samples was determined using a microplate reader (Molecular Device LLC, Sunnyvale, CA). For quantifying uranium (VI) concentrations, a standard curve was constructed using uranium acetate (VI) in the presence of 40 µL buffer and 10 µL 1 mM NADH (Sigma), but in the absence of bacteriophage.
2.11. Batch Sorption of U(VI) on Phages
To determine U(VI) sorption characteristics on various phages as a function of aqueous U(VI) concentration, batch sorption experiments were performed in duplicates at room temperature (22˚C ± 1˚C) in 15-mL Ultrafree-MC centrifugal filter units having a microporous membrane with a molecular cutoff value at 50 kDa (Millipore Inc, Billerica, MA). Ultrapure water was used as a background solution. The suspension in the tubes consisted of 5 mL phages (1 × 109 pfu/mL) solution and an aqueous U(VI) concentration ranging from 5.15 × 10−3 to 12.4 × 10−9 M. The mixture samples were equilibrated for 24h on a reciprocal shaker, and then centrifuged for 30 min (3500 rpm) for phase separation using centrifuge filter tubes with a molecular cutoff of 50 k Da. The liquids that passed through the filter were collected from the bottom of the filter tubes for chemical analysis. Uranyl concentration in the filtrates was analyzed using kinetic phosphorescence analyzer (KPA, Chemchek Instruments, Richland, WA). The adsorbed uranyl was calculated from the difference between the total U(VI) added and the measured final aqueous U(VI) concentrations in the suspensions. The mass of the T7 phages is based on a T7 molecular mass of 50 × 106 g/mol [23]. The uranyl [U(VI)] absorption distribution coefficient Kd was measured as previously described [1,24].
2.12. TEM (Transmission Electron Microscope) Experiments
Purified phage particles (1 × 109 pfu/mL) were prepared for TEM in an anaerobic glove bag (Ar/H2, 95:5) using anoxic solutions. 1 mL of phage particle suspension that had been incubated for 3 days with 0.5 mM uranium acetate was centrifuged (2300 × g, 5 min). The phage particles were then mounted on 200 mesh copper grids with formvar support film that was coated with carbon. Unstained sections were examined at 200 kV using JEOL 2010 high-resolution TEM equipped with LaB6 filament, utlizing a resolution of 1.9 Å. Images were digitally collected and analyzed using DigitalMicrograph software (Gatan Inc, Pleasanton, CA). The elemental composition of precipitates was determined using Energy Dispersive X-ray spectroscopy (Oxford Instruments, Fremont, CA) equipped with SiLi detector and analyzed with ISIS software. The selected area diffraction patterns were evaluated using the Desktop Microscopist software (Lacuna Laboratories, Tempe, AZ).
2.13. XANES Experiments
To verify the production of U(IV) on the phage surfaces, selected phage particles were incubated with 0.5 mM uranium acetate solution in water for 3 days at room temperature. Phage viability was confirmed before and after incubation with U(VI) solution in an anaerobic chamber. Phage particles were then purified by PEG8000 precipitation twice, and dried completely after the second precipitation. Phage particles were resuspended thoroughly with 0.5 mL EpoThin epoxy (Buehler Inc, Lake Bluff, IL) in an aluminum tube that was 12.2 mm inner diameter × 12.2 mm length. The tube was sealed at one end using Parafilm (American Can Company, Greenwich, CT), and the samples were dried overnight. Blocks were sectioned anaerobically to 100 mm with a Diatome 45- degree diamond knife using an Ultracut UCT ultramicrotome (Leica. Inc., Bannockburn, IL). These cut solidified samples were then polished and mapped with a stainless steel spatula. Seal thin section samples were sent to the Advanced Photon Source at Argonne National Laboratory in Chicago, IL. Uranium hot spots were chosen for XANES analysis in order to determine of U valence states.
3. Results
3.1. Construction of T7 Phage Library
mRNA was isolated from Geobacter sulfurreducens (ATCC strain 51573) and Shewanella oneidensis MR-1 (ATCC strain 70050), and cDNA was synthesized using random hexanucleotide primers. This was performed essentially as previously described for the construction of genomic libraries [15,25]. The vast majority of synthesized cDNA fragments were between 500 and 2000 base pairs in length, and were apparent as a large smear following agarose gel electrophoresis and DNA staining (Figure S2). This suggests that the cDNA contains randomized pools of genes representative of the genomes of these microbes. These cDNA fragments were ligated into T7 Select 10-3 vector arms between EcoRI and HindIII restriction sites, and incorporated into T7 bacteriophage in essentially the same manner as previously described [15,25].
3.2. High-Throughput Identification of Chromate Reductase Activities
A fast and efficient selection methodology was deployed to screen the functional activity of the phage library based on the ability to reduce chromate. Three rounds of selection involved infection of 1.5 mL of freshly cultured E. coli strain BLT-5403 with phage (500 pfu) using 96 deep-well microplates (Figure S1). The chromate reduction was measured using the absorbance spectrum at 370 nm (ε370 = 4700 M−1·cm−1) [21] in the first round of screening, and measuring the NADH consuming by absorbance at 340 nm in the second round of screening. Finally in the third round, phages from wells to reduce chromate (<60% of control wells) were rescreened following the introduction of infection of E. coli BLT 5403 cells and an isolation of phages based on the NADHdependent reduction of chromate by colorimetric development using 1,5-diphenylcarbazide (Figure S3), as previously described [11,12]. On the basis of these three rounds of screening, there were some individual T7 plaques picked from the chromate responsive wells that continued to show significant chromate and uranyl reduction when compared to the wild-type phage that underwent no insertion (Figure 1). Only these phage mutations were sent for DNA sequencing.
