American Journal of Plant Sciences
Vol.3 No.2(2012), Article ID:17592,12 pages DOI:10.4236/ajps.2012.32034
Solanaceae Evolutionary Dynamics of the I2-NBS Domain
![]()
Department of Soil, Plant, Environmental and Animal Production Sciences, University of Naples Federico II, Portici, Italy.
Email: ercolano@unina.it
Received October 3rd, 2011; revised November 15th, 2011; accepted November 29th, 2011
Keywords: DNA Polymorphism; Phylogenetic Analysis; R Genes; Selection Mechanism; Tomato; Wild Species
ABSTRACT
In Solanaceae family several plant resistant genes to pathogen (R genes) have been mapped and cloned. Most of them encode Nucleotide Binding Site Leucine Rich Repeat domain (NBS-LRR) protein. However, little is known about the resistance genes variability pattern and the evolutionary process acting on different species belonging to the same family. The aims of the present work, was to genotype and study the evolutionary relationship of fifty wild tomato accessions using the I2 resistance gene sequences. Thirty-three new candidate homologues I2 resistance gene nucleotide sequence were obtained from wild tomato species. Nucleotide polymorphisms in I2-NBS domain was detected in wild tomato species: diversity could have accumulated over a long time and species sorting could have produced new variants. In order to study the NBS-LRR domain variability we analyzed the evolution process acting on the amino acid sequence. The FEL method (codon Model) based on dN/dS, was used to estimate the presence of positive, negative and neutral selection acting on each codon. The I2-NBS domain sequence data studied seems to be under a general purification process of evolution. However, intermittent bouts of positive selection sites were detected in high variable regions. Phylogenetic analysis conducted within the Solanaceae family shows that the Solanum genus is under a rapid adaptative divergence process and Nicotiana and Capsicum clustered separately; Solanum peruvianum, in particular, displayed to be the most polymorphic specie. These results might be important for the identification of new sources of resistance genes to tomato pathogens.
1. Introduction
Resistance to pathogens attack in plant is regulated by genes that confer resistance to a specific pathogen. The relationship between plant and pathogen involves plant resistance genes (R genes) and pathogen avirulence genes (Avr-genes) [1]. Plant resistance R genes, enable the plant to recognize the presence of specific pathogens and initiate defense responses [2]. The R gene structure plays a key role in resistance, encoding, sometimes, specialized receptors that recognize the corresponding avirulence (Avr) produced by pathogens [3,4]. Most of the R genes belong to an ancient family that encodes proteins with nucleotide-binding site (NBS) and Leucine-Rich-Repeat (LRR) domains [5-7] and, therefore, can also be detected in silico [8]. The NBS shows to be quite useful to localize, identify and study resistance genes through cloning strategies based on the PCR approach allowing them to be retrieved in public databases [9,10].
The ability of plants to evolve under biotic pressure requires in-depth understanding of R gene organization and evolution. Domestication and early breeding led to the loss of some important features including resistances to diseases and pests. Therefore the attention of breeders turned to the wild relatives of cultivated plants [11]. Important sources of resistance genes to tomato pathogens are reported in wild species [12]. The I2 gene, identified in S. pimpinellifolium (L. pimpinellifolium), encodes an NBS-LRR protein that confers resistance to race 2 of Fusarium oxysporum f. sp. lycopersici, the causative agent of vascular wilt disease in many Solanum species [13]. The I2-locus is localized in the long arm of chromosome 11 and contains at least 7 members. One member (I2C-1) confers partial resistance to race 2 of pathogen and another member (I2C-K) complete resistance [13,14]. It is likely that other Solanaceae species harbor R genes related to I2. For example, the R3a gene on S. demissum chromosome 11 is an I2 homolog conferring resistance to Phytophthora infestans [15]. The exploration of the natural biodiversity in wild tomato species could have a critical role in discovering other genetic source of resistance gene to overcome the new pathogen variants. This work was designed to achieve several aims: identify resistance gene homolog (RHGs) in wild tomato species, analyzing the polymorphism level of the I2-NBS; secondly, to evaluate models of I2 gene evolution and selection acting on specific amino acids; finally, to identify essential residues that might be involved in the plant evolution of new specificities or in regulating defense activation.
2. Material and Methods
2.1. Plant Material and DNA Extraction
Fifty wild tomato accessions, belonging to six different species were analyzed for the presence of I2 gene homologs. In particular the Solanum lycopersicon species explored in this study are: Solanum peruvianum (L. peruvianum), Solanum pimpinellifolium (L. pimpinellifolium), Solanum habrochaites (L. hirsutum), Solanum chilense (L. chilense), Solanum corneliomulleri (L. glandulosum) and Solanum neorickii (L. parviflorum) (Table 1). In Table 1 is also reported the nomenclature recently revisited in 2005 [16]. Seeds were kindly provided by the CNR-IGV. About ten plants per accession were planted. The DNA was extracted from leaves 4- to 5-week-old plants [17].
2.2. PCR and Sequencing of the I2-NBS Domain in Wild Tomato Species
The I2 gene (AF118127) was analyzed with InterProScan tool (http://www.ebi.ac.uk/InterProScan/) to identify resistance gene domains. The NBS domain-specific primers were designed with Primer3 (http://frodo.wi.mit.edu/cgibin/primer3/primer3_www.cgi):
I2F: CTGAAGGATTTGATGCTTTG
I2R: GTCTTCCGACCTCTTCAAGT
PCR was executed with 25 ng of genomic DNA, 10 pmol primers, 1 U of Taq DNA polymerase Kit (Invitrogen), 10 pmol dNTPs, 2 mM MgCl2 in 25 μl reaction volumes. Amplification was performed using the following cycling conditions: 1 min 94˚C, followed by cycles 30 of 1 min at 94˚C, 1min and 30 sec at 60˚C and 2 min at 72˚C and 7 min at 72˚C. The PCR products were separated on 1.5% agarose gels and purified using High Pure PCR product purification Kit (Roche). Sequence reaction was made according to ABI PRISMR BigDye™ Terminator v 3.1 Ready reactions Cycle Sequencing Kit protocol. The samples were purified using 1/10 vol of Sodium acetate 3 M, 2.5 vol of ethanol (95%). ABI PRISMR 3100-Avant Genetic Analyzer was used for the sequenceing. All sequences were repeated three times to avoid any PCR artifacts. Low quality sequences were excluded. Identical sequences were recorded and one representative was used for phylogenetic analysis. All the unique sequences have been deposited into GenBank with accession numbers HM101253-HM101274.
2.3. Analysis of Intraspecific/Interspecific Level of Variability
Divergence between different population/species and the I2 reference gene was evaluated using DnaSP 3.0 [18]. The divergence is expressed as the number of net nucleotide substitutions per site among different populationns (Da) along the nucleotide alignment sequence.
