American Journal of Plant Sciences
Vol.4 No.4(2013), Article ID:30083,12 pages DOI:10.4236/ajps.2013.44108
QTL Mapping for Partial Resistance to Southern Corn Rust Using RILs of Tropical Sweet Corn
![]()
1Interdisciplinary Graduate Program in Genetic Engineering, Kasetsart University, Bangkok, Thailand; 2Department of Agonomy, Faculty of Agriculture, Kasetsart University, Nakhon Pathom, Thailand; 3Rice Science Center, Kasetsart University, Nakhon Pathom, Thailand; 4Genome Institute, National Center for Genetic Engineering and Biotechnology, Pathum Thani, Thailand.
Email: *somvong.tra@gmail.com
Copyright © 2013 Kitti Wanlayaporn et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received February 28th, 2013; revised March 31st, 2013; accepted April 10th, 2013
Keywords: Sweet Corn; Southern Corn Rust; Partial Resistance; Quantitative Trait Loci (QTLs); Combined across Locations
ABSTRACT
Southern corn rust is one of destructive diseases in maize caused by Puccinia polysora Undrew. A mapping population of tropical sweet corn recombinant inbred lines (RILs) derived from a cross between hA9104 and hA9035 inbred lines were set up to detect quantitative trait loci (QTLs) involved in partial resistance to southern corn rust. Eighty nine RILs were used to evaluate resistance levels using nine-point relative scale (1-9) at Sweet Seeds, Suwan Farm, Thailand include combined analysis. A genetic linkage map was constructed with 157 SSR markers, with a total length of 2123.1 cM, covering 10 chromosomes. Broad-sense heritability of individual location ranged from 0.76 and 0.82 and combined across locations was 0.87. Multiple QTL mapping (MQM) was applied for the identification of the QTLs. Fifteen QTLs were detected on chromosome 1, 2, 5, 6, 9 and 10 in both locations and combined across locations. QTLs on chromosome 1, 5 and 6 were contributed by alleles of resistant parent hA9104 while others were contributed by alleles from the susceptible parent, hA9035. Phenotypic variance of each QTL explained ranged from 6.1% to 41.8% with a total of 69.8% - 81.9%. QTL on chromosome 1, 6 and 10 were stable QTLs detected in both locations.
1. Introduction
Southern corn rust is caused by Puccinia polysora Undrew, a biotrophic fungus of foliar disease in corn. It is classified as an obligate parasite affecting corn production all over the world and is normally found in warmtemperate and tropical cultivating zones. It affected the corn production in the US [1], Asia [2] and Africa [3]. The disease causes damages to the whole corn plant: leaves, stalks, leaf sheath and husks especially on leaf surface. Symptoms initially appear on leaves and expand throughout an entire plant in mature stage resulting in leaf necrosis and complete destruction of photosynthetic areas until the plant eventually dies [4].
In 1949, southern corn rust was first identified in West Africa and it caused yield loss of up to 50% [5]. In 1972- 1974, the disease continually occurred in USA corn fields, and 45% of all products were damaged [6,7]. A similar phenomenon took place in Australia in 1959 [8] In Asia, 80% - 84% of yield loss in Philippines was reported [9]. In China, the disease was first identified in 1972 [10], and the outbreak in 1998 caused of yield loss between 42% and 53% of all corn products [11].
The disease has emerged in Thailand since 1967 [12] it caused a severe damage of corn varieties in Nakhonratchasima, Saraburi and Tak provinces. Biodiversity and classification of the pathogens in Thailand were studied using morphology and molecular markers. The pathogen that caused the disease in Thailand was identified as P. polysora Undrew strain and the occurrences of genetic variations were revealed. [13] reported that epidemic of disease in sweet corn affecting 6% - 6.5% of fresh ear weight and ear quantity decreased as the severity of the symptoms.
Generally, chemical control has been applied to manage southern corn rust. Nevertheless, chemical measure was used to against the disease by fungicide which had side effects to environment. Breeding for resistance in elite varieties as appears to be a better method for controlling the southern corn rust disease with minimal effects on the environment.
Classifications and identifications of P. polysora Undrew races have previously been studied. Races EA1, EA2 and EA3 were found in East Africa [14-16]. Another six races, PP.3, PP.4, PP.5, PP.6, PP.7 and PP.8 isolated from North and Central American maize germplasm are distinguishable from the East African races [17]. The last one, PP.9 was isolated from South African maize [18].
Race-specific resistance genes, Rpp1 and Rpp2 were identified from Mexican and Columbian lines by [19] which provided resistance to P. polysora race EA1 and EA2, respectively. These two genes were shown to be loosely linked to each other but their positions on genome have not been determined [20]. The major gene for southern corn rust resistance Rpp9 was isolated from PI186208, South African maize and conferred resistance to PP.9. This gene is closely linked to Rpp1, resistance gene to common rust, located on short arm of chromosome10 [21]. Rpp10 and Rpp11 were isolated from Colombian and Mexican corn germplasm, respectively [22]. Rpp10 exhibited a complete dominant inheritability with highly resistance to races EA1 and EA2 which differed from Rpp1 which conferred resistance to only race EA1. Rpp11 revealed partial dominance of resistance to both of these races with large necrotic areas observed on plant. There was no evidence for linkage between Rpp10 and Rpp11. Other major genes were B1138TRpp gene from B1138T, South Africa corn [5], RppP25 from P25 line [2], in Qi319 corn carried resistance gene, RppQ, [10,23] and RppD isolated from W2D corn [24]. Their allelic relationship to Rpp9 has not been studied.
Even though race-specific resistance provides a high level of resistance to disease and easier to work with in breeding programs, race-nonspecific or partial specific resistance last longer and provide protection against pathogens of broader specificity [25-27]. The study of race-specific resistance of southern corn rust in tropical regions constantly failed because the tropical area has a condition that allows the existence of multiple races of P. polysora [28]. In addition, the race specific resistance is ineffective, unstable and short-term because of the rapid evolution of the pathogens [29]. [30] suggested that genes conferring race-specific resistance were not effective in preventing the spread of southern rust disease in Africa, a tropical region. Partial resistance or slow-rusting resistance has been used instead of race-specific resistance that could be achieved to study in case of appearance of multiple races, race mutation to new race or even across environments study [31,32].
The first quantitative trait loci (QTL) mapping was studied in crop plant [33] and afterward a number of QTL mapping for resistance to plant diseases have been conducted. Combination of molecular mapping techniques and marker-assisted selection may be a tool for breeders to identify the genetic of resistance in plant efficiently [34]. Major QTLs of resistance to southern corn rust were located on short arm of chromosome 10 using different sets of corn germplasms [2,10,23,35-37] and these QTLs were tightly linked to Rpp9, however, the relationship of allelic or among races and racial specificity of these genes has not been determined.
QTL of partial resistance for southern corn rust have been mapped on 6 [12], 3 and 4 [36], 4, 8, 9 and 10 [37], 3, 4 and 9 [38] and 9 [39] and co-localization of their QTLs across studies have been done [40].
