Food and Nutrition Sciences
Vol. 2 No. 6 (2011) , Article ID: 6621 , 10 pages DOI:10.4236/fns.2011.26087
Influence of the Backslopping Practice on the Microbial Diversity of the Lactococcus Population in a Model Cheesemaking
![]()
1Isara-Lyon, Agrapôle, Laboratoire de microbiologie alimentaire, Lyon, France; 2INRA-UR342 Technologie et Analyses Laitières, Poligny, France; 3Comité Interprofessionnel du Gruyère de Comté R & D Unit, Poligny, France.
Email: ydemarigny@isara.fr
Received May 10th, 2011; revised July 5th, 2011; accepted July 12th, 2011.
Keywords: backslopping, natural whey starter, model cheesemaking, Lactococcus
ABSTRACT
The objective of this work was to study the microbial diversity of natural whey starters (NWS), with a special focus on Lactococcus strains, during 10 successive days of backslop-ping. Each day, whey samples were enumerated for their Lactococcus, Enterococcus, Leuconostoc and Lactobacillus levels. The diversity and dynamics of the Lactococcus population in whey were investigated using phenotypic methods such as acidifying aptitude and enzymatic activities. Molecular approach using REP-PCR, ERIC-PCR, plamid profiles, phages and prophages research and PFGE was also applied. Lactococci were the main population in whey. Strains from raw milk were sub-dominant. Based on PFGE and phenotypic results lactococci in whey displayed a more heterogeneous phenotype and pulsotype which may reflect greater variations than previously observed within starter. No phages and prophages were spotted on. Plasmids did not seem to be exchanged from strain to strain. The backslopping practice seemed to allow the strains of the starter to rapidly acquire a specificity of their own. The changes observed presumed a slow adapta-tion of the strains to the “backslopping environment”. The study of NWS diversity constitutes an important step for the comprehension of acidification defects that recurr-ently occur in cheesemaking technologies using the backslopping practice.
1. Introduction
In cheesemaking technologies, the use of natural whey starters (NWS) is inherited from the ancient backslopping practice. It is still run through to process traditional farm-house goat cheeses—Rocamadour, Picodon—or some Swiss-type and hard cooked cheeses such as Parmigiano Reggiano and Comté. NWS are complex ecosystems. They are composed of a great number of lactic acid bacteria (LAB) split between several genera, species, subspecies and strains. The LAB originates mainly from previous cheese making operations but also from the plant environment and the raw milk microflora [1]. The use of NWS brings many technological advantages to the cheese. LAB contribute to the acidification and the formation of the curd. They also release numerous enzymes in the cheese core, favouring ripening and the development of desired organoleptic features [2,3]. Moreover the LAB metabolism is known to influence the cheese structure—production of exopolysaccharides, proteolysis—or its biopreservation—production of antimicrobial substances [4]. NWS are also depicted as phage infection resistant ecosystems [5].
It is difficult to cultivate NWS over several weeks. Cheese makers using backslopping have frequently to cope with acidification defects. This situation is gotten worse by the scarce number of published data on NWS. This is probably due to the complexity of NWS and to the fact that traditional scientific methods are not entirely appropriate to study these ecosystems [6]. Thus, it is necessary to acquire knowledge about NWS diversity in order to maintain the backslopping practice.
Recently, a model of cheesemaking based on the backslopping practice has been elaborated at the Isara-Lyon food microbiology laboratory. Based on the statistical analysis of the physico-chemical and microbial data, this model has proven to be efficient for studying the backslopping practice [7]. In this article, the microbial results collected in the 10 successive whey starters are presented and hypotheses are advanced to explain the microbial dynamic and diversity, with a special focus on Lactococcus.
2. Materials and Methods
2.1. Model Cheesemaking
“Tomme de Savoie” type cheeses were made following the 10-day model cheesemaking technology as previously described [7]. The principle of this model cheesemaking was as follows. First day, milk was inoculated with the starter (2%, v/v) made of equal ratios of three characterized lactic acid bacteria (LAB) strains—two Lactococcus lactis subsp. lactis (LL1 and LL2) and one L. lactis subsp. cremoris (LC). During the nine following days, the whey was used from one day to the other as NWS. The milk used was pasteurized and re-seeded with four identified LAB strains from the genera Lactococcus (L. lactis subsp. lactis), Enterococcus, Lactobacillus and Leuconostoc to mimic the contribution of the raw milk flora. These strains were added to a maximum final level of 4 log(cfu)/mL. The pasteurized milk seeded with these known raw milk LAB will be later designated in the text as “Re-seeded Model Milk” (RMM). All the strains added to the raw milks had been previously identified and characterized [7,8].
Each day, whey samples were collected for further microbiological analyses.
2.2. Enumeration and Lactococci Isolation
In the 10 whey samples, lactococci, enterococci, lactobacilli and leuconostocs were checked respectively on M17 agar (30˚C, 24 h, aerobiosis, Biokar diagnostics, 60,000 Beauvais, France), BEA agar (37˚C, 48 h, aerobiosis, Biokar diagnostics), FH agar [9] and MSE agar [10]. After enumeration, 16 to 20 isolates per whey were collected on M17 agar, purified on the same medium and stored at −80˚C. A total of 184 presumed lactococci isolates was collected in the 10 whey samples.
2.3. Phenotypic Characterisation, Acidification Aptitude and Enzymatic Activities of Lactococci Isolates
Salt resistance (40 g/L NaCl), growth at 15˚C and 37˚C, sugar fermentations and arginine dihydrolase capability were studied as described by Demarigny [11].
Isolates were all analysed for their acidification ability in milk according to Demarigny et al. [12]. The acidification aptitude of the strains (dpH) is expressed in pH units. The results are reported as mean values expressed by all the strains isolated each day.
Some lactococci were tested for several enzymatic activities by means of APIZYM kit (Biomérieux, 69280 Marcy l’Etoile, France).