3.3. Chromate Reduction Is Encoded by Small Polypeptides Homologous to Aβ-Like Peptides
DNA sequencing of ten different phage mutants capable of mediating chromate reduction were identified by the presence of short peptides derived from the noncoding DNA of Geobacter (6 clones) or Shewanella (4 clones) (Table 1). These short peptide sequences have motifs predicted to be structurally similar to portions of the 42 amino acid Aβ peptide, which is known to bind and reduce transition metals in the presence of a range of different biological reductants, including NADH [26-30]. Based on the PASTA algorithm, these sequences encode β-pairings known to participate in cross-β-fibrillar aggregates that are important for high-affinity metal binding [26,27,29,30].
3.4. Uranium Precipitation Observed on the Aβ Displayed Bacteriophage T7
To explore the potential of the 42 amino acid Aβ peptide displayed on bacteriophage to reduce chromate and uranyl metals ions, we created a phage construct encoding the 42 amino acid Aβ peptide using synthesized DNA. The phage construct that displays the 42 amino acid Aβ peptide actively reduced both chromate and uranyl metal
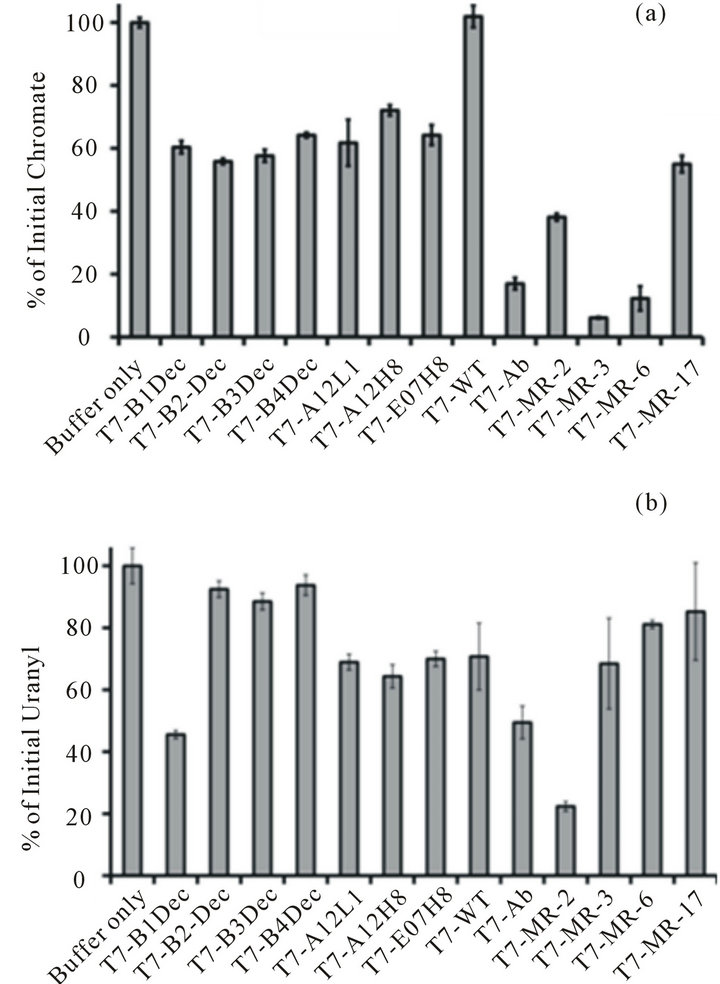
Figure 1. Different T7 phage mutations tested for chromate [Cr(VI)] (a) and uranyl [U(VI)) (b) reduction. The histogram represents the percentage of chromate [Cr(VI)] and uranyl [U(VI)] reduction after incubation with the purified T7 phage mutations with 0.1 mM NADH and 0.5 mM chromate [Cr(VI)] (a) or 0.5 mM uranyl [U(VI)] (b) for three days at RT. All experiments were done triplicate. The average values were presented here, with the error bar representing the standard derivation with three independent experiments.
ions (Figure 1). To further confirm the uranium reduction, the purified phage particles were incubated with chromate or uranyl solution for three days. Through washing and PEG precipitation, the phage particles were then analyzed with TEM. The black uranium precipitation was observed in unstained TEM micrographs for phage expressing the Aβ peptide. By comparison, wildtype phages incubated with uranyl and NADH were not visible prior to staining with Nano-W (Figures 2(a), (b)). There was no obvious chromate precipitation in the unstained TEM micrographs for phages expressing the Aβ peptide, however phage shape was clearly visible in those phage particles after chromate reduction experiments were performed, while wild-type phage incubated with chromate and NADH were not visible without staining. Control staining experiments indicate that similar phage abundances (i.e., 1 × 109 pfu/mL) were present in both wt phage T7 and Aβ-T7 phage samples. Clear phage-like particles with diameters around 50 - 100 nm, were visible in all stained images. Analysis of the precipitation by EDS (electron dispersive spectroscopy) confirmed the presence of large abundance of uranium elements (Figure 2(c), full spectrum in Figure S4). These results indicated that the 42 amino acid Aβ peptide was capable of using an available reducing agent (i.e. NADH) to promote chromate and uranyl reduction. Furthermore, the ability to detect precipitated uranium on the surface of a phage sample after uranium reduction and following repeated washes suggests a strong association of uranyl with the phage particles.
3.5. Uraninite Valence States of Aβ-T7 Phage Samples
U(VI) reduction was quantified colorimetrically using 2-(2-thiazolyazo-p-cresol) (wavelength at 625 nm) in 0.1% Triton X-100,N-acetyl-N,N,N-trimethylammonium bromide (125 nM), and 12.5 mM triethanolamine buffer (pH 6.5) with Aβ-T7-bacteriphage. Valence states of the reduction product precipitants were further quantitatively identified using XANES measurements of Aβ-T7 phage. The purified Aβ-T7 phage particles resulting from uranyl reduction experiments were further thoroughly washed and PEG precipitated. Then the samples were sealed, dried and blocked, and polished prior to shipment to the Advanced Light Source at Argonne National Laboratory for analysis. Uranium hot spots were chosen for XANES analysis for determination of the U valence state (Figure 3(a)). Based on the spectrum of standards (uranyl nitrate and UO2), the uranium U valence state from the sample of the T7-phage expressing Aβ peptide was matched to [U(IV)] standard (Figure 3(b)). This result indicated that uraninite [U(IV))] was reduced from uranyl [U(VI)] by the bacteriophage using the reducing agent (i.e. NADH in our experiments).