2.4. Dataset Building and Multiple Alignments
A nucleotide Solanaceae species dataset was produced by downloading all the published Resistance Gene Analogues sequences (RGA) related to I2 gene, removing all marker sequences, repeat sequences, low-quality sequences, pseudo-genes and sequences not closely linked to the NBS-LRR resistance gene domain. Putative homolog genes were identified aligning the sequences with I2 reference gene. Only DNA sequences without stop codons were used for subsequent analysis. The distribution of I2 sequences was assessed using Megablast. Searches were performed on 23 May 2007. GenBank I2 sequences of Solanaceae species showed an identity >90%. The new I2 sequences were added to the RGA Solanaceae dataset (Table 2). A two letters code was added to the GenBank accession number, to simplify species identity (NT = Nicotiana tabacum, SM = Solanum melongena, LE = Lycopersicum esculentum, CA = Capsicum annuum,

Table 1. List of wild lycopersicon species planted and studied for the I2-NBS domain. The traditional and revised nomenclature [35] is also reported. Data related to the geographic origin and altitude are also described.
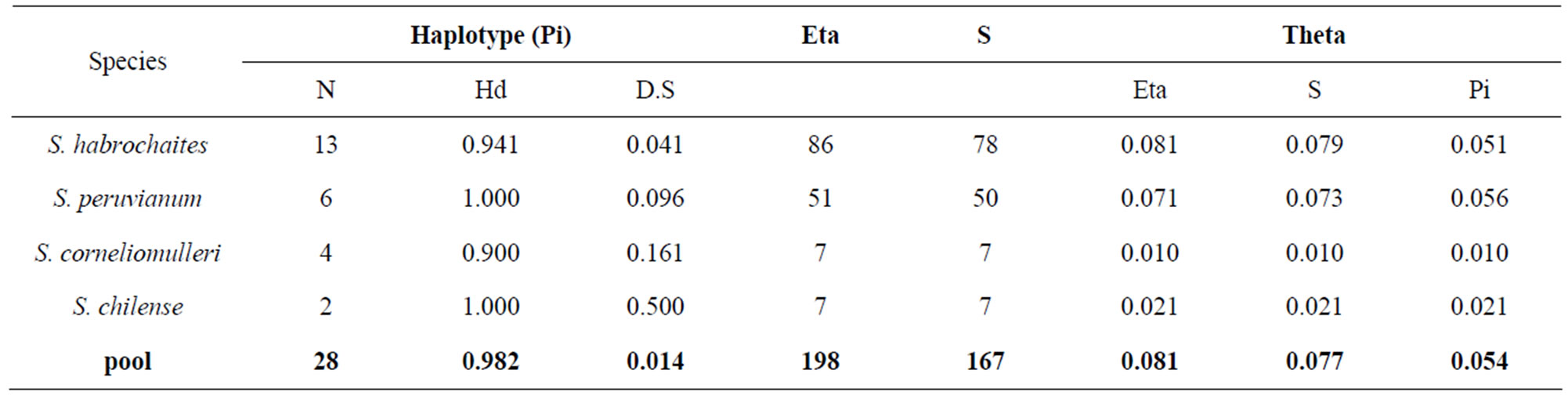
Table 2. Intra-specific polymorphism analysis of the I2-NBS domain. Variability is expressed as N (number of haplotype), Hd (haplotype diversity), Eta (number of single nucleotide polymorphisms), S (number of segregating sites), and Pi (nucleotide differences per site). The standard deviation is also indicated (D.S.) S. neorickii and S. pimpinellifolium were not included in this analysis because not enough sequences were collected.
ST = Solanum tuberosum, SC = Solanum caripense). ExPASy (Expert Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics (SIB) was used to translate all nucleotide sequences into amino acid sequences. The I2 amino acid sequence (reference gene) was used as template to identify the correct gene frame shift.
2.5. Multiple Alignments
All multiple alignments were generated using ClustalW [19] applying the default settings and manually edited with Bioedit 7.0 [20].
The nucleotide multiple alignment derived from the homolog I2 genes identified in wild tomato species was used as a backbone to align the corresponding amino acid datasets. Pairwise comparisons and multiple alignment were performed using MEGA3 [21] and Bioedit 7.0. A unique sequence dataset for nucleotide and amino acid sequences was generated. DnaSP 4.0 [18,22] was used to explore nucleotide polymorphisms in our wild tomato sequences.
2.6. Phylogenetic Analysis
Nucleotide phylogenetic analysis was conducted on I2 dataset. A unique sequences dataset was obtained using phylogenetic nucleotide trees analysis. GTR evolutionary model [23] using the PHYML v2.4.4 program [24] was applied. Non-parametric bootstrap [23] was performed to test the robustness of the tree topologies (1000 replicates). Trees were visualized with the Geneious software (Copyright © 2005-2007 Biomatters Ltd.). The JTT + T model was selected by Protest [25] and was used as the evolutionary model setting for the PHYML v 2.4.4 program [24] to create the phylogenetic amino acid tree (1000 bootstrap replicates).
2.7. Amino Acid Site Evolution
Selective pressure acting on the NBS-I2 resistance gene domain was investigated evaluating non-synonymous to synonymous substitution ratio, dN/dS. Tests were conducted to estimate the evolution of each codon: positive (ω > 1); neutral (ω = 1); and negative (ω < 1). The likelihood approach [26] was used. Codon substitution analysis [27] was calculated using DataMonkey-FEL model (codon Model, Log Likewood of –3649.6670) (http://ww w.datamonkey.org/) [28] using the 3rd frame (according to the resistance gene frame).
3. Results
3.1. Intraspecific and Interspecific I2 Variability in Wild Tomato Species
A specific PCR approach on I2-NBS domain was chosen to identify homolog genes in wild tomato species. A total of 33 wild tomato accessions were successfully sequenced and the data were used for the following analysis. Twenty-two unique sequences were submitted in GenBank database as reported in material and methods. A total of 11.187 nucleotides for I2 gene were obtained. BlastN analysis reveals for each wild tomato species at least 96% identity with the tomato reference gene. Multiple alignment of single fragments 339 bp long was performed evidencing several polymorphic sites (data not showed). The initial alignment required manual editing in order to minimize gaps. Figure 1 shows the nucleotide variability trend of each species compared to the I2 reference gene. All species (except S. neorickii and S. pimpinellifolium) showed a low level of variability trend from the nucleotide 0 to 143 and a general ipervariable nucleotide region (from nucleotide 163 to 323). In particular, two maximum peaks at 163 and 263 were identified. Intraspecific polymorphism level was evaluated for 4 species. Table 2 reports the following data: number of haplotypes, haplotype diversity (Hd), Eta (n. of single nucleotide polymorphisms), number of segregating sites (S) and finally the number of nucleotide differences per site (Pi). According to our results S. peruvianum showed
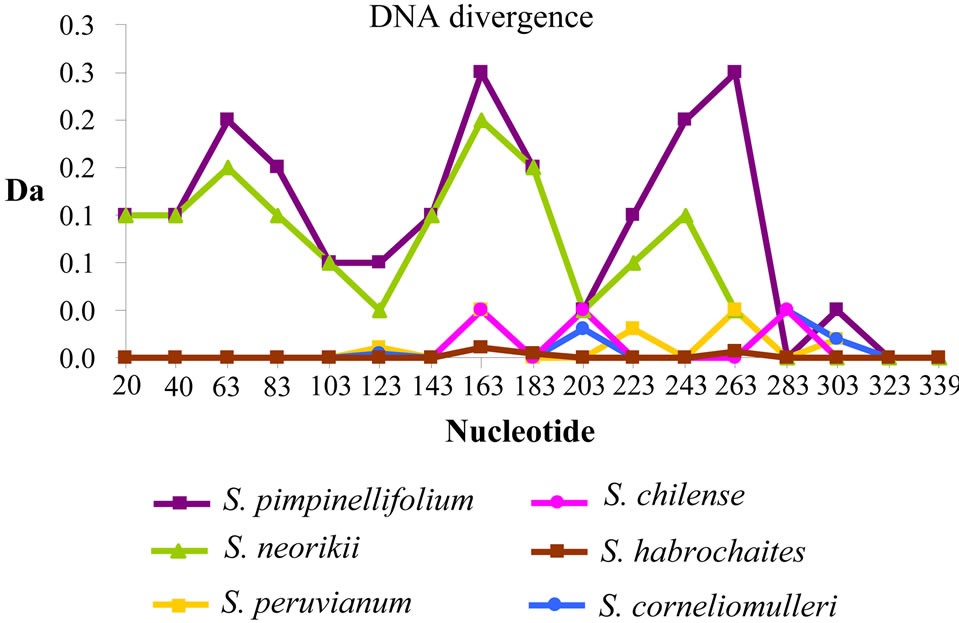
Figure 1. Analysis of DNA divergence among wild tomato species sequences. Each species is compared with the I2 reference gene sequence. The divergence is expressed as the number of net nucleotide substitutions per site between populations (Da). The nucleotide positions are shown in ordinate.