2.4. DNA Analysis
2.4.1. Extraction of Total DNA
From a single culture incubated at 30˚C for 24 h, total DNA was extracted by using the Nucleospin tissue kit (Macheray-Nagel, 67722 Hoerdt, France).
2.4.2. PCR Amplifications, Gel Electrophoresis and Computer Analyses
Specific fragments of the histidine operon were recovred using primers His1 and His2 (Sigma-Aldrich, Illkirch, France) [13]. Inter-Repetitive Extragenic Palindromic sequences were amplified by means of two 18-mer primers, Rep1R-Dt and Rep2-D (Sigma), as previously described by Versalovic et al. [14]. PCR amplifications and gel electrophoresis were performed following the procedure of Dalmasso et al. [8]. The band patterns were normalized and processed using GelCompar 3.1 software (Applied Maths, Kortrijk, Belgium) as previously described by Demarigny et al. [12]. To ensure reproducible REP-PCR results, total DNA from two strains was prepared independently six times and essayed by REP-PCR. All profiles of a strain had similarity coefficient higher or equivalent to 80%. Primer sets ERIC1R/ERIC2 were also used for ERIC-PCR amplifications as previously described by Depouilly et al. [15].
2.4.3. Pulsed Field Gel Electrophoresis (PFGE) and Computer Analyses
Lactococcus strains were grown in M17 broth (Biokar diagnostics) supplemented with 0.5 mol·l−1 sucrose. The preparation of genomic DNA in agarose plugs was performed as previously described by Bouton et al. [16]. Genomic DNA digestion was carried out with 25 U restriction endonuclease SmaI (Sigma) for 4h at 25˚C, in the buffer indicated by the supplier. Samples were electrophoresed through 10 g·l−1 agarose (Sigma) with TBE buffer (Sigma) according to the conditions described by Murchan et al. [17]. Electrophoresis were performed in a CHEF-DR® III electrophoresis cell (Biorad). Staphylococcus NCTC 8325 DNA fragments obtained after digestion with SmaI were used as size standard.
DNA fragments were visualized by UV illumination (312 nm) after ethidium bromide staining and photographed.
2.4.4. Plasmid Extraction and Computer Analyses
Plasmids were extracted from cells following the methodology developed by O’Sullivan and Klaenha-mmer [18]. Gel electrophoresis was performed in an agarose gel (0.7%) under the following conditions: 100 V for 4 h in Tris acetate EDTA buffer. Staining and analyses were then performed as indicated for PCR fragments. According to this technique, the size of the plasmids recovered did not exceed 35 kb.
2.5. Phage and Prophage Research
The presence of phages in the wheys was tested by means of the technique developed by Moineau et al. [19]. This methodology is aimed at the detection of the three main types of phages found in dairy products, C2, P335 and 936.
According to Brüssow [20], in dairy lactic acid bacteria, prophages belong mainly to the P335 type. The PCR methodology applied to detect P335 prophages was the same as described by Labri and Moineau [21], with primers: P335A 5’-GAAGCTAGGCGAATCAGTAA-3’ and P335B 5’-GATTGCCAT-TTGCGCTCTGA-3’. The presence of the P335 sequence in the bacterial genome was checked by the appearance of a 196 kb fragment.
2.6. Statistical Analysis
Lactococci acidification aptitudes were compared performing the dot box methodology. ANOVA, NewmanKeuls’ test and correlation were made with the STATITCF software (5th version, Institut Techniques des Céréales et des Fourrages, 1995).
3. Results
3.1. Enterococci, Leuconostocs and Lactobacilli
Enumeration results are presented on Figure 1. No leuconostocs were counted in the whey samples. When they were detected, enterococci levels ranged from 1 to 4 log(cfu)·ml−1. They were inferior to the detection level—
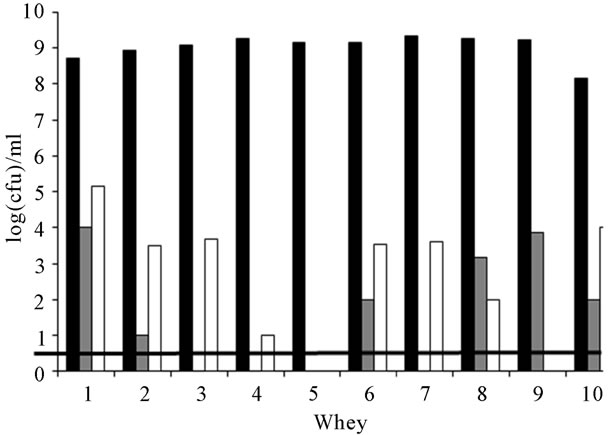
Figure 1. Cellular levels of lactococci (■), enterococci (■) and lactobacilli (□) in the 10 successive whey starter samples. The microflora detection level (▬) was 1 log(cfu)·ml−1.
1 log(cfu)·ml−1—in wheys 3, 4, 5 and 7. No significant correlation was observed between the enterococci from RMM and their presence in wheys (r = 0.25). Lactobacilli were also counted at low levels in whey. Their levels never exceeded 5.2 log(cfu)·ml−1. A weak correlation was observed between the lactobacilli levels in RMM and whey (r = 0.52).
3.2. Enumeration Results and Phenotypic Identification of Lactococci Strains
Lactococci prevailed on the other populations in whey. Their levels ranged from 8.15 to 9.32 log(cfu)·ml−1 as observed on Figure 1. M17 medium being not a specific medium, it implied the verification of the taxonomic status of the enumerated colonies. A total of 184 colonies was then isolated and characterized, that is 16 to 20 isolates from day to day—around 30% of the colonies counted on a Petri dish. All the isolates were assigned to the Lactococcus genus. More precisely they displayed the phenotypic features of the subspecies L. lactis subsp. lactis. For example, they showed positive arginine dihydrolase capability and they were able to grow in a 40 g·l−1 NaCl culture broth. They were also able to use both maltose and ribose.