3.6. Batch Sorption of U(VI) to Phages
Equilibrium adsorption of uranyl onto the phage surface was measured using a kinetic phosphorescence analyzer. Measured absorption capabilities varied by an order of magnitude, but there was no displayed correlation with the rates of uranyl ion reduction by different phage mutants (Table 2, Figure S5). For example, MR-2 mutant showed very strong uranyl reduction (Figure 1), while the absorption distribution coefficient Kd value was the lowest: 2 × 109 (µmol/kg)/(µmol/L). The wild type T7 phage had Kd value 7 × 109 (µmol/kg)/(µmol/L), while other phage mutations had the Kd value at similar levels of around 109 - 1010 (µmol/kg)/(µmol/L). Indeed, wildtype bacteriophages bind uranyl metal ions with a high absorption distribution coefficient Kd (Table 2, Figure S5). This is a much higher absorption rate in comparison to other nanomaterials [31-33]. The natural affinity of phage for U(VI) is so strong that the uranyl absorbed on the phage surface is proximity to the catalytic peptides, thus the absorption may facilitate uranyl reduction by
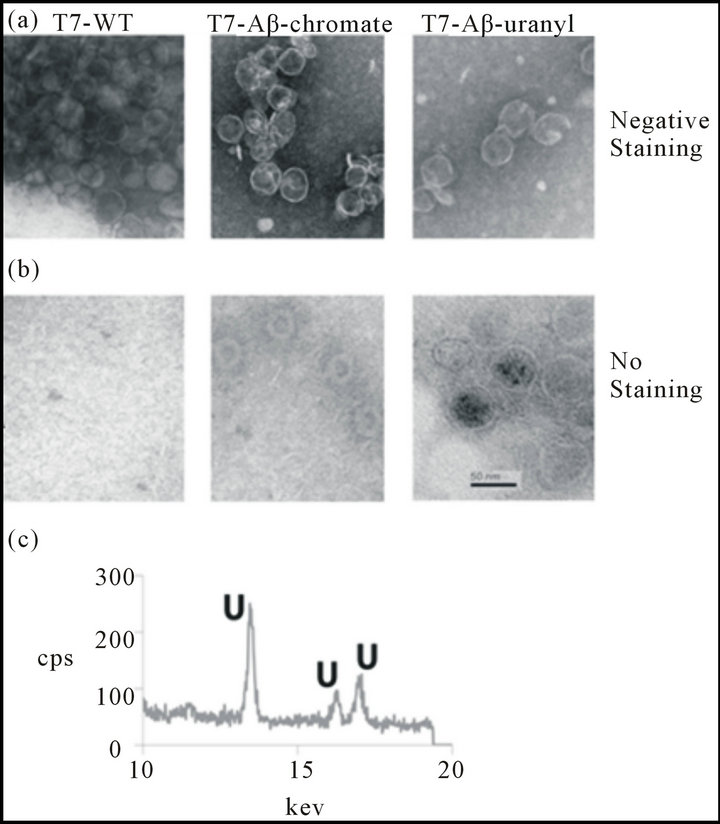
Figure 2. TEM-EDS experiments for wild type and Aβ-T7 phage after chromate and uranyl reduction experiments. (a) Wild type T7 and Aβ-T7 phage particles after chromate reduction experiments (incubated with chromate [Cr((VI)] and NADH three days) were washed, purified and stained with Nano-W for TEM experiments. (b) Wild type T7 and Aβ-T7 phage particles after uranyl reduction experiments (incubated with U(VI) and NADH three days) were washed and purified for TEM without staining. (c) Spectrum of EDS from the dark precipitation on the surface of Aβ-T7 phage. The unique spectrum peaks of EDS were identical for uranium element.
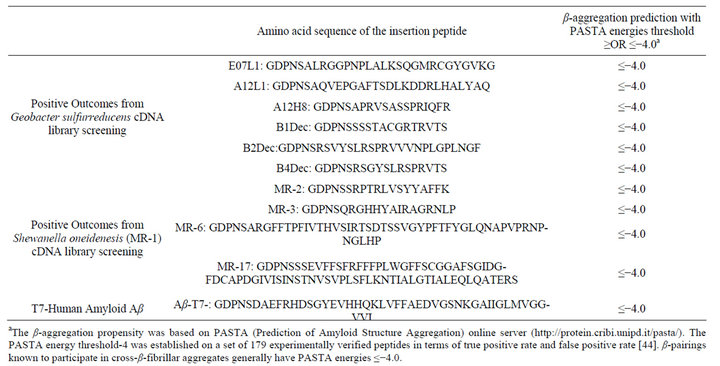
Table 1. Positive mutations bearing β-aggregation peptides.
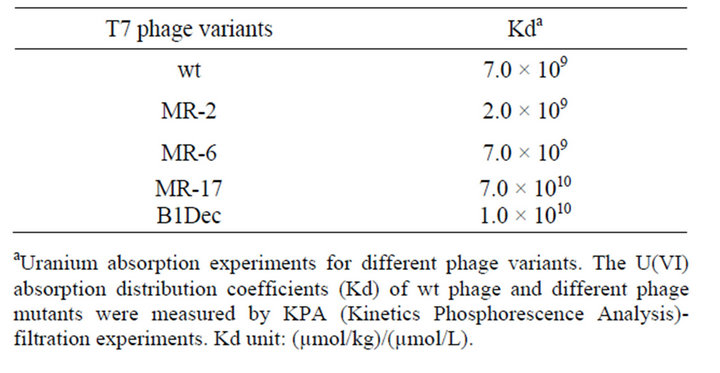
Table 2. Phage variants uranyl absorption distribution coefficients Kda.
aUranium absorption experiments for different phage variants. The U(VI) absorption distribution coefficients (Kd) of wt phage and different phage mutants were measured by KPA (Kinetics Phosphorescence Analysis)- filtration experiments. Kd unit: (µmol/kg)/(µmol/L).