the highest score of intra-specific variability and S. habrochaites showed the highest score for variability expressed as number of segregating sites and number of single nucleotide polimorphisms.
3.2. Solanaceae I2-NBS Sequences Catalogues
BlastN analysis against the NCBI database was conducted in order to collect the Solanaceae sequences with the highest level of identity closely related to the I2-NBS reference gene. A total of 84 published Solanaceae RGAs were retrieved. Table 3 reports the homologs I2-NBS identified in 13 Solanaceae species, included 6 wild tomato species. The 84 Solanaceae nucleotide sequences related to the I2 resistance gene, downloaded from the NCBI database, comprised: 15 sequences from L. esculentum (S. lycopersicum), 6 from S. melongena, 37 from N. tabacum, 8 from S. tuberosum, 5 from S. caripense, 9 from C. annuum, 19 from S. habrochaites (L. hirsutum), 5 from S. corneliomulleri (L. glandulosum), 6 from S. peruvianum (L. peruvianum), 2 from S. pimpinellifolium (L. pimpinellifolium), 2 from S. chilense (L. chilense), 1 from S. neorickii (L. parviflorum). From the GenBank dataset we detected just three wild tomato I2 resistance sequences from S. pimpinellifolium sequences, (the wild donor species of the I2 resistance gene; AF118127; AF408704), and S. habrochaites (AF534287). With our work we produced 33 new wild tomato I2-NBS domain sequences: 18 new sequences of S. habrochaites (11 of them were unique); 6 unique sequences of S. peruvianum; 5 of S. corneliomulleri (4 were unique); 2 of S. chilense, 1 of S. neorickii; and finally 1 sequence of S. pimpinellifolium. All the I2-NBS Solanaceae homolog sequences have an identity of at least 90% with the reference gene. In particular, the sequence of S. pimpinellifolium I2C-5
(AF408704) and S. lycopersicum I2C-2 resistance (AF00 4879) showed 99% identity with the reference gene previously described as members of the I2 cluster locus, as did the potato late blight resistance protein R3a gene (AY849382). A nucleotide alignment of the NBS domain (NBS-I2-ALN) unique sequences, 25 derived from our work and 56 from NCBI database, was created. The NBS-I2-ALN is 336 positions long with 22 gaps. In particular 6 gaps (2 codons) are typically shown only in 4 C. annuum genotypes (DQ205996; DQ206012; DQ205 998; DQ205980) from position number 38 to 44. Two tomato accessions (AF004879 and AF534287) showed 3 gaps at the same positions (51 - 53); one N. tabacum and 2 S. melongena genotypes (respectively DQ206210, DQ20 6026 and DQ206073) displayed 6 gaps at positions 69 - 78 (data not shown). Nucleotide sequences were translated into amino acid sequences according to the frame of the reference resistance gene. The new dataset of 67 unique protein sequences was used for further analysis.
3.3. Phylogenetic Analysis of Nucleotide Sequences
Phylogenetic analysis based on nucleotide sequences was conducted on NBS-I2-ALN using a likelihood test. Phylogenetic nucleotide trees were inferred using the GTR evolutionary model. A low level of distance in term of nucleotide substitution among all accessions was found (as indicated in the bar). The reliability of tree was then established by conducting 1000 bootstrap replications. The resulting phylogenetic tree can be studied, designnating 9 main groups (A-I) (Figure 2). Most of the wild tomato sequences cluster in group A. This dataset contains only Solanum sequences (including the reference functional gene I2). Group B is defined by five wild tomato species sequences originating herein. Cluster C includes potato late blight resistance protein R3a gene (STAY849382) and the tomato I2 paralogues (LEAF 004879) that cannot be easily positioned. The cluster D is characterized by only pepper (C. annuum) sequences, (bootstrap value 999). Group E includes few Solanum sequences difficult to cluster and one of N. tabacum. Finally, groups F, G, H, I contain: only N. tabacum sequences.