3.3. Genotypic Diversity of the Lactococcus Population in Whey
To evaluate the diversity of the Lactococcus population in whey, the 184 isolates were analyzed by REP-PCR (Figure 2). No RMM lactococci were isolated, meaning that this population was only subdominant. The limit of detection for RMM lactococci was estimated by dividing the number of colonies sampled by the total count of colonies on the medium from which the isolates were recovered. It was obtained that the maximum level of the RMM population did not exceed a ratio of 1:7 of the total number. Besides, the majority of isolates (whey 2 to whey 10) exhibited a profile identical to the LL1 and LL2 starter strain (cluster 1). It is noteworthy that REPPCR did not allow separating strains LL1 and LL2, although they displayed different phenotypic features—in particular, acidification activity. Fifteen strains belonging to the cluster 1 were also tested by ERIC-PCR. They all displayed similar profiles even if some slight differences are visible, their similarity coefficient being higher or equivalent to 80% (Figure 3). It is interesting to note that the strains isolated from the first whey displayed a different REP profile.
Twenty isolates covering the 9 REP-PCR clusters were randomly tested for the presence of a polymorphism in the histidine operon. It was observed that some of them such as strains L5 and L12, gave a band 200 pb larger
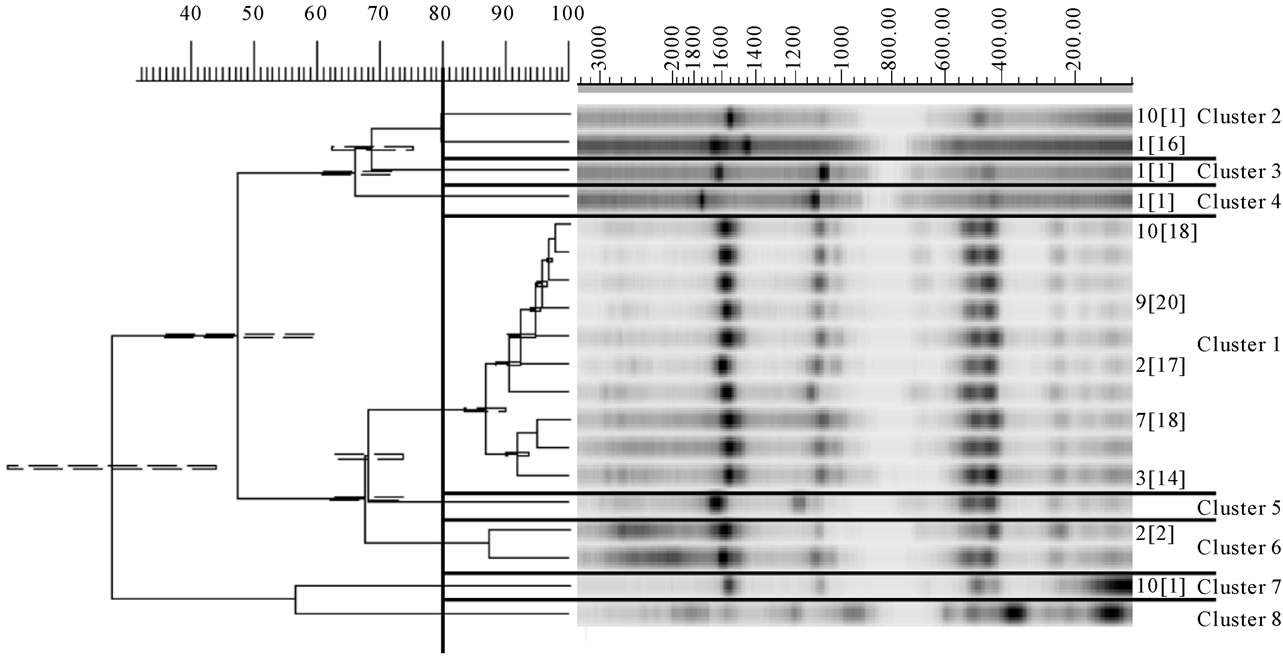
Figure 2. Dendrogram drawn by UPGMA of correlation value of normalized REP-PCR patterns from lactococci obtained with primers Rep1R-Dt and Rep2-D. Each pattern is identified by a whey number indicating the strain origin and by a number between brackets refering to the number of strains which displayed this profile. Nine clusters from 1 to 9 are defined at a coefficient of similarity of 80%. LL: L. lactis subsp. lactis; LC: L. lactis subsp. cremoris; RMM: lactococci from re-seeded model milks.