In this study, evaluation of resistance to southern corn rust was performed in tropical sweet corn under two environment conditions in tropical area: Sweet Seeds research station (Sweet Seeds), Saraburi and National Corn and Sorghum Research Center (Suwan Farm), Nakhonratchasima where epidermic of southern corn rust has appeared in Thailand. The objective of this research was to detect the position and number of QTLs associated with partial resistance to southern corn rust in 89 RILs under different two locations including combined across locations from a cross between the inbreds hA9104 and hA9035 using molecular markers (SSRs). Gaining a better understanding of genetic basis of resistance of southern corn rust would provide useful knowledge for maize breeder and plant pathologies for breeding programs.
2. Materials and Methods
2.1. Plant Materials and Field Experiments
A mapping population of tropical sweet corn 133 F5:6 recombinant inbred lines (RILs) derived from a cross between parent inbred hA9104 (resistance) and hA9035 (susceptible). Two parents were developed and obtained from Sweet Seeds Co., Ltd., (Sweet Seeds) Saraburi. ATS-5, susceptible hybrid line obtained from Sweet Corns Products Co., Ltd. was first grown around the experimental field for one month to be spreader row for source of pathogen. RILs and parents were grown at Sweet Seeds station field (Sweet Seeds), Saraburi, and experiment station of National Corn and Sorghum Research Center (Suwan Farm), Nakhonratchasima, Thailand in May to September, 2008. Experiment design was randomized blocks design (RBD) with two replicates. Kei0504 and Ki11 inbred line received from Suwan Farm were used as resistant and susceptible checking line to southern corn rust, respectively.
Each individual of RILs, parents and checking lines were sown in a 4 meters long row with 15 - 20 plants per row. The planting space was 0.75 × 0.25 meters. Chemical fertilizer of 15-15-15 (312.5 kg/ha) was immediately used after planting and 46-0-0 (156.25 kg/ha) was applied at 20 days after planting in both locations. Adequate herbicide was reasonably used to control weeds.
2.2. Evaluation of Resistance for Southern Corn Rust
The 133 progenies of F5:6 RILs, parents inbred line hA9104 and hA9035 and checking line Kei0504 and Ki11 were grown at experiment field of Sweet Seeds and Suwan Farm in May to September, 2008. ATS-5 was grown as spreader row to enhance for spreading the inoculums of southern rust pathogen by natural epiphytotic method or spreader row technique in two locations. Resistance level assessment was performed in individual plant of each line (15 plants per line) and mean score was calculated for each plot at 80 days after planting.
Severity of rust were scored as rust rating score using a nine-point relative scale of [36] in which one indicated as highly susceptible (leaves completely covered with southern rust pustules) and nine designated as highly resistant (entirely free of pustules).
2.3. Statistical Analysis
Multiple range test, analysis of variance or ANOVA, genetic variance components and correlation coefficients of rust rating between two locations and combined analysis for southern corn rust resistance in RILs population were analyzed using STATGRAPHICS Plus [41] and IRRISTAT 5.0 software programs [42]. Broad-sense heritability of southern corn rust resistance was assessed using analysis method of genetic variation components in ANOVA.
2.4. DNA Preparation
Fresh leaves of each individual RIL and parent plants at 7 - 8 leaf stage were collected for isolate genomic DNA using CTAB method [43]. Quantity and quality of extracted DNA samples were analyzed with 1% agarose gel electrophoresis and ethidium bromide staining.
2.5. SSR Analysis
Sequence of these simple sequence repeat (SSR) primers were obtained from MaizeGDB database (http://www. maizegdb.org) and PCR primers flanking those loci were synthesized by DNA Technology Laboratory, Nakhonpathom, Thailand. A total of 461 primer pairs were screened for polymorphism between parental lines. Distinguishable markers were used in genotyping. Amplification of SSRs was performed as described by [44]. PCR amplifications were processed on Mastercycler pro384 (Eppendorf®) with following PCR conditions: initial denaturing was done at 95˚C for 5min, and then 35 cycles of 95˚C for 30 s, 55˚C for 30 s and 72˚C for 1 min with a final 72˚C for 10 min. All chemicals for PCR were from Fermentas®. PCR products were detected with 4.5% gel electrophoresis (PAGE) and silver staining was performed according to the protocol described by [45]. Alleles of hA9104, hA9035 and heteorozygous (hA9104/ hA9035) were designated as A, B and H, respectively.
2.6. Linkage Map Construction
The Chi-square goodness-of-fit test was used to identify expected Mendelian segregation ratios 1:1 of all SSR markers [46]. The linkage map was constructed by MapMaker 3.0b software [47]. Group of linkage was created by command “group” following with LOD of 3.0 and recombination fraction of 0.5. Ripple function was used to estimate the order of markers [48]. Finally, the genetic distances were calculated by the Kosambi mapping function [49] which recombination frequencies parameters was applied to centiMorgans (cM). The missing data, distorted segregation and similarity of all SSR markers were assessed and removed if any marker showed high level of segregation distortion or more than 10% missing data. MapChart 2.2 for windows was used to draw linkage map [50].
2.7. QTL Analysis and Mapping
Identification of QTLs for southern corn rust under two locations and combined was analyzed using software MapQTL® v4.0 with multiple-QTL mapping or MQM mapping model, cofactor selection for analyzing QTL was automatically selected by option of cofactor selection [51]. QTL associated with resistance of southern corn rust, phenotypic variance explain (PVE) or R2 and parent allele additive effect of each QTL were calculated. The permutation test at 1000-time permutations was used to estimate LOD threshold scores [52]. LOD scores of equal and more than 3.0 was determined to major QTL. LOD score > 2.2 was acceptable to minor QTL.
3. Results
3.1. Evaluation for Resistance of Southern Corn Rust in Two Locations
Assessment for resistance of southern corn rust was conducted in two locations. First, research station at Sweet Seeds Co., Ltd. (Sweet Seeds), Saraburi, Thailand and second, National Corn and Sorghum Research Center (Suwan Farm), Nakhonratchasima, Thailand in 2008 were studied and analyzed in late rainy season, May to September in 2008 by natural epiphytotic method and resistance of southern corn rust was evaluated by nine-point relative scale (1 = highly susceptible, 9 = highly resistance). Both two locations always have extremely epidemic of southern corn rust. Unfortunately, Downy mildew disease occurred during seedling stage at Suwan Farm caused the destruction of the sweet corn plants estimated for 44 lines. Therefore, 89 of 133 inbred lines could be tested while at Sweet Seeds, problem of seed germination of 10 progeny lines has arisen influenced to remain 123 of 133 inbred lines for analysis. Destroyed and unavailable inbreds in both locations were cut off. Rust rating between Sweet Seeds and Suwan Farm were used to analyze combined across locations with amount of 89 RILs.