3.4. Analysis of Selection on Individual Codons within the NBS Domain
In order to study the NBS-LRR domain variability we analyzed the type of evolution process acting on the amino acid sequence. The FEL method (codon Model) was used to estimate the presence of positive, negative and neutral selection acting on each codon. The analysis of selection on individual codons suggested a general process of purification. Figure 3 reports a graphical representation of the ω trend. Most sites are under negative
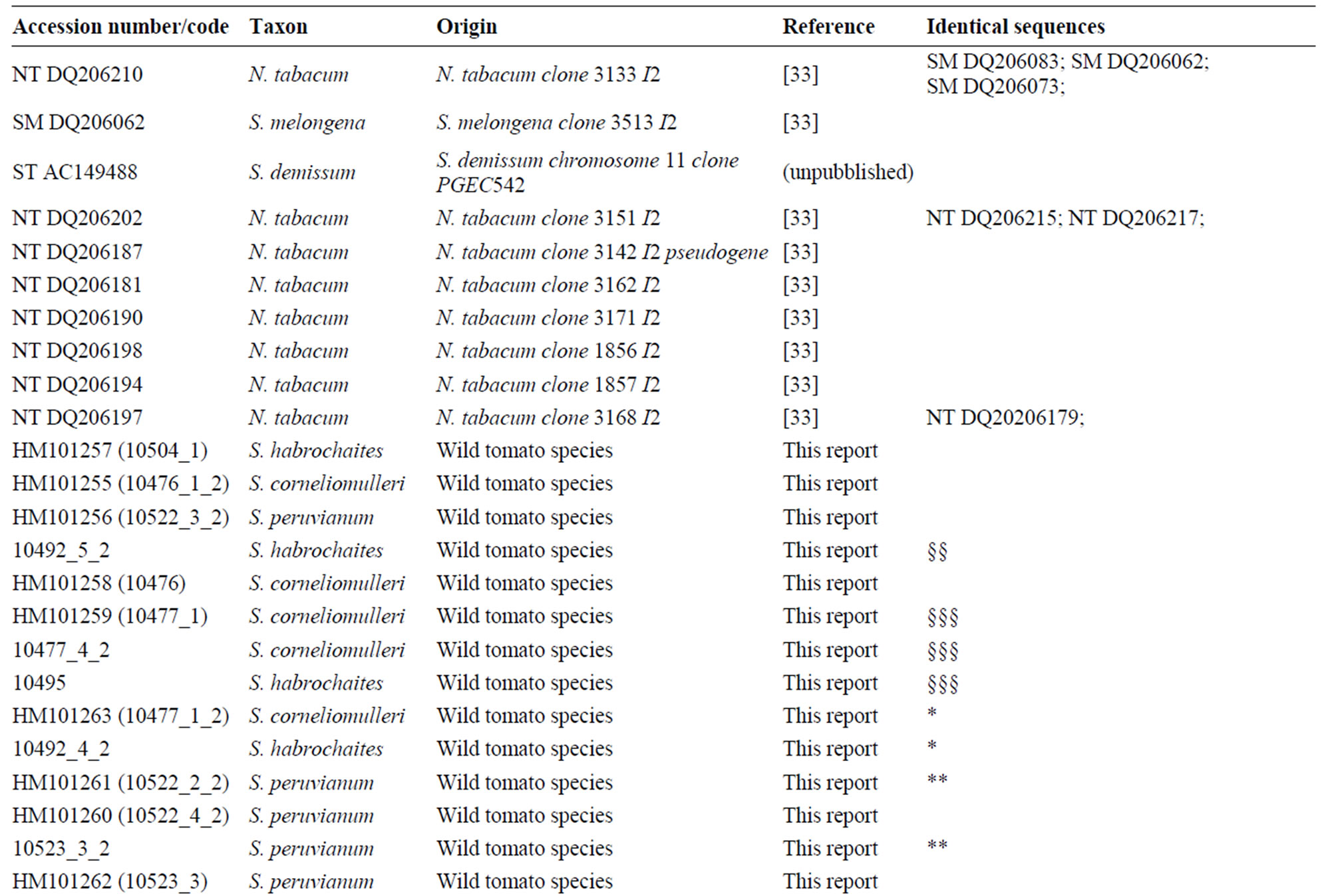