Aβ-like peptides, although the tight binding is insufficient for catalysis. These results are consistent with prior suggestions that the structures of β-amyloid like peptides are required for the catalytic reduction of metal ions in the presence of biological reductants [29,30,34].
4. Discussion
In searching for feasible uranium reductases for bacteriophage mediated bioreduction, a cDNA library of G. sulfurreducens and S. oneidenesis (MR-1) have been successfully subcloned into phage vectors T7Select10-3. High-throughput screening has produced mutations bearing short peptides on the bacteriophage surface. The bacteriophage bearing Aβ showed promising uranyl association. Both the wild type and the mutant bacteriophage strongly absorbed U(VI) (Figure 2 and S5), and the adsorption distribution coefficient Kd reached 109 - 1010 (mmol/kg)/(mmol/L) (Table 2). This is a very strong absorption as compared to other materials.
From the phage cDNA library screening, we did not identify any genes from G. sulfurreducens that have been reported to play an important role in bioremediation, such as hydrogenase enzymes or cytochromes [2,4,6,35,36]. Very unexpectedly small peptides encoded by DNA sequence from non-coding regions of the G. sulfurreducens and S. oneidenesis genomes were shown to reduce chromate and uranium. After DNA sequencing of these short regions, amino acid sequences were used to infer the likely formation of β-fibrillar aggregation with all 10 peptides (Table 1). This β-sheet rich secondary structure may explain why these peptides reduce chromate and uranium. To verify if the phage mediated uranium reduction indeed produce uranium (IV), the purified T7 bacteriophage bearing Aβ peptide were tested in the XANES. The results suggest that U(IV) was produced from the purified phage samples (Figure 3).
In addition to the bacterial reduction and precipitation of metal ions through extracellular cytochromes, a unique metalloprotein mechanism has been found to be associated with neurodegenerative diseases in the human brain [26,30,34,37]. Evidence shows that metal ions such as zinc, copper, and iron can be reduced with amyloid-β proteins and precipitated in the brain because amyloid βpeptide complexes possess a metalloprotein function by binding metal ions either with N-τ of the histidine imidazole ring and main chain amide nitrogens or N-Π atom [27,28,38,39]. Spectrophotometer experiments have established that the amyloid peptide reduces Fe(III) and Cu(II) to Fe(II) and Cu(I) when reducing reagents are available [29,30]. In this study, several small peptides
 (a)
(a) (b)
(b)
Figure 3. XANES analysis U(IV) presented on Aβ-T7 phage surface. XANES analysis was completed with three total replicates. The data shown is the average spectra of those replicates. (a) The uranium map from the mount section solid samples scan; (b) The integration and comparison to standards: uranyl nitrate (U(VI), blue line) and UO2, (U(IV), red line), the U valence state of the uranium on T7-phage bearing Aβ peptide (green line) were identified to be uranium (IV).
were identified from the phage selection system that may form amyloid aggregations. These may also be capable of reducing chromate and uranyl via similar mechanism.
Although the phage selection system did not find any cytochromes or hydrogenase genes, the identified small peptides are nevertheless unique findings relevant to phage-mediated bioremediation. The smaller the size of the peptides that are expressed on the phage surface, the higher the copy number of the peptides that can be expressed [40]. The cytochromes or hydrogenases are relatively large, and normally one copy number can be achieved with the T7 Select 10-3 display system. The copy number for those small peptides can be as high as 50 per phage particle. It is very likely that the higher copy number may facilitate the aggregation of Aβ-like peptides, and contribute to the reduction. In this consideration, small amyloid peptides will have more advantages than larger genes such as cytochromes and hydrogenases for the purpose of bioremediation using bacteriophage.
Extracellular electron acceptors within soils include humic compounds (such as extrocellular quinones) that can serve as electron shuttles to promote environmental restoration through the catalytic reduction of heavy metals and organic pesticides by means of abiotic catalytic mechanisms that do not require living microbes. Extracellular protein complexes of certain bacteria, intracellular NADH dependent reductase, and amyloid peptide from this study suggest that the reduction of opportunistic metals bound to a range of proteins is possible. The bacteriophage mutants that contained short peptides were able to enhance bioreduction of uranium and chromium by a strong absorption effect and a bioreduction property. In this study we only tested the T7 bacteriophage for chromate and uranium remediation in a laboratory scale. Further testing is required to determine whether the engineered bacteriophage can be used for field-deployment. Although many different types and phage strains have been isolated from the environment, and while the application of these bacteriophage for the purposes of gene typing and water quality indication have been well studied [41-43], the application of isolated bacteriophage or engineered bacteirophage for bioremediation has not yet explored. Therefore the study reported here offers initial data and analysis useful to the applications of bacteriophage for uranium bioremedia tions at Cr and U contaminated sites such as those managed by the US Department of Energy.
5. Acknowledgements
We thank Alice Dohnalkova, Charles T. Resch, Chongmin Wang, Brandy N. Gartman, Nikolla P. Qafoku, and Steve Heald for technical support. We thank Stephen Rutherford for proofreading and professional English edition of the manuscript. This research is supported by Pacific Northwest National Laboratory (PNNL) Laboratory Directed Research and Development Funds (LDRD #90001). Part of the research was conducted at the W.R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the US Department of Energy’s Office of Biological and Environmental Research (BER) and located at PNNL. PEL and HJ were also partly supported by BER’s Integrated Field Research Challenge Site (IFRC) at Rifle, CO, a multidisciplinary, multi-institutional project managed by PNNL. Battelle operates PNNL for the US Department of Energy under Contract DE-AC05-76RL01830.