The Kei0504 inbred line was used as a resistant checking line showed rust rating mean of 8.0 referred to highly resistance which the symptom of chlorosis on leaves surface appeared because chlorophyll on parenchyma tissue was destroyed and pustule was not observed. Ki11, the susceptible checking inbred line revealed a mean rust rating of 4.0 and a number of pustules were observed on leaf surface. Our parent inbred line, hA9104, showed a large number of pustules dispersed on leaf surface indicating that it is moderately resistant with average rust rating of 5.25 (5.0 and 5.5 for Sweet Seeds and Suwan Farm, respectively). On the other hand, hA9035, the susceptible line showed rust rating of 2.5 in both locations exhibiting necrosis of leaf caused by the domination of pustules. It seemed that hA9104 and hA9035 were moderately resistant and highly susceptible, respectively when compared to the checking inbred lines, Kei0504 and Ki11.
Population mean of rust rating in RILs were adjacent to mid-parent values with ranging from 3.34 to 3.75 for Sweet Seeds and Suwan Farm, respectively and combine across locations was 3.78. Genetic variation components of rust rating in 89 individuals were estimated using numeric values of mean square from Table 1 comprising Sweet Seeds, Suwan Farm and combined analysis across two locations. Phenotypic variances (VP) of RILs from Sweet Seeds, Suwan Farm and combined locations are 2.2086, 2.7013 and 1.8189, respectively. The genotypic variance (VG) of the Sweet Seeds, Suwan Farm and combined locations are 1.8073, 2.0448 and 1.5913 and environmental variance (VE) are 0.4013, 0.6565 and 0.6236, respectively. Interaction of genotypic variance and environmental variance (VGE) is 0.1353. Broad-sense heritability (H2) was also relatively high, 82%, 76% and 87% in Sweet Seeds, Suwan Farm and combined across locations, respectively (Table 1).
Frequency distributions of rust rating average of RILs population in two locations include combined across locations were continuous distribution (Figure 1) that means resistance for southern corn rust was quantitative inheritance controlled by polygenes [53]. Transgressive segregation was observed in RILs in both locations caused by mode of new gene recombination of RILs [54]. Complementary action of genes from the two parental species is a cause of occurrence of transgressive segregation [55] that means resistance for southern corn rust in our population was controlled by contribution of hA9104 and hA9035.
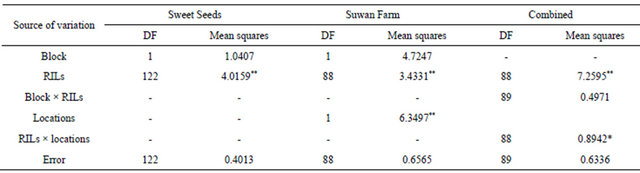
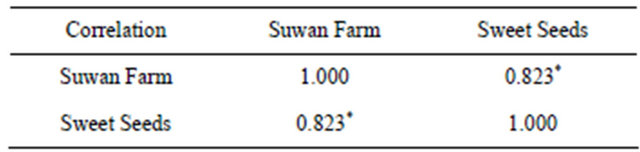
Mean squares of ANOVA revealed significant difference between RILs in both locations at 5% confidence level for all field trials (Table 2). For combined across locations, there were significant locations and RILs × locations interaction at 5% and 1% confidence level, respectively. The spearman rank correlation was chosen to analyze the relationship of rust rating in 89 RILs of RILs population in two locations. Rust rating of Sweet Seeds and Suwan Farm was positively correlated (P < 0.05, r = 0.82) (Table 3).
3.2. Genetic Linkage Map
A number of 461 SSR markers were used to construct a linkage map from 89 RILs F5:6 population. Seventy-two of 461 (15.63%) revealed distortion segregation tested by goodness-fit-test or Chi-square test by expected ratio 1:1 were excluded from the analysis. Two hundred and thirty-two of 461 markers (53.14%) showed poor quality of amplification or non-polymorphic amplification. The remaining 157 SSR markers (34.05%) were polymorphic and used in the generation of a genetic map of 89 F5:6 progenies derived from a cross between hA9104 and hA9035. The linkage map consisted of 10 chromosomes with a total length of 2123.1 cM and an average of 16.18 cM between markers. Chromosome 1 had the highest
Table 1. Mean of rust rating scores for parental lines (hA9104 and hA9035) and RILs population along with variance components consisted of phenotypic (VP), genotypic (VG), environmental (VE) and genotype X environment interaction (VGE) and broad-sense heritability (H2) at Sweet Seeds Research Station, Suwan Farm and combined across two locations in 2008.
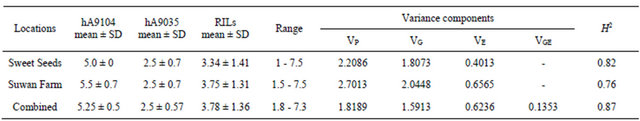
Table 2. ANOVA analysis for southern corn rust ratings in F5:6 population evaluated at Sweet Seeds Research Station, Suwan Farm and combined across locations in 2008.
* and ** significant difference at 5% and 1% confidence level, respectively.
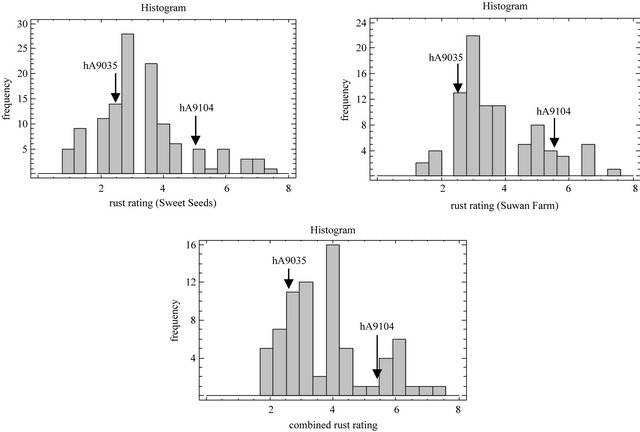
Figure 1. Histogram of rust ratings resistance based on 1 to 9 scale in which 1 = highly susceptible and 9 = highly resistant for a population of hA9104 × hA9035 sweet corn recombinant inbred lines evaluated at Sweet Seeds Research Station, Suwan Farm and combined across locations in 2008.
Table 3. Spearman rank correlation coefficients of rust rating between Sweet Seeds Station field and Suwan Farm in 89 RILs derived from a cross hA9104 × hA9035 in 2008.
P < 0.001; *Correlations at the 95.0% confidence level.
number of markers (23 markers) while chromosome 3 had the lowest number marker (10 markers). The order of mapped SSR markers was verified with the MaizeGDB map database.
3.3. Detected QTLs for Southern Corn Rust Resistance in RILs
To identify QTLs for southerm corn rust, we evaluated the RIL population in two locations, Sweet Seeds station field (Sweet Seeds), Saraburi and National Corn and Sorghum Research Center (Suwan Farm), Nakhonratchasima include combined analysis across these two locations. Analysis of linkage map and data of resistance for southern corn rust by MapQTL® v4.0 mapping software with MQM model was performed. Major and minor QTLs were classified by LOD score threshold: LOD score higher or equal to 3.0 is referred to as major QTL and LOD score higher than 2.2 but lower of 3.0 was designated as minor QTL.