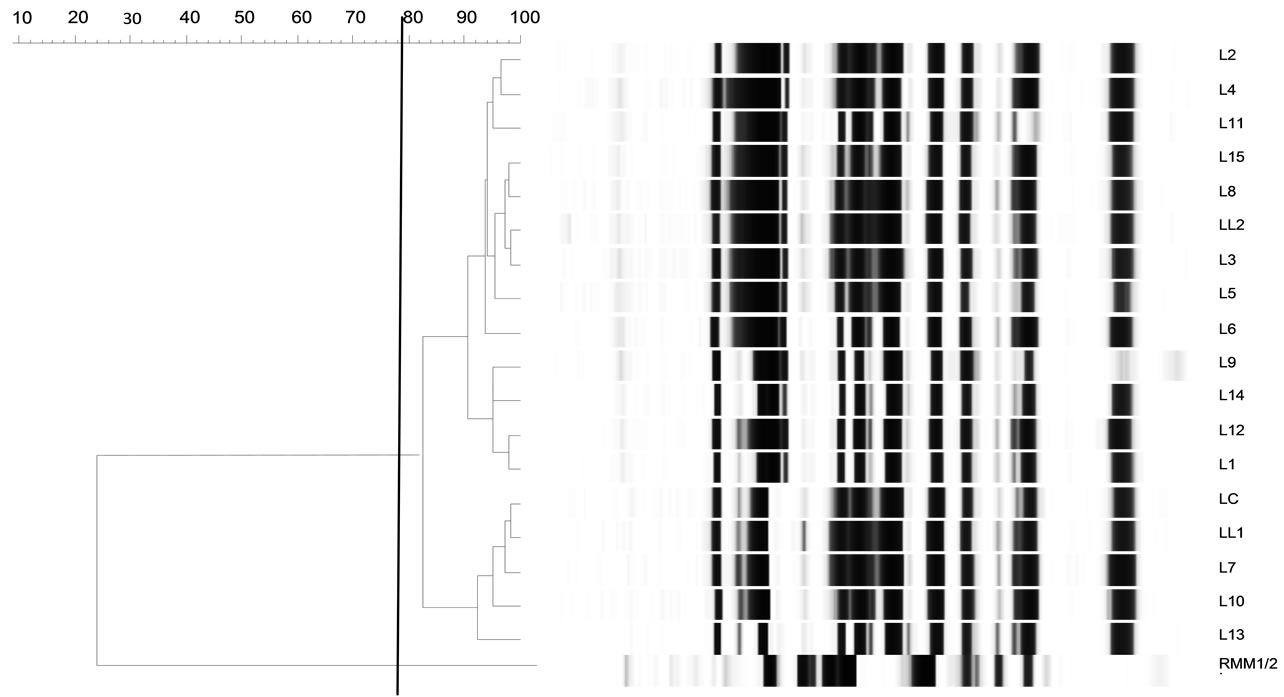
Figure 3. Dendrogram drawn by UPGMA of correlation value of normalized ERIC-PCR patterns from lactococci obtained with primers ERIC1 and ERIC2. LL1 and LL2: L. lactis subsp. lactis; LC: L. lactis subsp. cremoris; L1 to L15: lactococci isolated from wheys and belonging to cluster 1 (REP-PCR). RMM1 and RMM2: strains from re-seeded model milk; NCTC 8325: size standard.
than for the subspecies lactis. These results suggested that these strains belonged to the subspecies cremoris instead of the subspecies lactis as we had been able to observe it from the dendrogram (Figure 1, cluster 1). This observation was confirmed by PFGE analysis.
An example of some Lactococcus strain patterns obtained by PFGE is exposed on Figure 4. Except for strains RMM1, RMM2 and LC, all the strains belonged to the cluster 1 observed on Figure 2. Four groups (P1 to P4) can be observed. The first group included two RMM strains. Their SmaI patterns were clearly different from the other strains confirming the observations made before. Groups P2 and P3 included strains respectively related with starter strains LL2 and LL1. It is noteworthy that upholding the phenotypic tests, these two strains appeared different. Strain LL2 presented a large DNA fragment near 262 kb replaced on the LL1 pattern by a 210 kb fragment.
L5 and L12 strains included in the cluster P4, presented indistinguishable SmaI pattern with LC starter strain. This result confirmed the PCR identification. L5 and other associated isolates were assigned to the subspecies L. lactis subsp. cremoris, although these strains displayed similar phenotypic and REP-PCR profiles with the subspecies L. lactis subsp. lactis.
All these results show that the Lactococcus population of the whey seemed dominated by the three starter strains added the first day of cheese making, except for the strains isolated from the first whey. However, each population changed as a consequence of the backslopping. For instance, L5 and L12 L. lactis subsp. cremoris acquired phenotypic features from the subspecies lactis.
3.4. Plasmids Profiles
The strains isolated from the different wheys and milks were compared on the basis of their plasmid profiles (Figure 5). Lactococci included from 4 to 7 plasmids, their size ranging between 3 and 25 kb. The method used did not allow getting back larger plasmids. Lactococcus strains originating from wheys or from the initial starter showed the same profile. Compared with RMM strains and enterococci, this profile was clearly different. It means that there was probably no plasmid exchange between the dominant strains and RMM strains or enterococci.
3.5. Phage and Prophage Research
The presence of the phages C2, P335 and 936 was checked in the 10 successive wheys. They were never detected. The prophage P335 was searched in the ge-
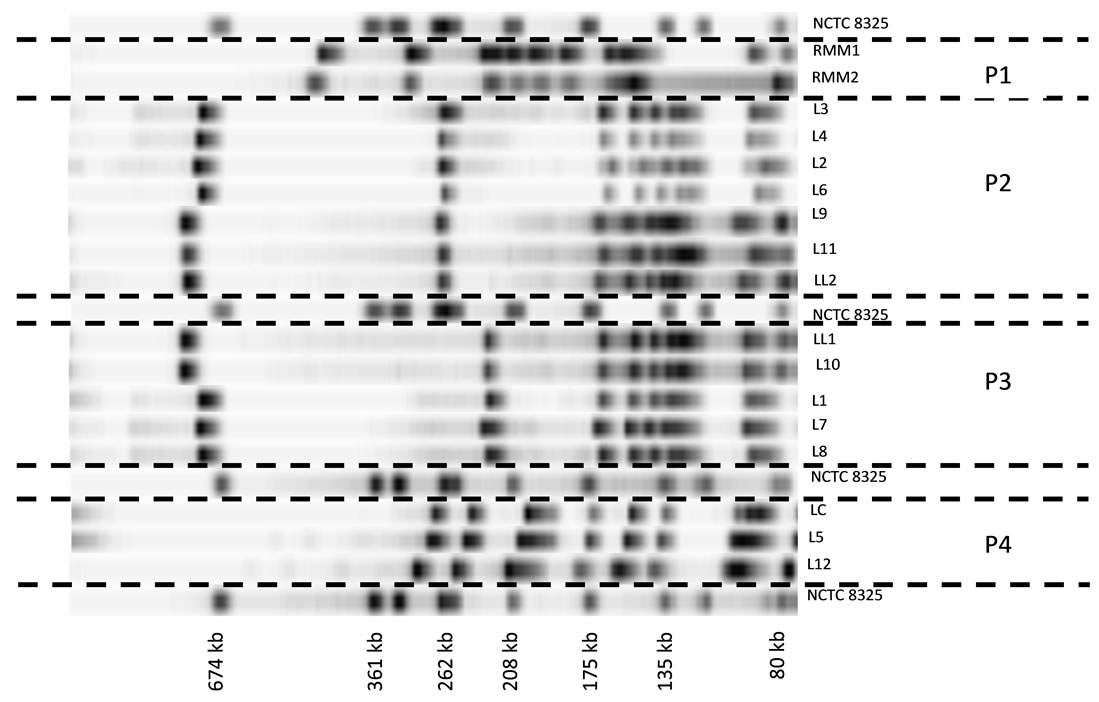
Figure 4. PFGE patterns of several lactococci DNA digested with SmaI. The clusters are numbered from P1 to P4. LL1 and LL2: L. lactis subsp. lactis; LC: L. lactis subsp. cremoris; L1 to L12: lactococci isolated from wheys. RMM1 and RMM2: strains from re-seeded model milk; NCTC 8325: size standard. Running conditions were 200 V at 14˚C for 10 h with 5 - 15 s pulse time then for 13 h with 15 - 60 s pulse time.