REFERENCES
- C. Liu, J. M. Zachara, L. Zhong, S. M. Heald, Z. Wang, B.-H. Jeon and J. K. Fredrickson, “Microbial Reduction of Intragrain U(VI) in Contaminated Sediment,” Environmental Science & Technology, Vol. 43, No. 13, 2009, pp. 4928-4933. doi:10.1021/es8029208
- A. L. N’Guessan, H. A. Vrionis, C. T. Resch, P. E. Long and D. R. Lovley, “Sustained Removal of Uranium From Contaminated Groundwater Following Stimulation of Dissimilatory Metal Reduction,” Environmental Science & Technology, Vol. 42, No. 8, 2008, pp. 2999-3004. doi:10.1021/es071960p
- J. K. Fredrickson and J. M. Zachara, “Electron Transfer at the Microbe-Mineral Interface: A Grand Challenge in Biogeochemistry,” Geobiology, Vol. 6, No. 3, 2008, pp. 245-253. doi:10.1111/j.1472-4669.2008.00146.x
- E. S. Shelobolina, M. V. Coppi, A. A. Korenevsky, L. N. DiDonato, S. A. Sullivan, H. Konishi, H. Xu, C. Leang, J. E. Butler, B. C. Kim and D. R. Lovley, “Importance of c-Type Cytochromes for U(VI) Reduction by Geobacter Sulfurreducens,” BMC Microbiology, Vol. 7, 2007, p. 16. doi:10.1186/1471-2180-7-16
- J. D. Wall and L. R. Krumholz, “Uranium Reduction,” Annual Review of Microbiology, Vol. 60, 2006, pp. 149- 166. doi:10.1146/annurev.micro.59.030804.121357
- R. B. Payne, D. M. Gentry, B. J. Rapp-Giles, L. Casalot and J. D. Wall, “Uranium Reduction by Desulfovibrio desulfuricans Strain G20 and a Cytochrome c3 Mutant,” Applied and Environmental Microbiology, Vol. 68, No. 6, 2002, pp. 3129-3132. doi:10.1128/AEM.68.6.3129-3132.2002
- D. R. Lovley and E. J. P. Phillips, “Bioremediation of Uranium Contamination with Enzymatic Uranium Reduction,” Environmental Science & Technology, Vol. 26, No. 11, 2002, pp. 2228-2234. doi:10.1021/es00035a023
- M. J. Wilkins, N. C. Verberkmoes, K. H. Williams, S. J. Callister, P. J. Mouser, H. Elifantz, L. N’Guessan A, B. C. Thomas, C. D. Nicora, M. B. Shah, P. Abraham, M. S. Lipton, D. R. Lovley, R. L. Hettich, P. E. Long and J. F. Banfield, “Proteogenomic Monitoring of Geobacter Physiology during Stimulated Uranium Bioremediation,” Applied and Environmental Microbiology, Vol. 75, No. 20, 2009, pp. 6591-6599. doi:10.1128/AEM.01064-09
- Y. Xiong, B. Chen, L. Shi, J. K. Fredrickson, D. J. Bigelow and T. C. Squier, “Targeted Protein Degradation of Outer Membrane Decaheme Cytochrome MtrC Metal Reductase in Shewanella oneidensis MR-1 Measured Using Biarsenical Probe CrAsH-EDT(2),” Biochemistry, Vol. 50, No. 45, 2011, pp. 9738-9751. doi:10.1021/bi200602f
- L. Shi, S. M. Belchik, A. E. Plymale, S. Heald, A. C. Dohnalkova, K. Sybirna, H. Bottin, T. C. Squier, J. M. Zachara and J. K. Fredrickson, “Purification and Characterization of the [NiFe]-Hydrogenase of Shewanella oneidensis MR-1,” Applied and Environmental Microbiology, Vol. 77, No. 16, 2011, pp. 5584-5590. doi:10.1128/AEM.00260-11
- Y. Barak, S. H. Thorne, D. F. Ackerley, S. V. Lynch, C. H. Contag and A. Matin, “New Enzyme for Reductive Cancer Chemotherapy, YieF, and Its Improvement by Directed Evolution,” Molecular Cancer Therapeutics, Vol. 5, No. 1, 2006, pp. 97-103. doi:10.1158/1535-7163.MCT-05-0365
- Y. Barak, D. F. Ackerley, C. J. Dodge, L. Banwari, C. Alex, A. J. Francis and A. Matin, “Analysis of Novel Soluble Chromate and Uranyl Reductases and Generation of an Improved Enzyme by Directed Evolution,” Applied and Environmental Microbiology, Vol. 72, No. 11, 2006, pp. 7074-7082. doi:10.1128/AEM.01334-06
- H. Jin, T. C. Squier and P. E. Long, “Dying for Good: Virus-Bacterium Biofilm Co-Evolution Enhances Environmental Fitness,” Biochemistry Insights, Vol. 5, 2012, pp. 1-9. doi:10.4137/BCI.S9553
- Z. Guo and D. Eisenberg, “The Structure of a FibrilForming Sequence, NNQQNY, in the Context of a Globular Fold,” Protein Science, Vol. 17, No. 9, 2008, pp. 1617-1623. doi:10.1110/ps.036368.108
- L. R. Krumpe, K. M. Schumacher, J. B. McMahon, L. Makowski and T. Mori, “Trinucleotide Cassettes Increase Diversity of T7 Phage-Displayed Peptide Library,” BMC Biotechnology, Vol. 7, 2007, p. 65. doi:10.1186/1472-6750-7-65
- P. Rekha, D. S. Suman Raj, C. Aparna, V. Hima Bindu and Y. Anjaneyulu, “Bioremediation of Contaminated Lake Sediments and Evaluation of Maturity Indicies as Indicators of Compost Stability,” International Journal of Environmental Research and Public Health, Vol. 2, No. 2, 2005, pp. 251-262. doi:10.3390/ijerph2005020008