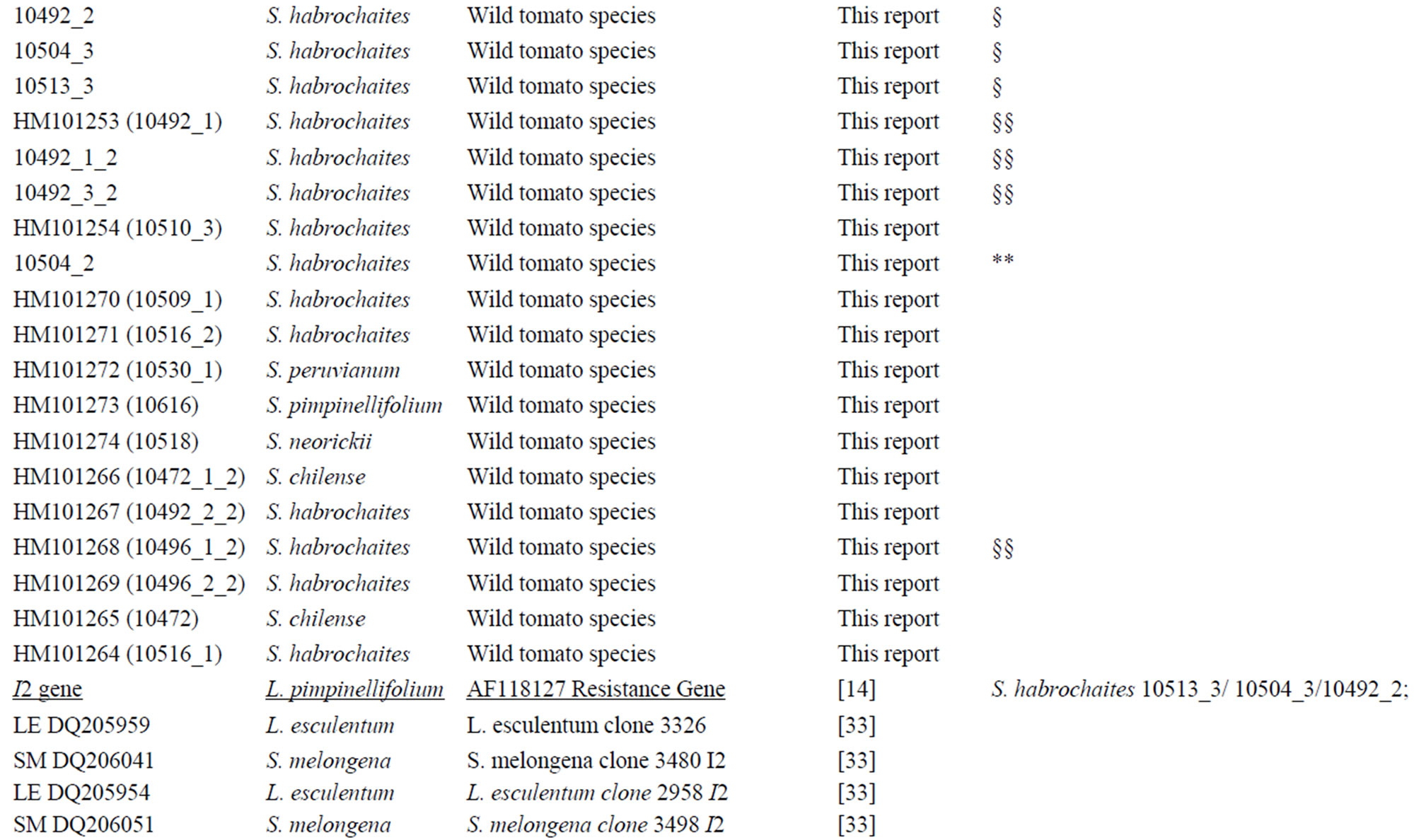
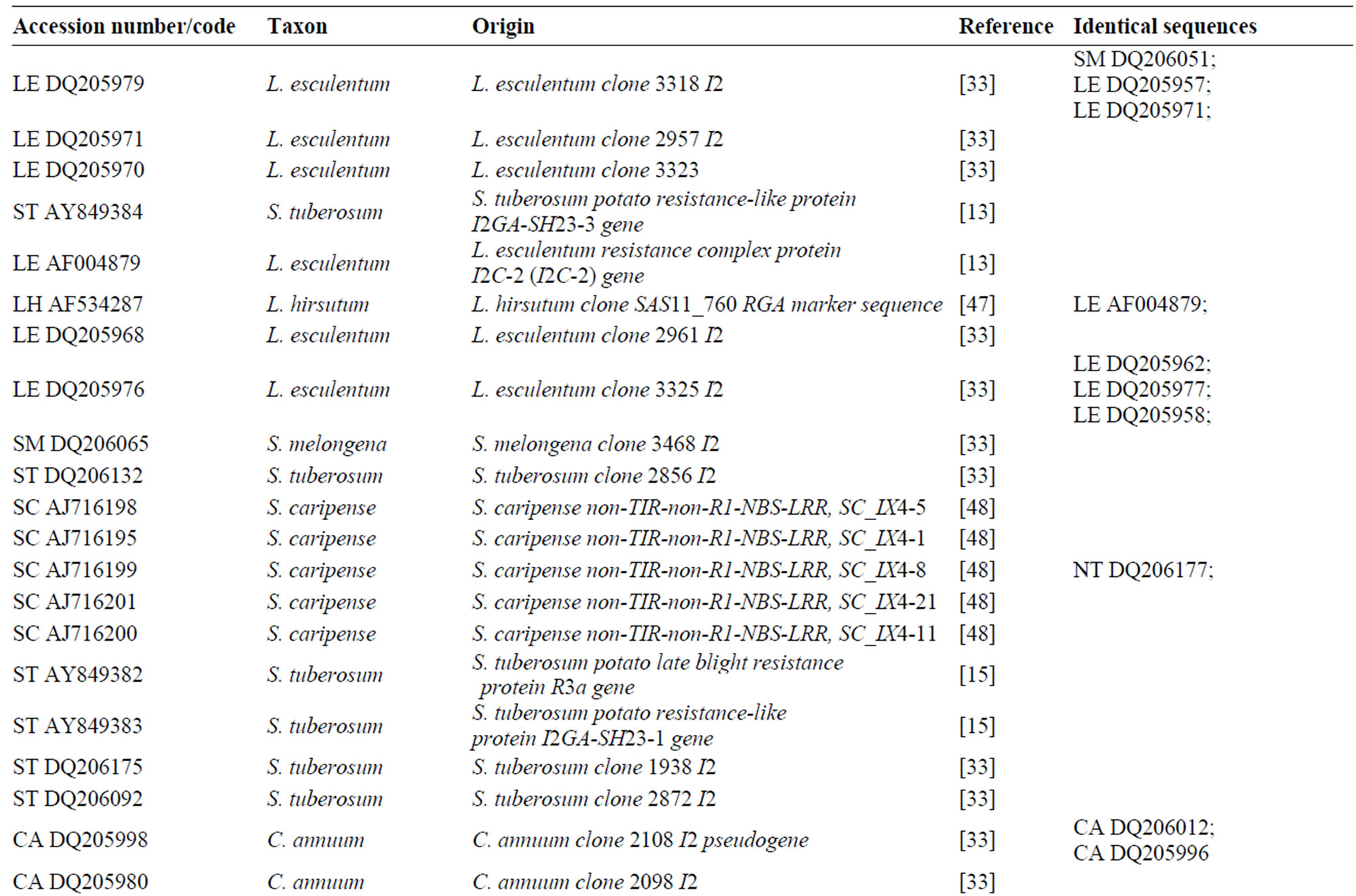

Table 3. List of wild lycopersicon accessions analyzed for the I2-NBS domain and Solanaceae sequences retrieved by BlastN analysis.
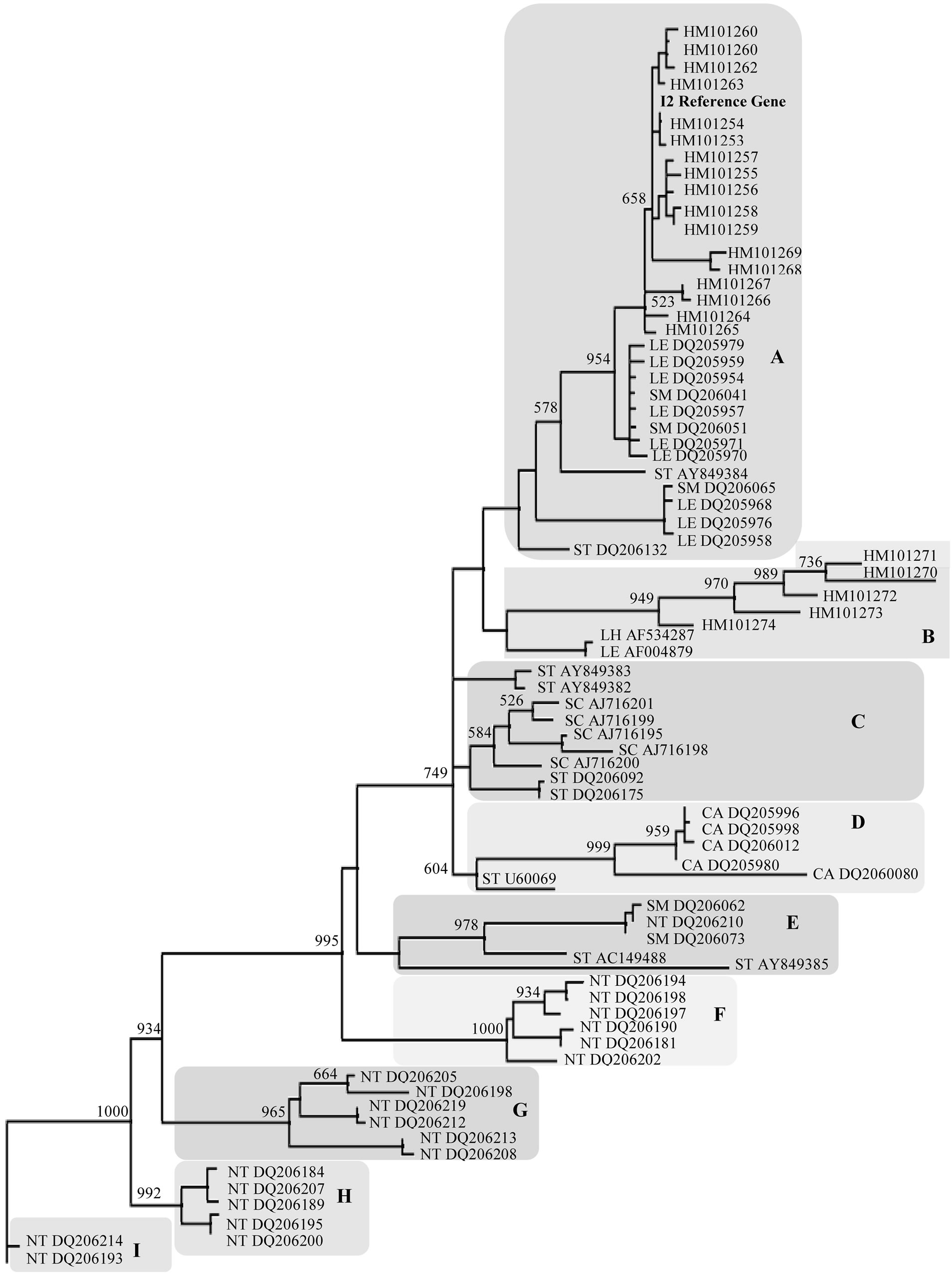
Figure 2. Evolutionary process in the I2-NBS domain in Solanaceae species. The trend of (dN/dS normalized) calculated according to the FEN model using the 0.05 significance level. The I2 resistance gene amino acid sequence was used as reference sequence.