Fifteen QTL regions associated with southern corn rust resistance were distributed over six chromosomes (chromosomes 1, 2, 5, 6, 9 and 10) (Figure 2). QTLs on chromosome 1, 5 and 6 were contributed by parent hA9104 whereas the other three chromosomes (2, 9 and 10) were contributed by parent hA9035. For Sweet Seeds, five QTLs were mapped on chromosomes 1, 2, 5, 6 and 10. In Suwan Farm, four QTLs were detected on chromosomes 1, 6, 9 and 10. When we combined the population from two locations, we found six QTLs located on chromosomes 1, 2, 5, 6, 9 and 10. A single QTL on
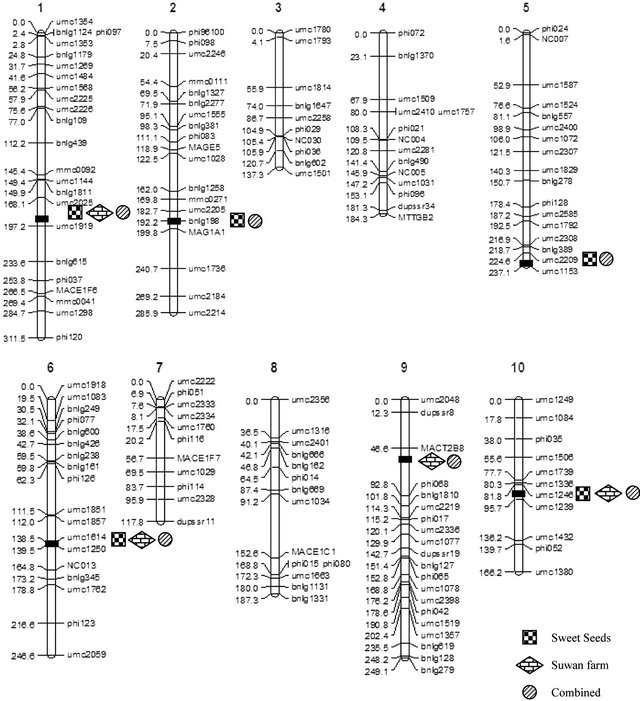
Figure 2. The position of QTLs conferring southern corn rust resistance of tropical sweet corn mapped from 89 RILs of F5:6 population from a cross hA9104 (resistant) and hA9035 (susceptible) under growth at Sweet Seeds, Suwan Farm include combined across two locations.
chromosome 9 showed the largest phenotypic variability with 41.8% under Suwan Farm field. The phenotypic variances of individual QTL ranged from 6.1% to 41.8%. Total phenotypic variances of detected QTLs in each location were estimated for 69.8%, 81.9% and 75.5% at Sweet Seeds, Suwan Farm and combined across locations. There were three stable QTLs were detected in both locations. One major QTL on chromosome 1 was flanked by markers umc2025 and umc1919 and explained 17.6% - 22.1% of phenotypic variation for rust rating of resistance. Two minor QTLs detected on chromosome 6 and 10 linked to the interval markers umc1614-umc1250 and umc1246-umc1239 and showed phenotypic variations of 7.0% - 7.4%and 15.1% - 22.0%, (Table 4).
4. Discussion
National Corn and Sorghum Research Center, Suwan Farm, located on Nakhonratchasima is an experimental cultivation area of corn has an average temperature of 25˚C - 30˚C and relative humidity of 75% - 95%, which is optimal for southern corn rust infection. Another oneat Sweet Seeds Co., Ltd or Sweet Seeds station field, located in Saraburi where cultivate commercial sweet corn is continually infected by southern corn rust. These two places have conditions that allow the spread of southern corn rust, so we decided to use them for the evaluation of resistance to southern corn rust in our RILS. We decided to use plants at 80 days after planting (DAP) for an investigation stage to evaluate the resistant level because the tendency of symptom virulence appeared maximum rate in mature stage. ATS-5, the hybrid line which is more susceptible to southern corn rust was sown as a spreader row to enhance the scattering of pathogen to experiment fields. Using of natural epiphytotic method without inoculation process could be done when the occurrence of disease has been frequently in experiment field [56,57].
Heritability of resistance for southern corn rust in Sweet Seeds and Suwan Farm were 82% and 76%, respectively. Combined analysis across two locations accounted for 87%. Interaction between genotype and environment (VGE) in combined across locations was 0.1353, suggesting that environmental variation of both
Table 4. Identification of QTLs for partial resistance to southern corn rust in hA9104 × hA9035 population in Sweet Seeds station field, Suwan Farm and combined across two locations.
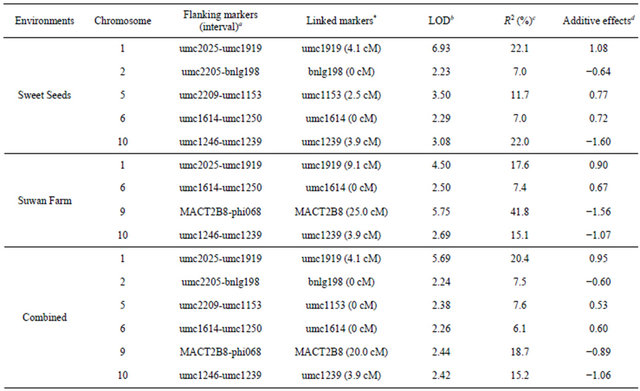
Total R2 of Sweet Seeds = 69.8%, total R2 of Suwan Farm = 81.9% and total R2 of combined across two locations = 75.5%; *The number showed in ( ) is genetic distance of QTL position placed to nearest marker; aNearest flanking markers linked with QTL regions; bDistance of chromosome position was measured from the closer marker to the maximum LOD peak of each QTL in CentiMorgan (cM) unit; cPercentage of phenotypic variance explain or PVE can explained by the given QTLs; dThe estimated additive effect calculated from estimated mean of the distribution (mu_) of the qualitative trait associated with genotype of A (hA9104) = mu_A and B (hA9035) = mu_B from formula (mu_A-mu_B)/2: “+” or positive = effect of allele of hA9104, “–” or negative = effect of allele hA9035.
locations slightly affected to genotypic variation. Heritability percentage of our mapping populations were categorized in high level [58]. The heritability of resistance for southern corn rust in maize has been studied and reported previously: 79% - 86% [36], 93% [37], 51% - 66% [12], and 53% - 68% [59]. We concluded that the inheritance of resistance for southern corn rust was attributable to the influence from genotypic variation more than environmental variation (VP = VG/VE). Breeders should be able to select suitable plant breeding method if they know heritability value of trait [60]. Selection e.g. mass selection, maternal line selection or even simple recurrent selection can be used in breeding of southern corn rust resistance.
In the past, Rpp9 gene was shown to effectively protect against the race PP.9 and other races. In fact, Rpp9, race-specific resistance gene was ineffective when multi races occurred. In 2010, there was a study to confirm the ineffectiveness of race-specific. Assessment of resistance was tested in Columbia, Hawaii, Nigeria, Philippines, Texas and Thailand using temperate Rpp-resistant line could not tolerance to southern corn rust in these areas [12].