- J. B. McKinlay and J. G. Zeikus, “Extracellular Iron Reduction Is Mediated in Part by Neutral Red and Hydrogenase in Escherichia coli,” Applied and Environmental Microbiology, Vol. 70, No. 6, 2004, pp. 3467-3474. doi:10.1128/AEM.70.6.3467-3474.2004
- J. Dec, K. Haider and J. M. Bollag, “Release of Substituents from Phenolic Compounds during Oxidative Coupling Reactions,” Chemosphere, Vol. 52, No. 3, 2003, pp. 549-556. doi:10.1016/S0045-6535(03)00236-4
- J. S. Kim, J. W. Park, S. E. Lee and J. E. Kim, “Formation of Bound Residues of 8-Hydroxybentazon by Oxidoreductive Catalysts in Soil,” Journal of Agricultural and Food Chemistry, Vol. 50, No. 12, 2002, pp. 3507- 3511. doi:10.1021/jf011504z
- H. Jin, G. L. Hayes, N. S. Darbha, E. Meyer and P. J. LiWang, “Investigation of CC and CXC Chemokine Quaternary State Mutants,” Biochemical and Biophysical Research Communications, Vol. 338, No. 2, 2005, pp. 987- 999. doi:10.1016/j.bbrc.2005.10.062
- G. J. Puzon, J. N. Petersen, A. G. Roberts, D. M. Kramer and L. Xun, “A Bacterial Flavin Reductase System Reduces Chromate to a Soluble Chromium(III)-NAD(+) Complex,” Biochemical and Biophysical Research Communications, Vol. 294, No. 1, 2002, pp. 76-81. doi:10.1016/S0006-291X(02)00438-2
- Y. H. Kwak, D. S. Lee and H. B. Kim, “Vibrio harveyi Nitroreductase Is Also a Chromate Reductase,” Applied and Environmental Microbiology, Vol. 69, No. 8, 2003, pp. 4390-4395. doi:10.1128/AEM.69.8.4390-4395.2003
- F. C. Bancroft and D. Freifelder, “Molecular Weights of Coliphages and Coliphage DNA. I. Measurement of the Molecular Weight of Bacteriophage T7 by High-Speed Equilibrium Centrifugation,” Journal of Molecular Biology, Vol. 54, No. 3, 1970, pp. 537-546. doi:10.1016/0022-2836(70)90124-5
- A. Kumar, R. K. Singhal, S. Rout, U. Narayanan, R. Karpe and P. M. Ravi, “Adsorption and Kinetic Behavior of Uranium and Thorium in Seawater-Sediment System,” Journal of Radioanalytical and Nuclear Chemistry, Vol. 295, No. 1, 2013, pp. 649-656. doi:10.1007/s10967-012-1825-8
- V. M. Chauthaiwale, A. Therwath and V. V. Deshpande, “Bacteriophage Lambda as a Cloning Vector,” Microbiology and Molecular Biology Reviews, Vol. 56, No. 4, 1992, pp. 577-591.
- A. I. Bush and R. E. Tanzi, “The Galvanization of BetaAmyloid in Alzheimer’s Disease,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 99, No. 11, 2002, pp. 7317-7319. doi:10.1073/pnas.122249699
- K. Suzuki, T. Miura and H. Takeuchi, “Inhibitory Effect of Copper(II) on Zinc(II)-Induced Aggregation of Amyloid Beta-Peptide,” Biochemical and Biophysical Research Communications, Vol. 285, No. 4, 2001, pp. 991- 996. doi:10.1006/bbrc.2001.5263
- T. Miura, K. Suzuki, N. Kohata and H. Takeuchi, “Metal Binding Modes of Alzheimer’s Amyloid Beta-Peptide in Insoluble Aggregates and Soluble Complexes,” Biochemistry, Vol. 39, No. 23, 2000, pp. 7024-7031. doi:10.1021/bi0002479
- X. Huang, M. P. Cuajungco, C. S. Atwood, M. A. Hartshorn, J. D. Tyndall, G. R. Hanson, K. C. Stokes, M. Leopold, G. Multhaup, L. E. Goldstein, R. C. Scarpa, A. J. Saunders, J. Lim, R. D. Moir, C. Glabe, E. F. Bowden, C. L. Masters, D. P. Fairlie, R. E. Tanzi and A. I. Bush, “Cu(II) Potentiation of Alzheimer Abeta Neurotoxicity. Correlation with Cell-Free Hydrogen Peroxide Production and Metal Reduction,” The Journal of Biological Chemistry, Vol. 274, No. 52, 1999, pp. 37111-37116. doi:10.1074/jbc.274.52.37111
- X. Huang, C. S. Atwood, M. A. Hartshorn, G. Multhaup, L. E. Goldstein, R. C. Scarpa, M. P. Cuajungco, D. N. Gray, J. Lim, R. D. Moir, R. E. Tanzi and A. I. Bush, “The A Beta Peptide of Alzheimer’s Disease Directly Produces Hydrogen Peroxide through Metal Ion Reduction,” Biochemistry, Vol. 38, No. 24, 1999, pp. 7609- 7616. doi:10.1021/bi990438f
- H. Zeng, A. Singh, S. Basak, K. U. Ulrich, M. Sahu, P. Biswas, J. G. Catalano and D. E. Giammar, “Nanoscale Size Effects on Uranium(VI) Adsorption to Hematite,” Environmental Science & Technology, Vol. 43, No. 5, 2009, pp. 1373-1378. doi:10.1021/es802334e
- A. Schierz and H. Zanker, “Aqueous Suspensions of Carbon Nanotubes: Surface Oxidation, Colloidal Stability and Uranium Sorption,” Environmental Pollution, Vol. 157, No. 4, 2009, pp. 1088-1094. doi:10.1016/j.envpol.2008.09.045
- T. Arnold, S. Utsunomiya, G. Geipel, R. C. Ewing, N. Baumann and V. Brendler, “Adsorbed U(VI) Surface Species on Muscovite Identified by Laser Fluorescence Spectroscopy and Transmission Electron Microscopy,” Environmental Science & Technology, Vol. 40, No. 15, 2006, pp. 4646-4652. doi:10.1021/es052507l