Figure 3. Evolutionary process in the I2-NBS domain in Solanaceae species. The trend of ω (dN/dS normalized) was calculated according to the FEN model using the 0.05 significance level. The I2 resistance gene amino acid sequence was used as reference sequence.
selection and few sites (characterized by purification) had a dN/dS normalized >0. In particular, using a 0.05 significance level, we found 65 sites under negative selection (ω < 1) and a few codons under a moderate positive selection process. The amino acid sequence between positions 32 to 52 seems to be characterized by a large number of codons under negative selection. The next region, instead, is a more variable sequence. Using a tighter parameter (a probability value of 0.1%) three amino acids under positive selection were evidenced in positions 16.55 and 80.
4. Discussion
The growing number of cloned R genes in the last 20 years offers the opportunity to study the evolutionary dynamics of this gene class. The ability of plant species to survive over evolutionary time might depend on their ability to maintain and usefully generate diversity at resistance loci [29]. R genes are often members of tightly linked multigene families, which can be functionally diversified. There has been speculation on the forces that play a key role in the evolution of R genes, allowing plants to generate novel resistance to match the changing patterns of pathogen virulence. Sequence comparisons among these genes have revealed remarkable similarities in general structure, and variability of specific domains, that participate in protein-protein interaction and signal transduction [30]. Evolution of resistance genes remains largely unexplored, but useful information has recently been gained from molecular and genetic analysis [10, 31,32]. Slight nucleotide variation in some strategic positions could have a very dramatic effect on protein function in intra or inter-molecular activity and hence in resistance response to pathogens and pests [33].
In our work we explored the putative presence of I2 alleles in 6 tomato species. The PCR approach showed that at least an amplicon is reproducible in all analyzed species. Considering the geographic distribution and the mating system of each species, we were not surprised to detect nucleotide polymorphism among species tested and versus the reference gene. S. neorickii and S. Pimpinellifolium displayed the highest net nucleotide substitution per site. The nucleotide diversity in S. habrochaites, S. peruvianum, S. chilense and S. corneliomulleri, instead, showed a conserved nucleotide sequence-core and a more variable core. The biological function of the conservative nucleotide sequence as well as the hypervariable nucleotide spot could be an important base for further plant-pathogen research [34]. Nucleotide polymorphism heterogeneity along I2 sequences implies that diversity is also important in single species evolution history.
Our investigation revealed a high level of diversity within S. peruvianum accessions. This population displayed the highest variability in terms of mutational sites and diversity (substitutions, insertions, deletions) and haplotype diversity (Hd) whereas S. habrochaites showed the highest value of Theta (S, Pi, Eta) that is probably related to the mating system. S. peruvianum is a selfincompatible species and S. habrochaites an outcrossing facultative species and hence are subjected to a major genetic shuffle. The high level of polymorphisms identified in these species for R genes is consistent with molecular phylogenetic data previously reported [35]. Molecular analysis of wild tomato species have shown that genetic variation within species decreases with an increasing degree of selfing [36-38]. S. peruvianum is reported to be the ancestral species of wild S. lycopersicon species [39]. Consequently, the huge potential for R gene diversity maintenance within a population could have accumulated over a long period of time. Lineage sorting of the polymorphism of S. peruvianum and the emergence of new variants after speciation events could have shaped Rgene diversity found in other species. The I2 locus originating from S. pimpinellifolium consists of a cluster of seven paralogous sequences on chromosome 11 [14]. An orthologous member of S. pennellii that confers partial resistance to F. oxysporum, lycopersici race 2 was also identified [40].
I2 paralogous sequences were also found on chromosomes 8 and 9 and three regions along chromosome 11 [10,13]. Recombination hot spots are reported in several R gene loci. This may be partially responsible for scatter of nucleotide polymorphism diversity among loci and species [41]. R gene polymorphism is an important component of variation for resistance to pathogens, and new insights can be gained by investigating the genealogies of these genes. Hence our next efforts were to increase the amount of information related to the architecture of diversity in I2 genes in the Solanaceae family. We identified putative I2 orthologues in several Solanaceae species hosts of F. oxysporum (S. demissum, S. tuberosum, S. caripense, S. melongena, C. annuum, N. tabacum) by in silico analysis. Multiple alignments obtained both for nucleotide and amino acid sequences presented enough sequence similarity to design phylogenetic trees. In general, phylogenetic relationships evidenced by the I2 tree reflect Solanaceae species division [42]. N. tabacum is the most distant Solanaceae species, followed by Capsicum (a separate group) and by tomato and potato, always clustering together. The R3a gene may have originated slighter before the I2 functional gene that is evolving rapidly in all Solanum species. The few S. melongena sequences were spread along the dendrogram. The fact that incompatible species generally consist of numerous heterogeneous populations [43] could explain this behavior. Moreover, a small group of five accessions including two S. peruvianum, two S. habrochaites and one S. pimpinellifolium species evidenced specific nucleotide features. Previous studies on phylogenetic analysis of I2 genes reported that evolutionary forces act on the NBS domain of this gene in agreement with the “birthand-death hypotesis” pattern of evolution [7] showing gene clustering between related species as well between distant species [34]. Our data support this hypothesis, as clearly showed in the phylogenetic tree where, three main clades are evidenced: one clade containing only ancestral Nicotiana sequences, one containing only Capsicum sequences and a wider clade containing sequences from different Solanum species indicating that a more recent divergent selection is shaping the I2 domain evolution. Identification of genes going through adaptation plays a key role in understanding evolutionary biology [44]. Sequence variability could be the result of random drift or could involve an “evolutionary selection process”. In our paper we applied all strategies required to avoid false positive results and we applied models to maximize the detection of sites subject to selection. The ω is, for its simplicity and robustness, one of the most widely used evolutionary tests [32,45-47]. The portion of I2 protein characterized in this study generally presents a ω below 1. This indicates that purifying selection is a functional constraint on the evolution of the DNA sequence. In the global purifying background selection three residues with a positive value of ω were found. Couch et al. (2006) [34] identified in a different Solanaceae dataset a higher number of codon under positive selection in the I2-NBS domain. In the three-dimensional conformation the three positively selected sites are located in external areas. They could be involved in inter or intra-molecular interactions necessary for appropriate binding activities or in the negative regulation of defences in the absence of a pathogen effector [32]. Purifying selection may be operating to remove or keep deleterious substitutions at low frequencies; single amino acid variation could be maintained by neutral selection and by intermittent bouts of positive selection. Translated amino acid sequences did not present stop codons suggesting that all the variants could be functional if a recognition event acts as switch. In conclusion R gene allelic diversity in plant wild populations is part of a complex evolutionary process for species survival. Selection for novel or diverse pathogen recognition capabilities is an important factor in species success. In the evolution of wild tomato species other important factors such as environmental conditions and mating should be taken into account. Tomato wild species evidence single accession peculiarities. The pattern of evolved difference suggests that the I2 gene is undergoing under a process of rapid adaptive divergence in the Solanum genus. Purifying selection serves to maintain the NBS cores stable, and neutral or positive selection ensures single amino acid variation. Elucidation of the selection mechanism acting on this domain could help to design new crop improvement strategies for the future.