In case of our study, we focused on partial resistance or race-nonspecific to disease in subtropical and tropical zone including Thailand, which are the source of multitude races of P. polysora [28]. The ineffectiveness of race-specific resistance to southern corn rust in tropical areas have been identified previously [25-26,29-32]. Moreover, partial resistance is more stable and and lasts longer than race-specific resistance [29].
QTL on chromosome 1 (1.06) showed the strongest signal in both locations referred to major QTL associated with resistance to southern corn rust. Our major QTL conferred partial resistance to southern corn rust on chromosome 1 distinguished to the results of earlier reports which major QTL of resistance to southern corn rust was located on short arm of chromosome 10 (bin 10.0) in different populations [2,10,23,32,36-37]. These QTLs were tightly linked with Rpp9 gene which resistance to southern corn rust race PP.9 and also adjacently linked to Rp1, common rust resistance gene, located on short arm of chromosome10 with 1.6 cM of genetic distance between these two resistance genes [18]. This discrepancy could be explained in a study previously reported [61]. They found that major QTL for resistance to common rust in F2:3 sweet corn (IL731 × W6786), IL731 derived from a cross IL677a and Golden Sensation mapped on chromosome 3 contradictory to the results of a study which determined that resistance to common rust in IL677a was controlled by a gene closely linked to Rp1 locus on chromosome 10 [62]. They proposed that due to different reactions of plant resistance mechanism between P. sorghi and RILs. IL677a and IL731 might have a broader based resistance to common rust than previously reported.
Mapping QTLs for partial resistance to southern corn rust was studied previously (Table 5) to be compared to our detected QTLs based on bin position on maize genome. QTL on bin 9.01 linked with phi 068 was similar QTL location linked with phi 28 [39], and on bin 10.03 linked with umc 1293 that there was one linked marker located on identical QTL region: umc130 [36]. Some of identified QTLs in this study were mapped to the same regions earlier described and some QTLs were located in different regions. The QTLs mapped to different loci possibly because of the difference in parents, mapping population, molecular markers, experimental environment and different mechanism of partial resistance. In the future, to use our identified markers accompany with earlier reported markers to screen other maize cultivars for resistance varieties to test the efficiency of markers could be actually useful in breeding program for sweet corn improvement to resistance southern corn rust disease.
Consideration of detected QTLs influencing to southern corn rust resistance and other published studies of QTL involved to resistance plant diseases were analyzed in each chromosome based on bin position which collected and reviewed [40] using a consensus map of IBM2 (B73 × Mo17) from MaizeGDB database. Maize chromosome 1 bin 1.06 is a region where a number of other QTLs for disease resistance genes were identified. For instance, QTL for resistance to Maize streak virus [63, 64], common smut caused by Ustilago maydis [65,66], common rust [67], grey leaf spot, Cercosspora zeaemaydis [68], and Northern corn leaf blight (NCLB), Exserohilum turcicum [69] were all located on chromosome 1. On bin 2.08, QTLs for resistance to Ustilago maydis, caused of common smut disease [66] and ear and stalk rots caused by Aspergillus flavus [70]. QTL located on chromosome 6 (6.04) which is also the location for other QTLs involved in diseases and pest resistance. For examples, two QTLs for Southwestern corn borer resistance (SWCB) on bin 6.04 [71], and on bin 6.06 [72], two QTLs for resistance to Fusarium moniliforme, an ear rot disease detected on bin 6.04 and 6.06 [73] or identified resistance genes, mdm1 for dwarf mosaic virus resistance (MDMV) [74], wsm1 resistance to wheat streak mosaic virus (WSMV) [75] and rhm1 conferring to resistance to Cochliobus heterostrophus, a fungul [76] etc. QTL for resistance to Aflatoxin (Aspergillus flavus) was detected on bin 9.01 [77]. On chromosome 10 (10.03) which this bin has been not reported for gene associated with resistance to disease.
Overall, some of our QTLs associated with resistance to southern corn rust were located on same QTL regions that have previously been reported while others were
Table 5. QTLs identified for resistance to southern corn rust were studied and reported previously.
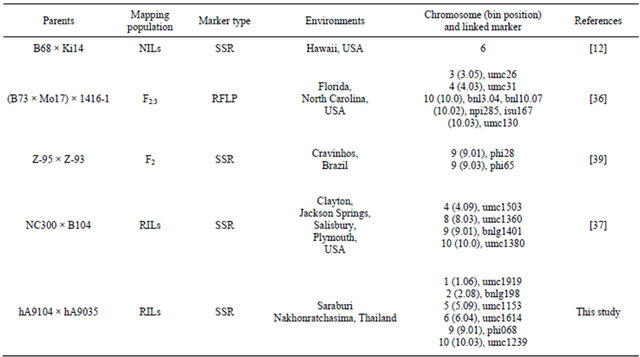
mapped to the same position of chromosomal segments involved in other plant disease resistance described earlier. The clustering of genes and QTLs involved in disease and insect resistance in maize are distributed over ten chromosomes. It is common to find two QTLs linked to different disease resistance in maize located on same chromosomal segments [78]. Studying the co-localization of disease QTLs in maize led to a better understanding of the genetic architecture of disease resistance in maize to analyze how relationship of resistance to disease among plant diseases.
The investigation of QTL conferring resistance to southern corn rust in maize through publication in the database showed that the genes were previously mapped to chromosome 2, 3, 4, 6, 8, 9 and 10 compared to our QTLs detected on chromosome 1, 2, 5, 6, 9 and 10. It seemed that novel major QTL on chromosome 1 was observed in this study. SSR markers linked with these QTLs associated with resistance to southern corn rust in maize were umc1919, umc1614 and umc1239. These markers will be useful in marker-assisted selection for improving resistance to southern corn rust across environments. Furthermore, the-development of gene specific marker such as single nucleotide polymorphism (SNP) could be performed in the future by narrowing down the region of interest through fine-scale mapping.
5. Acknowledgements
The research was financially supported by Thailand Research Fund (TRF) through The Royal Golden Jubilee Ph.D. Program (RGJ) and CPMO, NSTDA, Thailand.
REFERENCES
- R. N. Raid, S. P. Pennypacker and R. E. Stevenson, “Characterization of Puccinia polysora Epidemics in Pennsylvania and Maryland,” Phytopathology, Vol. 78, No. 5, 1988, pp. 579-585. doi:10.1094/Phyto-78-579
- Z. X. Liu, S. C. Wang, J. R. Dai, L. J. Huang and H. H. Cao, “Studies of Genetic Analysis and SSR Linked Marker Location of Gene Resistance to Southern Rust in Inbred Line P25 of Maize,” Acta Genetica Sinica, Vol. 30, No. 8, 2003, pp. 706-710.
- P. C. Agarwal, R. K. Khetarpal and M. M. Payak, “Polysora Rust of Maize Caused by Puccinia Polysora,” Indian Journal of Agricultural Sciences, Vol. 71, No. 4, 2001, pp. 275-276.