Figure 5. Example of plamid patterns of lactococci and enterococci. Each Lactococcus strain was selected on the basis of the PFGE clutters.
nome of the starter strains, one RMM strain and three lactococci coming from the wheys (L4, L5, L10). The results were always negative.
3.6. Phenotypic Diversity of the Lactococcus Population in Whey Phage and Prophage
Acidification aptitude of lactococci from the 10 successive NWS were presented in Figure 6. Each box is the mean of the acidification values of all the strains isolated each day. The results of the ANOVA test showed a significant difference in time (p < 0.1%). The NWS could be classified into three groups according to the Newman-Keuls’ test: 1 and 2 (a), 3, 4, 5, 6, 8, 9, 10 (b) and 7 (c).The acidification capability of the lactococci strains seemed to decrease from day to day. In the first whey,
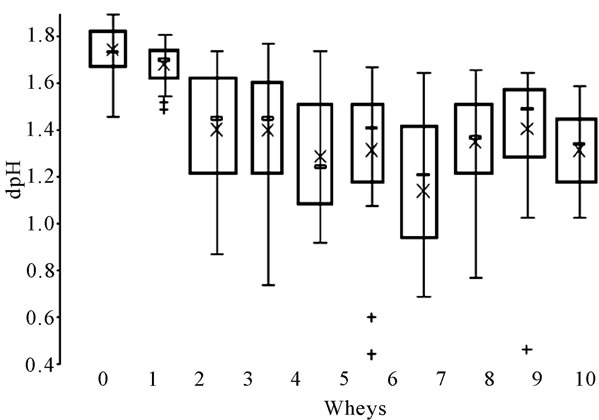
Figure 6. Representation of the mean acidification aptitude of lactococci isolated each day from the 10 successive wheys. ´ mean value; ▬ median value; + single strain. Letters correspond with the results of the Newman-Keuls’ test.
mean dpH value was close to 1.75 ± 0.11 whereas it was only equal to 1.14 ± 0.40 in whey 7. This represented a 50% loss compared with the aptitude of the initial starter. The Lactococcus population was consequently less and less acidifying as a consequence of the backslopping practice. Then, the acidification aptitude of lactococci increased again in wheys 8, 9 and 10 to settle around 1.37 ± 0.17.
To complete these observations, other phenotypic features were investigated. The enzymatic profiles of strains representative of the REP-PCR and PFGE clusters are displayed in Table 1. Compared with the starter strains (LL1, LL2 and LC), whey isolates did not display identical enzymatic activities. For example, L. lactis subsp. lactis L1, L2, L3 and L4 did not present β-galactosidase and valine arylamidase activities, and except for L1, C4-esterase activity. L. lactis subsp. cremoris L5 did not present valine arylamidase, C4-esterase and lipase activities. But, among the whey isolates, some differences were also observed confirming the former genotypic data. L1, L2, L3, L4 and L5 strains were all classified in the same REP-PCR cluster (cluster 1). However they did not display the same enzymatic profile. L1 strain possessed a C4-esterase whereas the other strains did not. As a general observation and keeping in mind the relatively low number of enzymatic activities tested, whey isolates seemed to lose enzymatic activities rather than to acquire new ones.
4. Discussion
NWS inherited from the ancient backslopping practice are still employed around the world for traditional cheesemaking. They provide a wide range of techno-
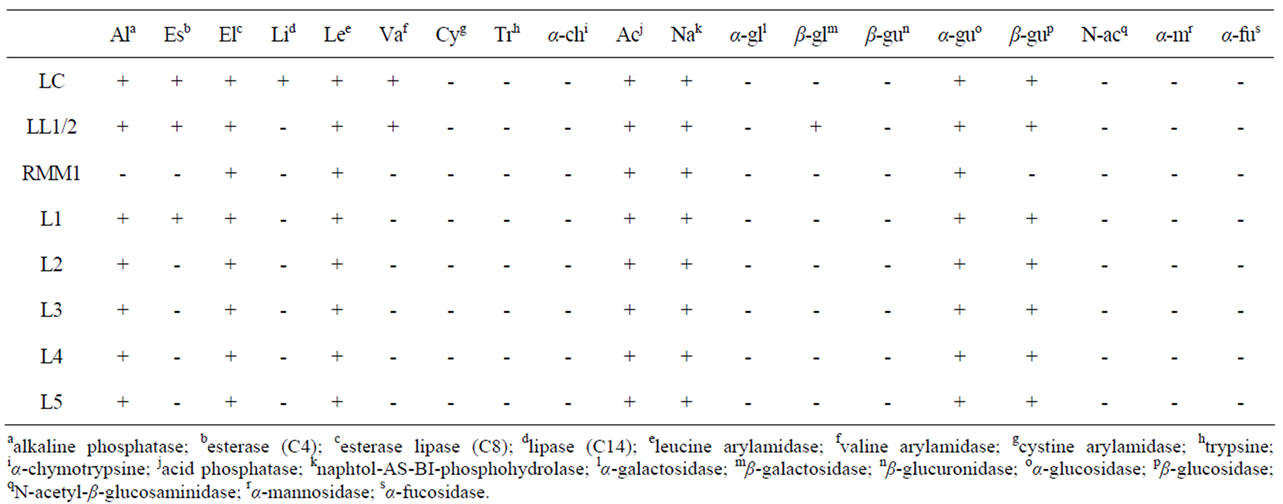
Table 1. Enzymatic profiles of some lactococci strains. LC: L. lactis subsp. cremoris; LL1/2: L. lactis subsp. lactis; RMM: strain from a re-seeded model milk; L1 to L5: lactococci from whey.