5. Acknowledgements
We thank Dr. Enrico Negrisolo for his help in phylogenetic analysis using Datamonkey and PHYML, Mark Walters for editing the manuscript, and Michele De Martino for the technical support. This work was performed in the framework of the projects PROM (Ministry of Agricultural, Food and Forestry Policies) and GenoPOM (Ministry of Education, University and Research).
REFERENCES
- H. H. Flor, “Current Status of the Gene-for-Gene Concept,” Annual Review of Phytopathology, Vol. 9, No. 1, 1971, pp. 275-276. doi:10.1146/annurev.py.09.090171.001423
- J. L. Dangl and J. D. G. Jones, “Plant Pathogens and Integrated Defense Responses to Infection,” Nature, Vol. 411, No. 6839, 2001, pp. 826-833. doi:10.1038/35081161
- I. R. Crute, “Gene-for-Gene Recognition in Plant-Pathogen Interactions,” Philosophical Transactions of the Royal Society of London, Series B, Biological Sciences, Vol. 346, 1994, pp. 345-349. doi:10.1098/rstb.1994.0151
- E. A. Van der Vossen, J. N. Van der Voort, K. Kanyuka, A. Bendahmane, H. Sandbrink, D. C. Baulcombe, J. Bakker, W. J. Stiekema and R. M. Klein-Lankhors, “Homologs of a Single Resistance Gene Cluster in Potato Confer Resistance to Distinct Pathogens: A Virus and a Nematode,” Plant Journal, Vol. 23, No. 5, 2000, pp. 567-76. doi:10.1046/j.1365-313x.2000.00814.x
- J. Bai , L. A. Pennill, J. Ning, S. W. Lee, J. Ramalingam, C. A. Webb, B. Zhao, Q. Sun, C. Nelson, J. E. Leach and S. Hulbert, “Diversity in Nucleotide Binding Site-Leucine Rich Repeat Genes in Cereals,” Genome Research, Vol. 12, No. 12, 2002, pp. 1871-1884. doi:10.1101/gr.454902
- S. B. Cannon, H. Zhu, A. M. Baumgarten, R. Spangler, G. May, D. R. Cook and N. D. Young, “Diversity, Distribution, and Ancient Taxonomic Relationships within the TIR and Non-TIR NBSLRR Resistance Gene Subfamilies,” Journal of Molecular Evolution, Vol. 54, No. 4, 2002, pp. 548- 562. doi:10.1007/s00239-001-0057-2
- R. W. Michelmore and B. C. Meyers, “Clusters of Resistance Genes in Plants Evolve by Divergent Selection and a Birth-and-Death Process,” Genome Research, Vol. 8, 1998, pp. 1113-1130.
- V. Kanazin, L. F. Marek and R. C. Shoemaker, “Resistance Gene Analogs Are Conserved and Clustered in Soybean,” Proceeding of the National Academy Science of United State of America, Vol. 93, No. 21, 1996, pp. 11746-11750.
- B. C. Meyers, A. W. Dickerman, R. W. Michelmore, S. Sivaramakrishnan, B. W. Sobral and N. D. Young, “Plant Disease Resistance Genes Encode Members of an Ancient and Diverse Protein Family within the Nucleotide-Binding Superfamily,” Plant Journal, Vol. 20, No. 3, 1999, pp. 317-332. doi:10.1046/j.1365-313X.1999.t01-1-00606.x
- Q. Pan, Y. S. Liu, O. Budai-Hadrian, M. Sela, L. Carmel-Goren, D. Zamir and R. Fluhr, “Comparative Genetics of Nucleotide Binding Site-Leucine Rich Repeat Resistance Gene Homologs in the Genomes of Two Dicotyledons: Tomato and Arabidopsis,” Genetics, Vol. 155, No. 1, 2000, pp. 309-322.
- B. Mieslerova, A. Lebeda, Y. S. Chetelat, “Variation in Response of Wild Lycopersicon and Solanum spp. against Tomato Powdery Mildew (Oidium lycopersici),” Journal Phytopathology, Vol. 148, No. 5, 2000, pp. 303-311. doi:10.1046/j.1439-0434.2000.00492.x
- H. Laterrot, “Breeding Strategies for Disease Resistance in Tomatoes with Emphasis to the Tropics: Current Status and Research Challenges,” 1st International Symposium Tropical Tomato disease, Recife, 21-22 November 1996, pp. 126-132.
- N. Ori, Y. Eshed, I. Paran, G Presting, D. Aviv, S. Tanksley, D. Zamir and R Fluhr, “The I2C Family from the Wilt Disease Resistance Locus I2 Belongs to the Nucleotide Binding, Leucine-Rich Repeat Superfamily of Plant Resistance Genes,” Plant Cell, Vol. 9, No. 4, 1997, pp. 521-32.
- G. Simons, J. Groenendijk, J. Wijbrand, M. Reijans, J. Groenen, P. Diergaarde, T. Van der Lee, M. Bleeker, J. Onstenk, M. Both, M. Haring, J. Mes, B. Cornelissen, M. Zabeau and P. Vos, “Dissection of the Fusarium I2 Gene Cluster in Tomato Reveals Six Homologs and One Active Gene Copy,” Plant Cell, Vol. 10, No. 6, 1998, pp. 1055-1068.
- S. Huang , V. G. Vleeshouwers, J. S. Werij, R. C. Hutten, H.J. van Eck, R. G. Visser and E. Jacobsen, “The R3 Resistance to Phytophthora infestans in Potato Is Conferred by Two Closely Linked R Genes with Distinct Specificities,” Molecular Plant Microbe Interaction, Vol. 17, No. 4, 2004, pp. 428-435. doi:10.1094/MPMI.2004.17.4.428
- I. E. Peralta and D. M. Spooner, “Morphological Characterization and Relationships of Wild Tomatoes (Solanum L. Section Lycopersicon),” Monographs in Systematic Botany from the Missouri Botanical Garden, Vol. 104, 2005, pp. 227-257.
- M. T. Fulton, J. Chunwongse and S. D. Tanksley, “Microprep Protocol for Extraction of DNA from Tomato and Other Herbaceous Plants,” Plant Molecular Biology Reporter, Vol. 13, No. 3, 1995, pp. 207-209. doi:10.1007/BF02670897
- J. Rozas and R. Rozas, “DNASP Version 3: An Integrated Program for Molecular Population Genetics and Molecular Evolution Analysis,” Bioinformatics, Vol. 15, No. 2, 1999, pp. 174-175. doi:10.1093/bioinformatics/15.2.174
- J. D. Thompson, D. G. Higgins and T. J. Gibson, “CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position- Specific Gap Penalties and Weight Matrix Choice,” Nucleic Acid Research, Vol. 22, No. 22, 1994, pp. 4673-4680. doi:10.1093/nar/22.22.4673
- T. A. Hall, “BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT,” Nucleic Acids Symposium Series, Vol. 41, 1999, pp. 95-98.