- R. H. Cammack, “Studies on Puccinia polysora Undrew. I: The World Distribution of Forms of Puccinia polysora,” Transactions of the British Mycological Society, Vol. 41, No. 1, 1959, pp. 89-94. doi:10.1016/S0007-1536(58)80011-X
- D. Rhind, J. M. Waterston and F. C. Deighton, “Occurrence of Puccinia polysora Underw. in West Africa,” Nature, Vol. 169, 1952, pp. 631-634. doi:10.1038/169631a0
- M. C. Futrell, “Puccinia Polysora Epidermics on Maize Associated with Cropping Practice and Genetic Homogeneity,” Phytopathology, Vol. 65, No. 9, 1975, pp. 1041- 1042. doi:10.1094/Phyto-65-1040
- R. Rodrigues-Ardon, G. E. Scott and J. F. Hennen, “Maize Yield Losses Caused by Southern Corn Rust,” Crop Science, Vol. 20, No. 6, 1980, pp. 812-814. doi:10.2135/cropsci1980.0011183X002000060035x
- S. J. Barker, “Testing of Maize Hybrids Resistant to Puccinia polysora on the Atherton Tableland, Queensland,” Journal of Agriculture and Animal Science, Vol. 26, No. 3, 1969, pp. 319-327.
- G. M. Reyes, “An Epidemic Outbreak of the Maize Rust in Eastern and Central Visayas, Philippines,” The Philippines Journal of Agriculture, Vol. 18, No. 1-4, 1953, pp. 115-128.
- C. X. Chen, Z. L. Wang, D. E. Yang, C. L. Ye, Y. B. Zhao, D. M. Jin, M. L. Weng and B. Wang, “Molecular Tagging and Genetic Mapping of the Disease Resistance Gene RppQ to Southern Corn Rust,” Theoretical and Applied Genetics, Vol. 108, No. 5, 2004, pp. 945-950. doi:10.1007/s00122-003-1506-7
- Y. Y. Liu and J. Wang, “Southern Corn Rust Occurred in Hebei Province in 1998,” Plant Protection, Vol. 25, 1999, pp. 53-58.
- J. L. Brewbaker, S. K. Kim, Y. S. So, M. Logrono, H. G. Moon, R. Ming, X. W. Lu and A. D. Josue, “Partial Resistance in Maize to Southern Rust (Puccinia polysora Undrew.),” Crop Science, Vol. 51, No. 4, 2011, pp. 1393- 1409. doi:10.2135/cropsci2010.06.0327
- J. K. Pataky and J. M. Headrick, “Relationships between Common Rust Incidence and Severity on a Susceptible and Partially Resistant Sweet Corn Hybrid,” Phytopathology, Vol. 78, No.9, 1988, pp. 1155-1160. doi:10.1094/Phyto-78-1155
- A. K. Ryland and H. H. Storey, “Physiological Races of Puccinia polysora Underw,” Nature, Vol. 176, 1955, pp. 655-656. doi:10.1038/176655b0
- H. H. Storey and A. K. Ryland, “Resistance to the Maize Rust P. polysora,” Nature, Vol. 173, 1954, pp. 778-779. doi:10.1038/173778a0
- H. H. Storey and A. K. Howland, “The Tropical Rust Disease of Maize Caused by Puccinia polysora Underw,” East African Agriculture and Forestry Research Organization, 1962, pp. 55-57.
- A. L. Robert, “Host Ranges and Races of the Corn Rusts,” Phytopathology, Vol. 52, 1962, pp. 1010-1012.
- A. J. Ullstrup, “Inheritance and Linkage of a Gene Determining Resistance in Maize to an American Race of Puccinia polysora,” Phytopathology, Vol. 55, 1965, pp. 425-428.
- H. H. Storey and A. K. Howland, “Resistance in Maize to the Tropical American Rust Fungus, Puccinia polysora Undrew. I: Genes Rpp1 and Rpp2,” Heredity, Vol. 11, 1957, pp. 289-301. doi:10.1038/hdy.1957.26
- H. H. Storey and A. K. Howland, “Resistance in Maize to the Tropical American Rust Fungus, Puccinia polysora Undrew. II: Linkage of Genes Rpp1 and Rpp2,” Heredity, Vol. 13, 1959, pp. 61-65. doi:10.1038/hdy.1959.4
- R. P. Scheffer and A. J. Ullstrup, “A Host-Specific Toxic Metabolite from Helminthosporium carbonum,” Phytopathology, Vol. 55, 1965, pp. 1037-1038.
- H. H. Storey and A. K. Howland, “Inheritance of Resistance in Maize to the Virus of Streak Disease in East Africa,” Annals of Applied Biology, Vol. 59, No. 3, 1967, pp. 429-436. doi:10.1111/j.1744-7348.1967.tb04459.x
- C. Zhou, C. Chen and P. Cao, “Characterization and Fine Mapping of RppQ, a Resistance Gene to Southern Corn Rust in Maize,” Molecular Genetics and Genomics, Vol. 278, No. 6, 2007, pp. 723-728. doi:10.1007/s00438-007-0288-z
- Y. Zhang, L. Xu, D. F. Zhang, J. R. Dai and S. C. Wang, “Mapping of Southern Corn Rust Resistance-Genes in W2D Inbred Line of Maize (Zea mays L.),” Molecular Breeding, Vol. 25, No. 3, 2010, pp. 433-439. doi:10.1007/s11032-009-9342-3
- J. E. Vanderplank, “Disease Resistance in Plants,” Academic Press, New York, 1968.
- P. Lindhout, “The Perspectives of Polygenic Resistance in Breeding for Durable Disease Resistance,” Euphytica, Vol. 124, No. 2, 2002, pp. 217-226. doi:10.1023/A:1015686601404
- J. E. Parlevliet, “Durability of Resistance against Fungal, Bacterial and Viral Pathogens: Present Situation,” Euphytica, Vol. 124, No. 2, 2002, pp. 147-156. doi:10.1023/A:1015601731446
- C. R. Carlos and A. S. Ferreira, “Variability in Isolates of Puccinia polysora in Brazil,” Fitopatologia Brasileira, Vol. 27, No. 4, 2002, pp. 414-416.
- B. Keller, C. Feuillet and M. Messmer, “Genetics of Disease Resistance,” In: A. J. Slusarenko, R. S. S. Fraser and L. C. Van Loon, Eds., Mechanisms of Resistance to Plant Diseases, Kluwar Academic Publishers, Dordrecht, Netherlands, 2001, pp. 101-160.
- A. Raoul and B. Robinson, “Horizontal Pathosystem Management,” In: A. Raoul and B. Robinson, Eds., Plant Pathosystems, Springer-Verlag, 1976, pp. 96-111.