logical advantages for the dairy industry. Strangely, ex-cept in hard cheese technology, NWS are still unknown. This work was undertaken to monitor the evolution of a model NWS microflora during ten successive days, with a focus on the Lactococcus genus. The contribution of the raw milk LAB flora was also taken into account, a “reseeded-model milk” (RMM) being used every day. The Enterococcus population was low in whey. The cellular levels never exceeded 4 log(cfu)·ml−1 and hovered around 2.6 log(cfu)·ml−1 most of the time. Lactobacilli formed a small group as well, their cellular levels never exceeding 5.1 log(cfu)·ml−1. These results are consistent with the observations made by Tormo et al. [22] and with our own former results.
Lactococci were the dominant population in whey, their level reaching a stable value centred around 9.2 - 9.3 log(cfu)·ml−1. This also tallies with our results [12]. Even if the Lactococcus population level was quantitatively stable, qualitative changes occurred. Indeed, although no differences were noticed by REP-PCR, ERICPCR and plasmid extraction, the presence of different pulsotypes and phenotypic behaviour from NWS isolates suggested that the composition of the Lactococcus population changed in time. On day 1 however, the dominant Lactococcus population was clearly different from the starter and from the other strains analysed after that. At that time, we cannot explain this observation.
The REP-PCR technique was selected for its convenience to discriminate lactococci strains according to their origin—RMM or initial starter [23]. No strains from RMM could be found in whey, although they could constitute a sub-dominant group masked by the dominant lactococci population. Raw milk strains are known to possess exceptional rustic capabilities which allow them to interfere with starter strains [16] and to express their technological aptitudes during cheese ripening [24]. However, the low level of this “rustic” microflora in this work is perhaps the consequence of the experimental design, which led to underestimate their effective role. Corsetti et al. [25] showed that minor populations in sourdoughs—Enterococcus faecium and Pediococcus pentosaceus—were essential for the establishment of propitious conditions for the growth of Lactobacillus sanfran-ciscensis. In our study, it could be assumed that RMM lactococci may have contributed to the development of other flora, even if their level was undetectable.
If the REP-PCR technique was appropriate for the discrimination between RMM and whey strains, it failed to depict the diversity of whey strains since most of the strains isolated from whey were identical to the L. lactis subsp. lactis starter strain (LL1 and LL2) used on the first day of cheesemaking. The use of PFGE and PCR identification allowed to reveal that a dominant part of the Lactococcus starter population also included L. lactis subsp. cremoris strains. The REP-PCR technique may therefore be inappropriate to study the lactococci diversity in this ecosystem as previously observed by Turpin et al. [26]. These authors indicated that this technique should be used with more reliable methods. These elements showed the necessity of a polyphasic approach for studying complex ecosystems [27]. In particular, the use of two sets of primers was used successfully by Depouilly et al. [15] to discriminate strains from hard cooked cheeses.
These results partially tally with those of Passerini et al. [28]. These authors indicated that MLST-PCR approach was interesting to discriminate lactococci following their ecotype. But among “domesticated” strains, the diversity was rather scarce compared with environmental (i.e. raw milk) strains.
In this current study, L. lactis subsp. cremoris displayed phenotypic features of the lactis subspecies, a phenomenon already described by Salama et al. [29]. According to these authors, this could be possible via transducing phages in dairy environments. NWS could then be a suitable location for genetic exchanges between LAB strains. Indeed, the cheese model used in this study was built in such a way that no undesired contamination could happen [7]. Phages were searched, especially the types C2, P335 and 936, which are the most frequent phages encountered in dairy products [21]. They were never detected even if it is always possible that other phage types contaminate the whey. Moreover, phage P335 is known to be the most frequent prophage able to integrate the genome of lactococci [20]. It was not detected any more. Based on these results, we can argue that there were probably no transduction phenomena during the 10 successive days of backslopping.
The changes observed in the Lactococcus population occurred rapidly. On the second day, LC strains had already acquired lactis features whereas LL strains had lost some phenotypic features. For instance, strains LL1 and LL2 possessed valine aylamidase activity which was no more present among the strains isolated from the successive wheys. Scarce published data exist on rapid mutations in bacteria such as those observed in this study. Massey et al. [30] described rapid definitive mutations in Salmonella in stress conditions. The whey acidity (pH < 4.7) constituted a stress condition which could generate rapid mutations in lactococci strains during backslopping. The disappearance of some enzymatic aptitudes such as the β-galactosidase activity could also result from the loss of several plasmids by lactococci strains [31]. The study of acidification capabilities showed that strains lost progressively a part of their acidifying potential during the 7 successive days of backslopping. Acquisitions and losses of physiological abilities via plasmid tranferts in LAB have been extensively studied. For example, the transfer of antibiotic resistant genes via plasmid exchanges between Lactococcus, Enterococcus and Lactobacillus strains in milk and during cheesemaking was reported by Kelly et al. [32]. Closeness appears to be an essential condition for genetic material exchanges between LAB. During cheese making, the whey certainly favour the connection between bacteria, since this “opened” liquid environment allows the movement of molecules and, possibly, of strains. In this case, we only observed losses of physiological abilities and no acquisitions. Moreover, we did not notice any plamid exchange among Lactococcus strains or between different genus, e.g. between enterococci and lactococci. However, the method used did not allow recovering large plasmids (>35 kb). Based on all these results, we propose a progressive selection of more adapted populations from day to day to explain phenotypic changes. This hypothesis was tested by Cretenet et al. [33]. These authors observed that in a cheese matrix, L. lactis was submitted to several stress conditions, some being counteracted by multiple efficient strategies. Clearly, in our case, the bacteria may have slowly adapted to the “backslopping environment”.