- F. G. Gervais, D. Xu, G. S. Robertson, J. P. Vaillancourt, Y. Zhu, J. Huang, A. LeBlanc, D. Smith, M. Rigby, M. S. Shearman, E. E. Clarke, H. Zheng, L. H. Van Der Ploeg, S. C. Ruffolo, N. A. Thornberry, S. Xanthoudakis, R. J. Zamboni, S. Roy and D. W. Nicholson, “Involvement of Caspases in Proteolytic Cleavage of Alzheimer’s Amyloid-Beta Precursor Protein and Amyloidogenic A Beta Peptide Formation,” Cell, Vol. 97, No. 3, 1999, pp. 395- 406. doi:10.1016/S0092-8674(00)80748-5
- Y.-J. Chang, P. E. Long, R. Geyer, A. D. Peacock, C. T. Resch, K. Sublette, S. Pfiffner, A. Smithgall, R. T. Anderson, H. A. Vrionis, J. R. Stephen, R. Dayvault, I. Ortiz-Bernad, D. R. Lovley and D. C. White, “Microbial Incorporation of 13C-Labeled Acetate at the Field Scale: Detection of Microbes Responsible for Reduction of U(VI),” Environmental Science & Technology, Vol. 39, No. 23, 2005, pp. 9039-9048. doi:10.1021/es051218u
- I. Bento, V. H. Teixeira, A. M. Baptista, C. M. Soares, P. M. Matias and M. A. Carrondo, “Redox-Bohr and Other Cooperativity Effects in the Nine-Heme Cytochrome C from Desulfovibrio desulfuricans ATCC 27774: Crystallographic and Modeling Studies,” The Journal of Biological Chemistry, Vol. 278, 2003, pp. 36455-36469. doi:10.1074/jbc.M301745200
- C. Opazo, X. Huang, R. A. Cherny, R. D. Moir, A. E. Roher, A. R. White, R. Cappai, C. L. Masters, R. E. Tanzi, N. C. Inestrosa and A. I. Bush, “Metalloenzyme-Like Activity of Alzheimer’s Disease Beta-Amyloid. Cu-Dependent Catalytic Conversion of Dopamine, Cholesterol, and Biological Reducing Agents to Neurotoxic H2O2,” The Journal of Biological Chemistry, Vol. 277, 2002, pp. 40302-40308. doi:10.1074/jbc.M206428200
- X. H. Zong, P. Zhou, Z. Z. Shao, S. M. Chen, X. Chen, B. W. Hu, F. Deng and W. H. Yao, “Effect of pH and Copper(II) on the Conformation Transitions of Silk Fibroin Based on EPR, NMR, and Raman Spectroscopy,” Biochemistry, Vol. 43, 2004, pp. 11932-11941. doi:10.1021/bi049455h
- J. Dong, C. S. Atwood, V. E. Anderson, S. L. Siedlak, M. A. Smith, G. Perry and P. R. Carey, “Metal Binding and Oxidation of Amyloid-Beta within Isolated Senile Plaque Cores: Raman Microscopic Evidence,” Biochemistry, Vol. 42, No. 10, 2003, pp. 2768-2773. doi:10.1021/bi0272151
- S. S. Sidhu, “Engineering M13 for Phage Display,” Biomolecular Engineering, Vol. 18, No. 2, 2001, pp. 57-63. doi:10.1016/S1389-0344(01)00087-9
- S. B. Santos, A. M. Kropinski, P. J. Ceyssens, H. W. Ackermann, A. Villegas, R. Lavigne, V. N. Krylov, C. M. Carvalho, E. C. Ferreira and J. Azeredo, “Genomic and Proteomic Characterization of the Broad-Host-Range Salmonella Phage PVP-SE1: Creation of a New Phage Genus,” Journal of Virology, Vol. 85, No. 21, 2011, pp. 11265-11273. doi:10.1128/JVI.01769-10
- M. Gourmelon, M. P. Caprais, R. Segura, C. Le Mennec, S. Lozach, J. Y. Piriou and A. Rince, “Evaluation of Two Library-Independent Microbial Source Tracking Methods to Identify Sources of Fecal Contamination in French Estuaries,” Applied and Environmental Microbiology, Vol. 73, No. 15, 2007, pp. 4857-4866. doi:10.1128/AEM.03003-06
- D. I. Kurtboke, “Actinophages as Indicators of Actinomycete Taxa in Marine Environments,” Antonie Van Leeuwenhoek, Vol. 87, No. 1, 2005, pp. 19-28. doi:10.1007/s10482-004-6535-y
- A. Trovato, F. Seno and S. C. Tosatto, “The PASTA Server for Protein Aggregation Prediction,” Protein Engineering, Design & Selection, Vol. 20, No. 10, 2007, pp. 521-523. doi:10.1093/protein/gzm042
Appendix
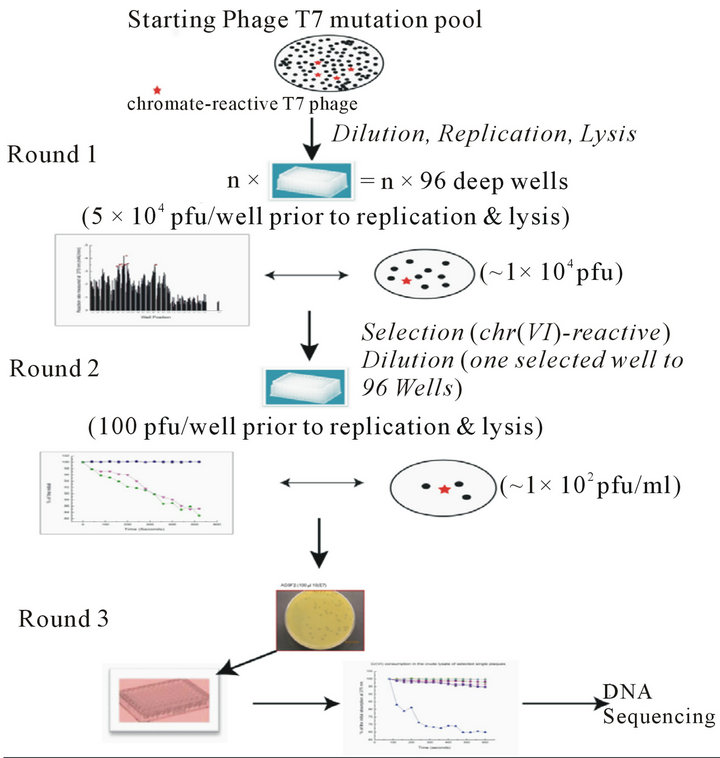
Figure S1.. Digraph of the high through-put phage screening system. The first round was overall dilution (5 × 104 pfu/well) of the pool library into multiple deep 96-well plates and the supernatants were collected from overnight culture and tested on the chromate reduction by measuring the absorbance at 370 nm wavelength. The second round of screening started from the dilation (1 × 102 pfu/well) of the responsive phage mutants pool from first round screening, chromate reduction was measured by A340 (NADH consuming). For the 3rd round of screening, the responsive phage mutants from 2nd round screening were cultured individually (1 pfu/well), and the chromate reduction was measured by DPC method. Only those phage mutants still showing responsive after passed three rounds of screenings were sent for DNA sequencing..