- B. A. Bailey, W. Schuh, R. A. Frederiksen, A. J. Bockholt and J. D. Smith, “Identification of Slow-Rusting Resistance to Puccinia polysora in Maize Inbreds and Single Crosses,” Plant Disease, Vol. 71, No. 6, 1987, pp. 518- 521. doi:10.1094/PD-71-0518
- G. E. Scott and N. Zummo, “Effect of Genes with SlowRusting Characteristics on Southern Corn Rust in Maize,” Plant Disease, Vol. 73, 1989, pp. 114-116. doi:10.1094/PD-73-0114
- A. H. Paterson, E. S. Lander, J. D. Hewitt, S. Petersons, E. Lincoln and S. D. Tanksley, “Resolution of Quantitative Traits into Mendelian Factors Using a Complete Linkage Map of Restriction Fragment Length Polymorphisms,” Nature, Vol. 335, No. 6192, 1988, pp. 721-726. doi:10.1038/335721a0
- N. D. Young, “QTL Mapping and Quantitative Disease Resistance in Plants,” Annual Review of Phytopathology, Vol. 34, 1996, pp. 479-501. doi:10.1146/annurev.phyto.34.1.479
- G. E. Scott, S. B. King and J. W. Armour, “Inheritance of Resistance to Southern Corn Rust in Maize Population,” Crop Science, Vol. 24, No. 2, 1984, pp. 265-267. doi:10.2135/cropsci1984.0011183X002400020013x
- J. B. Holland, D. V. Uhr, D. Jeffers and M. M. Goodman, “Inheritance of Resistance to Southern Corn Rust in Tropical-by-Corn-Belt Maize Populations,” Theoretical and Applied Genetics, Vol. 96, No. 2, 1998, pp. 232-241. doi:10.1007/s001220050732
- M. P. Jines, P. Balint-Kurti, L. A. Robertson-Hoyt, T. Molnar, J. B. Holland and M. M. Goodman, “Mapping Resistance to Southern Rust in a Tropical by Temperate Maize Recombinant Inbred Topcross Population,” Theoretical and Applied Genetics, Vol. 114, No. 4, 2007, pp. 659-667. doi:10.1007/s00122-006-0466-0
- J. C. Jiang, G. O. Edmeades, I. Armstead, H. R. LaWtte, M. D. Hayward and D. Hoisington, “Genetic Analysis of Adaptation Differences between Highland and Lowland Tropical Maize Using Molecular Markers,” Theoretical and Applied Genetics, Vol. 99, No. 7-8, 1999, pp. 1106- 1119. doi:10.1007/s001220051315
- K. R. Brunelli, H. P. Silva and L. E. Aranha-Camargo, “Mapeamento de Genes de Resistencia Quantitativa a Puccinia polysora em Milho,” Fitopatologia Brasileira, Vol. 27, No. 2, 2002, pp. 134-140. doi:10.1590/S0100-41582002000200003
- R. J. Wisser, P. J. Balint-Kurti and R. J. Nelson, “The Genetic Architecture of Disease Resistance in Maize: A Synthesis of Published Studies,” Phytopathology, Vol. 96, No. 2, 2006, pp. 120-129. doi:10.1094/PHYTO-96-0120
- StatPoint Technologies Inc, “STATGRAPHICS Centurion XVI User Manual,” StatPoint Technologies, USA, 2009.
- International Rice Research Institute, “IRRISTAT for Windows Version 5.0,” Makati City, 2005.
- S. O. Rogers and A. J. Bendich, “Extraction of DNA from Plant, Fungal and Algal Tissues,” In: S. B. Gelvin and R. A. Schilperoort, Eds., Plant Molecular Biology Manual, Kluwer Academic Publishers, Boston, 1994, pp. 1-8.
- H. Akagi, Y. Yokozeki, A. Inagaki and T. Fujimura, “Microsatallite DNA Markers for Rice Chromosomes,” Theoretical and Applied Genetics, Vol. 93, No. 7, 1996, pp. 1071-1077. doi:10.1007/BF00230127
- B. J. Bassam, G. C. Anollés and P. M. Gresshoff, “Fast and Sensitive Silver Staining of DNA in Polyacrylamide Gels,” Analytical Biochemistry, Vol. 196, No. 1, 1991, pp. 80-83. doi:10.1016/0003-2697(91)90120-I
- D. F. Austin and M. Lee M, “Comparative Mapping in F2:3 and F6:7 Generations of Quantitative Trait Loci of Grain Yield and Yield Components in Maize,” Theoretical and Applied Genetics, Vol. 92, No. 7, 1996, pp. 817-826. doi:10.1007/BF00221893
- S. E. Lincoln, M. Daly and E. S. Lander, “Constructing Genetic Maps with MAPMAKER/EXP 3.0,” 3rd Edition, Whitehead Institute for Biomedical Technical Report, Cambridge, Mass, 1992.
- X. Chen, S. Temnykh, Y. Xu, Y. G. Cho and S. R. McCouch, “Development of a Microsatellite Framework Map Providing Genome-Wide Coverage in Rice (Oryza sativa L.),” Theoretical and Applied Genetics, Vol. 95, No. 4, 1997, pp. 553-567. doi:10.1007/s001220050596
- D. D. Kosambi, “The Estimation of Map Distance from Recombination Values,” Annals of Eugenics, Vol. 12, No. 1, 1944, pp. 172-175.
- R. E. Voorrips, “MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs,” Heredity, Vol. 93, No. 1, 2002, pp. 77-78. doi:10.1093/jhered/93.1.77
- J. W. Van Ooien, M. P. Boer, R. C. Jansen and C. Maliepaard, “MapQTL® 4.0: Software for the Calculation of QTL Positions on Genetic Map,” Plant Research International, Wageningen, 2002.
- R. W. Doerge and G. A. Churchill, “Permutation Tests for Multiple Loci Affecting a Quantitative Epidemics in Pennsylvania and Maryland,” Genetics, Vol. 78, No. 1, 1996, pp. 579-585.
- Y. Cho, V. N. Njiti, X. Chen, K. Triwatayakorn, M. A. Kassem, K. Meksem, D. A. Lightfoot and A. J. Wood, “Quantitative Trait Loci Associated with Foliar Trigonelline Accumulation in Glycine max L.,” Journal of Biomedicine and Biotechnology, Vol. 2, No. 3, 2002, pp. 151-157. doi:10.1155/S1110724302204039
- S. Sriphadet, C. J. Lambrides and P. Srinives, “Inheritance of Agronomic Traits and Their Interrelationship in Mungbean Vigna radiata (L.) Wilczek,” Journal of Crop Science and Biotechnology, Vol. 10, No. 4, 2007, pp. 123-132.
- C. M. Rick and P. G. Smith, “Novel Variation in Tomato Species Hybrids,” The American Naturist, Vol. 87, No. 837, 1953, pp. 359-373.
- R. R. Nelson, “The Meaning of Disease Resistance in Plant,” In: R. R. Nelson, Ed., Breeding Plants for Disease Resistance: Concepts and Applications, The Pennsylvania State University Press, Harrisburg, 1973, pp. 13-25.
- G. E. Russell, “Plant Breeding for Pest and Disease Resistance,” Butterworths, London, 1978.
- H. F. Robinson, R. E. Comstock and P. H. Harvey, “Genotypic and Phenotypic Correlations in Corn and Their Implications in Selection,” Agronomy Journal, Vol. 43, 1949, pp. 282-287.