5. Conclusions
This work constitutes a new step in the understanding of NWS diversity and brings new insights concerning its evolution in time. Indeed, our results—obtained in a model system—mean that NWS diversity does not only seem to depend on the great number of LAB that composed them or on the multitude of possible strain sources. It also rests on the huge possibilities of adaptation of the strains that can occur during successive whey backslopping, this phenomenon being itself controlled by the influence of environmental parameters. The specific role of the environment would be interesting to analyse as a validation of our hypothesis. Since it is difficult to show the generation of rapid mutations inside the genome of the Lactoccocus population, other methods would be also interesting to be tested (MLST, MLSA).
6. Acknowledgements
The authors would like to thank Marc Chareyron for his help in the writing of this paper. They are also grateful to Philippe Guyot, Franck Dufresne and Fanny Moignard for their technical support during this study.
REFERENCES
- G. Bertoni, L. Calamari and M. Grazia Maianti, “Producing Specific Milks for Speciality Cheeses,” Proceedings of the Nutrition Society, vol. 60, No. 2, 2001, pp. 231- 246. doi:10.1079/PNS200080
- M. A. Herreros, J. M. Fresno, M. J. González Prieto and M. E. Tornadijo, “Technological Characterization of Lactic Acid Bacteria Isolated from Armada Cheese (a Spanish Goats’ Milk Cheese),” International Dairy Journal, vol. 13, No. 6, 2003, pp. 469-479. doi:10.1016/S0958-6946(03)00054-2
- G. Mauriello, L. Moio, A. Genovese and D. Ercolini, “Relationships between Flavoring Capabilities, Bacterial Composition, and Geographical Origin of Natural Whey Cultures Used for Traditional Water-Buffalo Mozzarella Cheese Manufacture,” Journal of Dairy Science, vol. 86, No. 2, 2003, pp. 486-497. doi:10.3168/jds.S0022-0302(03)73627-3
- L. Topisirovic, M. Kojic, D. Fira, N. Golic, I. Strahinic and J. Lozo, “Potential of Lactic Acid Bacteria Isolated from Specific Natural Niches in Food Production and Preservation,” International Journal of Food Microbiology, vol. 112, No. 3, 2006, pp. 230-235. doi:10.1016/j.ijfoodmicro.2006.04.009
- G. K. Y. Limsowtin, I. P. Powell and E. Parente, “Types of Starters,” In: T. M. Cogan and J. P. Accolas, Ed., Dairy Starter Cultures, Wiley-VCH, New York, 1996, pp. 101-129.
- G. Giraffa, “Studying the Dynamics of Microbial Populations during Food Fermentation,” FEMS Microbiology Reviews, vol. 28, No. 2, 2004, pp. 251-260.
- M. Dalmasso, D. Hennequin, C. Duc and Y. Demarigny, “Influence of Backslopping on the Acidifications Curves of ‘Tomme’ Type Cheeses Made during Ten Successive Days,” Journal of Food Engineering, Vol. 92, No. 1, 2009, pp. 50-55. doi:10.1016/j.jfoodeng.2008.10.019
- M. Dalmasso, S. Prestoz, V. Rigobello and Y. Demarigny, “Evolution of Raw Cow Milk Microflora, Especially Lactococci, Enterococci, Leuconostocs and Lactobacilli over a Successive 12 Day Milking Regime,” International Journal of Dairy Science, Vol. 3, No. 3, 2008, pp. 117-130. doi:10.3923/ijds.2008.117.130
- D. Isolini, M. Grand and H. Glättli, “Selective Medium for the Detection of Obligatory and Facultative Heterofermentative Lactobacilli,” Schweizerische Milchwirtschaftliche Forschung, vol. 19, No. 3, 1990, pp. 57-59.
- J. V. Mayeux, W. E. Sandine and P. R. Elliker, “A Selective Medium for Detecting Leuconostoc in Mixed-Strain Starter Cultures,” Journal of Dairy Science, vol. 45, 1962, p. 655.
- Y. Demarigny, “Rôle de la Flore Naturelle du Lait Cru et de Paramètres Technologiques (Teneur en Sel et Température d’Affinage) sur l’Evolution des Caracté- ristiques Biochimiques, Microbiologiques et Sensorielles des Fromages à Pâte Pressée Cuite,” PhD Thesis, Université de Bourgogne, Dijon, 1997.
- Y. Demarigny, C. Sabatier, N. Laurent, S. Prestoz, V. Rigobello and M. J. Blachier, “Microbial Diversity in Natural Whey Starters Used to Make Traditional Rocamadour Goat Cheese and Possible Relationships with Its Bitterness,” Italian Journal of Food Science, vol. 3, No. 18, 2006, pp. 251-266.
- D. Corroler, I. Mangin, N. Desmasures and M. Gueguen, “An Ecological Study of Lactococci Isolated from Raw Milk in the Camembert Cheese Registered Designation of Origin Area,” Applied and Environmental Microbiology, vol. 64, No. 12, 1998, pp. 4729-4735.