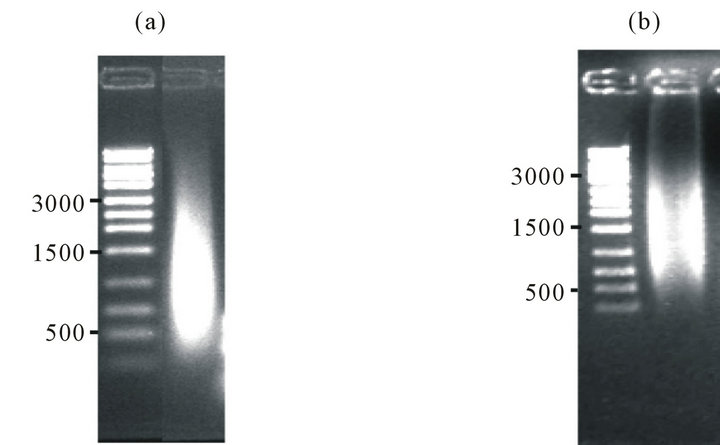
Figure S2.. Validation of the cDNA library. Size distribution of cDNA library constructed using RT-PCR for Shewanella oneidensis MR-1 (ATCC strain 70050) (panel (a)) or Geobacter sulfurreducens (ATCC strain 51573) (panel (b)) (right lanes) in comparison to standards (left lanes) resolved using 0.5% agarose gel electrophoresis.
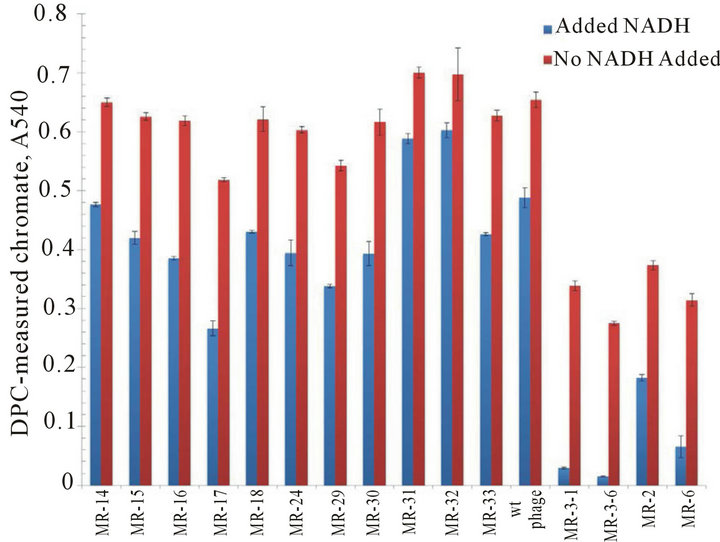
Figure S3.. Positive clones following NADH-dependent increases in chromate reduction for representative phages following third round of screening. Chromate reduction (measured by DPC method) was more significant when the NADH (100 μM) was present (blue bars) compared to without NADH (red bars) during the round 1 screening with 10 non-responsive phage mutations and 5 responsive phage mutations. The wild type phage (no-insertion) is a non-responsive for the chromate reduction.

Figure S4.. The full spectrum of the EDS for the uranium elementary analysis from the T7-Aβ phage experiment, as described in Figure 3.
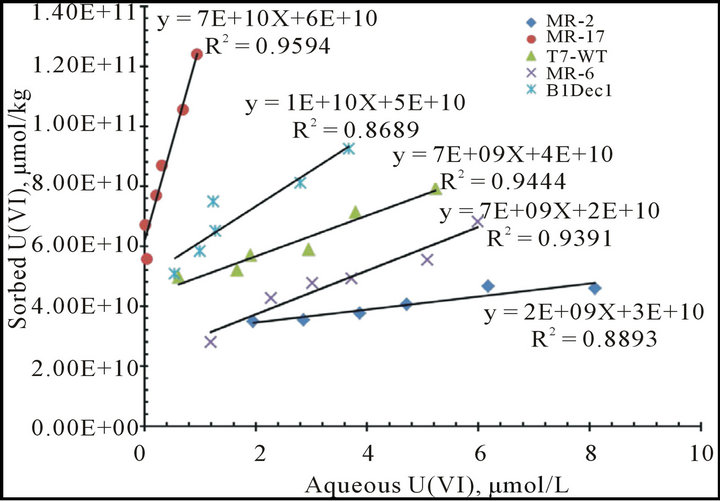
Figure S5.. Batch U(VI) absorption experiments and the absorption distribution coefficient Kd measurements with selected phage variants. Kd were determined from the slope of the liner curve fitting.
NOTES
*Corresponding author.