- C. J. Hee and Y. Takeo, “Cell Wall Contents and Estimation of Genetic Parameters for Polysora Rust Resistance in Tropical Maize (Zea mays L.),” Journal of the Faculty of Agriculture Kyushu University, Vol. 56, No. 2, 2011, pp. 231-236.
- [61] A. R. Hallauer and J. B. Mirinda, “Quantitative Genetics in Maize Breeding,” Iowa State University Press, Ames, 1988.
- [62] A. F. Brown, J. A. Juvik and J. K. Pataky, “Quantitative Trait Loci in Sweet Corn Associated with Partial Resistance to Stewart’s Wilt, Northern Corn Leaf Blight, and Common Rust,” Phytopathology, Vol. 91, No. 3, 2001, pp. 293-300. doi:10.1094/PHYTO.2001.91.3.293
- [63] S. K. Kim and J. L. Brewbaker, “Inheritance of Resistance of Sweet Corn Inbred IL677a to Puccinia sorghi Schw,” Hortscience, Vol. 22, No. 6, 1987, pp. 1319-1320.
- [64] A. Pernet, D. Hoisington, J. Franco, M. Isnard, D. Jewell, C. Jiang, J. L. Marchand, B. Reynaud, J. C. Glaszmann and D. González de León, “Genetic Mapping of Maize Streak Virus Resistance from the Mascarene Source I: Resistance in Line D211 and Stability against Different Virus Clones,” Theoretical and Applied Genetics, Vol. 99, No. 3-4, 1999, pp. 524-539. doi:10.1007/s001220051266
- [65] A. Pernet, D. Hoisington, J. Dintinger, D. Jewell, C. Jiang, M. Khairallah, P. Letourmy, J. L. Marchand, J. C. Glaszmann and D. González de León, “Genetic Mapping of Maize Streak Virus Resistance from the Mascarene Source II: Resistance in Line CIRAD390 and Stability across Germplasm,” Theoretical and Applied Genetics, Vol. 99, No. 3, 1999, pp. 540-553. doi:10.1007/s001220051267
- [66] M. R. Kerns, J. W. Dudley and G. K. Rufener, “QTL for Resistance to Common Rust and Smut in Maize,” Maydica, Vol. 44, 1999, pp. 37-45.
- [67] T. Lübberstedt, D. Klein and A. E. Melchinger, “Comparative QTL Mapping of Resistance to Ustilago maydis Across Four Populations of European Flint-Maize,” Theoretical and Applied Genetics, Vol. 97, No. 8, 1998, pp. 1321-1330. doi:10.1007/s001220051025
- [68] T. Lübberstedt, D. Klein and A. E. Melchinger, “Comparative Quantitative Trait Loci Mapping of Partial Resistance to Puccinia sorghi Across Four Populations of European Flint Maize,” Phytopathology, Vol. 88, No. 12, 1998, pp. 1324-1329. doi:10.1094/PHYTO.1998.88.12.1324
- [69] A. Lehmensiek, A. M. Esterhuizen, D. Van-Staden, S. W. Nelson and A. E. Retief, “Genetic Mapping of Gray Leaf Spot (GLS) Resistance Genes in Maize,” Theoretical and Applied Genetics, Vol. 103, No. 5, 2001, pp. 797-803. doi:10.1007/s001220100599
- [70] A. L. Dingerdissen, H. H. Geiger, M. Lee, A. Schechert and H. G. Welz, “Interval Mapping of Genes for Quantitative Resistance of Maize to Setosphaeria turcica, cause of Northern Leaf Blight, in a Tropical Environment,” Molecular Breeding, Vol. 2, No. 2, 1996, pp. 143-156. doi:10.1007/BF00441429
- [71] K. N. Busboom and D. G. White, “Inheritance of Resistance to Aflatoxin Production and Aspergillus Ear Rot of Corn from the Cross of Inbreds B73 and Oh516,” Phytopathology, Vol. 94, No. 10, 2004, pp. 1107-1115. doi:10.1094/PHYTO.2004.94.10.1107
- [72] M. M. Khairallah, M. Bohn, C. Jiang, A. Deutsch, D. C. Jewell, J. A. Mihm, A. E. Melchinger, D. Gonzalez-deLeon and D. A. Hoisington, “Molecular Mapping of QTL for Southwestern Corn Borer Resistance, Plant Height and Flowering in Tropical Maize,” Plant Breeding, Vol. 117, No. 4, 1998, pp. 309-318. doi:10.1111/j.1439-0523.1998.tb01947.x
- [73] S. Groh, D. Gonzalez-de-Leon, M. M. Khairallah, C. Jiang, D. Bergvinson, M. Bohn, D. A. Hoisington and A. E. Melchinger, “QTL Mapping in Tropical Maize III: Genomic Regions for Resistance to Diatraea spp. and Associated Traits in Two RIL Populations,” Crop Science, Vol. 38, No. 4, 1998, pp. 1062-1072. doi:10.2135/cropsci1998.0011183X003800040030x
- [74] S. Perez-Brito, D. Jeffers, D. Gonzalez-de-Leon, M. Khairallah, G. Cortes-Cruz, G. Velasquez-Cardelas, S. Azpiroz-Rivero and G. Srinivasan, “QTL Mapping of Fusarium moniliforme Ear Rot Resistance in Highland Maize, Mexico,” Agrociencia, Vol. 35, 2001, pp. 181- 196.
- [75] K. D. Simcox, M. D. McMullen and R. Louie, “Co-Segregation of the Maize Dwarf Mosaic Virus Resistance Gene, Mdm1, with the Nucleolus Organizer Region in Maize,” Theoretical and Applied Genetics, Vol. 90, No. 3-4, 1995, pp. 341-346. doi:10.1007/BF00221975
- [76] M. D. McMullen and R. Louie, “Identification of a Gene for Resistance to Wheat Streak Mosaic Virus in Maize,” Phytopathology, Vol. 81, No. 6, 1991, pp. 624-627. doi:10.1094/Phyto-81-624
- [77] D. Zaitlin, S. DeMars and Y. Ma, “Linkage of rhm, a Recessive Gene for Resistance to Southern Corn Leaf Blight, to RFLP Marker Loci in Maize (Zea mays L.) Seedlings,” Genome, Vol. 36, No. 3, 1993, pp. 555-564. doi:10.1139/g93-076
- [78] C. Paul, G. Naidoo, A. Forbes, V. Mikkilineni, D. White and T. Rocheford, “Quantitative Trait Loci for Low Aflatoxin Production in Two Related Maize Populations,” Theoretical and Applied Genetics, Vol. 107, No. 2, 2003, pp. 263-270. doi:10.1007/s00122-003-1241-0
- [79] M. D. McMullen and K. D. Simcox, “Genomic Organization of Disease and Insect Resistance Genes in Maize,” Molecular Plant Microbe Interaction, Vol. 8, No. 6, 1995, pp. 811-815.
NOTES
*Corresponding author.