- J. Versalovic, T. Koeuth and J. R. Lupski, “Distribution of Repetitive DNA Sequences in Eubacteria and Application of Fingerprinting of Bacterial Genomes,” Nucleic Acids Research, vol. 19, No. 24, 1991, pp. 6823-6831. doi:10.1093/nar/19.24.6823
- A. Depouilly, F. Dufrene, E. Beuvier and F. Berthier, “Genotypic Characterisation of the Dynamics of the Lactic Acid Bacterial Population of Comté Cheese,” Lait, Vol. 84, No. 1-2, 2004, pp. 155-167. doi:10.1051/lait:2003036
- Y. Bouton, P. Guyot, E. Beuvier, P. Tailliez, P. and R. Grappin, “Use of PCR-Based Methods and PFGE for Typing and Monitoring Homofermentative Lactobacilli during Comté Cheese Ripening,” International Journal of Food Microbiology, vol. 76, No. 1-2, 2002, pp. 27-38. doi:10.1016/S0168-1605(02)00006-5
- S. Murchan, M. E. Kaufmann, A. Deplano, R. De Ryck, M. Struelens, C. Elsberg Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. Van Leeuwen, A. Van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes and B. Cookson, “Harmonization of Pulsed-Field Gel Electrophoresis Protocols for Epidemiological Typing of Strains of Methicillin-Resistant Staphylococcus Aureus: A Single Approach Developed by Consensus in 10 European Laboratories and Its Application for Tracing the Spread of Related Strains,” Journal of Clinical Microbiology, vol. 41, No. 4, 2003, pp. 1574-1585. doi:10.1128/JCM.41.4.1574-1585.2003
- D. J. O’Sullivan and T. R. Klaenhammer, “Rapid MiniPrep Isolation of High-Quality Plasmid DNA from Lactococcus and Lactobacillus spp,” Applied and Environmental Microbiology, Vol. 59, No. 8, 1993, pp. 2730-2733.
- S. Moineau, J. Fortier, J. Ackermann and S. Pandian, “Characterization of Lactococcal Bacteriophages from Québec Cheese Plants,” Canadian Journal of Microbiology, Vol. 38, No. 9, 1992, pp. 875-882. doi:10.1139/m92-143
- H. Brüssow, “Phages of Dairy Bacteria,” Annual Reviews in Microbiology, vol. 55, No. 1, 2001, pp. 283-303.
- S. Labrie and S. Moineau, “Multiplex PCR for Detection and Identification of Lactococcal Bacteriophages,” Applied and Environmental Microbiology, vol. 66, No. 3, 2000, pp. 987-994. doi:10.1128/AEM.66.3.987-994.2000
- H. Tormo, A. Kodjo and P. Talliez, “Contribution d’un Levain Naturel à la Spécificité des Fromages Fermiers de Chèvre,” Proceedings Conference Produits Alimentaires Fermiers, Clermont-Ferrand, 2004, p. 137.
- E. Casalta, J. M. Cachenaut, C. Aubert, F. Dufrene, Y. Noël and E. Beuvier, “Application of Specific Starters for the Manufacture of Venaco Cheese,” Lait, vol. 85, No. 3, 2005, pp. 205-222. doi:10.1051/lait:2005019
- T. M. Cogan, T. P. Beresford, J. Steele, J. Broadbent, N. P. Shah and Z. Ustunol, “Advances in Starter Cultures and Cultured Foods,” Journal of Dairy Science, vol. 90, No. 9, 2007, pp. 4005-4021. doi:10.3168/jds.2006-765
- A. Corsetti, L. Settanni, S. Valmorri, M. Mastrangelo and G. Suzzi, “Identification of Subdominant Sourdough Lactic Acid Bacteria and Their Evolution during Laboratory-Scale Fermentations,” Food Microbiology, vol. 24, No. 6, 2007, pp. 592-600. doi:10.1016/j.fm.2007.01.002
- W. Turpin, C. Humblot, A. Hammoudi and J. P. Guyot, “Limite de l’Utilisation de la REP-PCR Comme Méthode Rapide d’Identification des Bactéries Lactiques,” Proceedings Conférence Club des Bactéries Lactiques, Toulouse, May 2009.
- I. Mainville, N. Robert, B. Lee and E. R. Farnworth, “Polyphasic Characterization of the Lactic Acid Bacteria in Kefir,” Systematic and Applied Microbiology, vol. 29, No. 1, 2006, pp. 59-68. doi:10.1016/j.syapm.2005.07.001
- D. Passerini, C. Beltramo, M. Coddeville, Y. Quentin and P. Ritzenthaler, “Genes but Not Genomes Reveal Bacterial Domestication of Lactococcus lactis,” PLoS ONE, Vol. 5, No. 12, 2010, pp. 1-12. http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0015306
- M. S. Salama, T. Musafija-Jeknic, W. E. Sandine and S. J. Giovannoni, “An Ecological Study of Lactic Acid Bacteria: Isolation of New Strains of Lactococcus Including Lactococcus lactis Subspecies Cremoris,” Journal of Dairy Science, Vol. 78, No. 5, 1995, pp. 1004-1017. doi:10.3168/jds.S0022-0302(95)76716-9
- R. C. Massey, P. B. Rainey, B. J. Sheehan, O. M. Keane and C. J. Dorman, “Environmentally Constrained Mutation and Adaptive Evolution in Salmonella,” Current Biology, vol. 9, No. 24, 1999, pp. 1477-1480. doi:10.1016/S0960-9822(00)80117-7
- S. Mills, O. E. McAuliffe, A. Coffey, A. G. F. Fitzgerald and R. P. Ross, “Plasmids of Lactococci—Genetic Accessories or Genetic Necessities?” FEMS Microbiology Review, vol. 30, No. 2, 2005, pp. 243-273. doi:10.1111/j.1574-6976.2005.00011.x
- B. G. Kelly, A. Vespermann and D. J. Bolton, “Gene Transfer Events and Their Occurrence in Selected Environments,” Food and Chemical Toxicology, Vol. 47, No. 5, 2009, pp. 978-983. doi:10.1016/j.fct.2008.06.012
- M. Cretenet, V. Laroute, V. Ulvé, S. Jeanson, S. Nouaille, S. Even, M. Piot, L. Girbal, Y. Le Loir, P. Loubière, S. Lortal and M. Cocaign-Bousquet, “Dynamic Analysis of the Lactococcus lactis Transcriptome in Cheeses Made from Milk Concentrated by Ultrafiltration Reveals Multiple Strategies of Adaptation to Stresses,” Applied and Environmental Microbiology, vol. 77, No. 1, 2011, pp. 247-257. doi:10.1128/AEM.01174-10