- S. Kumar, K. Tamura and M. Nei, “MEGA3: Integrated Software for Molecular Evolutionary Genetics Analysis and Sequence Alignment,” Briefings in Bioinformatics, Vol. 5, No. 2, 2004, pp. 150-163. doi:10.1093/bib/5.2.150
- J. Rozas, J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, “DnaSP, DNA Polymorphism Analyses by the Coalescent and Other Methods,” Bioinformatics, Vol. 19, No. 18, 2003, pp. 2496-2497. doi:10.1093/bioinformatics/btg359
- J. Felsenstein, “Phylip (Phylogeny Inference Package) Version 3.6. Distributed by the Author,” Department of Genomic Sciences, University of Washington, Seattle, 2004.
- S. Guindon, F. Lethiec, P. Duroux and O. Gascuel, “Phyml Online—A Web Server for Fast Maximum LikelihoodBased Phylogenetic Inference,” Nucleic Acid Research, Vol. 33, 2005, pp. W557-W559. doi:10.1093/nar/gki352
- J. A. Cuff, E. Birney, M. E. Clamp and G. J. Barton, “ProtEST: Protein Multiple Sequence Alignments from Expressed Sequence Tags,” Bioinformatics, Vol. 16, No. 2, 2000, pp. 111-116. doi:10.1093/bioinformatics/16.2.111
- P. Yang, K. N. Liou, M. I. Mishchenko and B. C. Gao, “Efficient Finite-Difference Time-Domain Scheme for Light Scattering by Dielectric Particles: Application to Aerosols,” Applied Optics, Vol. 39, No. 21, 2000, pp. 3727-3737. doi:10.1364/AO.39.003727
- Z. Yang and J. P. Bielawski, “Statistical Methods for Detecting Molecular Evolution,” Trends in Ecology & Evolution, Vol. 15, No. 12, 2000, pp. 496-503. doi:10.1016/S0169-5347(00)01994-7
- S. L. Pond and S. D. Frost, “Datamonkey: Rapid Detection of Selective Pressure on Individual Sites of Codon Alignments,” Bioinformatics, Vol. 21, No. 10, 2005, pp. 2531-2533. doi:10.1093/bioinformatics/bti320
- K. E. Hammond-Kosack and J. D. G. Jones, “Plant Disease Resistance Genes,” Annual Review of Plant Physiology and Plant Molecular Biology, Vol. 48, 1997, pp. 573-605. doi:10.1146/annurev.arplant.48.1.575
- W. Sanseverino, G. Roma, M. De Simone, L. Faino, S. Melito, E. Stupka, L. Frusciante and M. R. Ercolano, “PRGdb: A Bioinformatics Platform for Plant Resistance Gene Analysis,” Nucleic Acids Research, Vol. 38, No. 1, 2010, pp. D814-D821. doi:10.1093/nar/gkp978
- A. R. Friedman, “The Evolution of Resistance Genes in Multi-Protein Plant Resistance Systems,” Current Opinion in Genetics & Development, Vol. 17, No. 6, 2007, pp. 1-7. doi:10.1016/j.gde.2007.08.014
- P. Tiffin and D. A. Moeller, “Molecular Evolution of Plant Immune System Genes,” Trends in Genetics, Vol. 22, No. 12, 2006, pp. 662-670. doi:10.1016/j.tig.2006.09.011
- Y. Belkhadir, R. Subramaniam and J. L. Dangl, “Plant Disease Resistance Protein Signaling: NBS-LRR Proteins and Their Partners,” Current Opinion in Plant Biology, Vol. 7, No. 4, 2004, pp. 391-399. doi:10.1016/j.pbi.2004.05.009
- B. C. Couch, R. Spangler, C. Ramos and G. May, “Pervasive Purifying Selection Characterizes the Evolution of I2 Homologs,” Molecular Plant Microbe Interaction, Vol. 19, No. 3, 2006, pp. 288-303. doi:10.1094/MPMI-19-0288
- I. E. Peralta and D. M. Spooner, “Classification of Wild Tomatoes: A Review,” Kurtziana, Vol. 28, No. 1, 2000, pp. 45-54.
- C. M. Rick, J. F. Fobes and S. D. Tanksley, “Evolution of Mating Systems in Lycopersicon hirsutum as Deduced from Genetic Variation in Electrophoretic and Morphological Characters,” Plant Systematic and Evolution, Vol. 132, 1979, pp. 279-298. doi:10.1007/BF00982390
- A. L. Caicedo and B. A Schaal, “Population Structure and Phylogeography of Solanum pimpinellifolium Inferred from a Nuclear Gene,” Molecular Ecology, Vol. 13, No. 7, 2004, pp. 1871-1882. doi:10.1111/j.1365-294X.2004.02191.x
- G. Bonnema, D. Schipper, Van S. Heusden, P. Lindhout and P. Zabel, “Tomato Chromosome 1: High Resolution Genetic and Physical Mapping of the Short Arm in an Interspecific Lycopersicon esculentum × L. peruvianum Cross,” Molecular Genomics and Genetics, Vol. 253, 1997, pp. 455-462.
- M. B. Sela-Buurlage, O. Budai-Hadrian, Q. Pan, L. Carmel-Goren, R. Vunsch, D. Zamir and R. Fluhr, “Genome-Wide Dissection of Fusarium Resistance in Tomato Reveals Multiple Complex Loci,” Molecular Genetics and Genomics, Vol. 265, No. 6, 2001, pp. 1104- 1111. doi:10.1007/s004380100509
- K. Roselius, W. Stephan and T. Städler, “The Relationship of Nucleotide Polymorphism, Recombination Rate and Selection in Wild Tomato Species,” Genetics, Vol. 171, No. 2, 2005, pp. 753-763. doi:10.1534/genetics.105.043877
- M. Mazurek, E. T. Cirulli, S. M. Collier, L. G. Landry, B. C. Kang, E. A. Quirin, J. M. Bradeen, P. Moffett and M. M. Jahn, “The Fractionated Orthology of Bs2 and Rx/ Gpa2 Supports Shared Synteny of Disease Resistance in the Solanaceae,” Genetics, Vol. 182, 2009, pp. 1351- 1364. doi:10.1534/genetics.109.101022
- S. Knapp, L. Bohs, M. Nee and D. M. Spooner, “Comparative and Functional Genomics, Solanaceae a Model for Linking Genomics with Biodiversity,” Comparative and Functional Genomics, Vol. 5, No. 3, 2004, pp. 285- 291. doi:10.1002/cfg.393
- J. De Meaux and T. Mitchell-Olds, “Evolution of Plant Resistance at the Molecular Level: Ecological Context of Species Interactions,” Heredity, Vol. 91, No. 4, 2003, pp. 345-352. doi:10.1038/sj.hdy.6800342
- S. Kryazhimskiy and J. B. Plotkin, “The Population Genetics of dN/dS,” PLoS Genetics, Vol. 4, No. 12, 2008, Article ID: e1000304. doi:10.1371/journal.pgen.1000304
- M. Anisimova, J. P. Bielawski and Z. Yang, “Accuracy and Power of Bayes Prediction of Amino Acid Sites under Positive Selection,” Molecular Biology Evolution, Vol. 19, No. 6, 2002, pp. 950-958. doi:10.1093/oxfordjournals.molbev.a004152
- A. Devoto, H. A. Hartmann, P. Piffanelli, C. Elliott, C. Simmons, G. Taramino, C. S. Goh, F. E. Cohen, B. C. Emerson, P. Schulze-Lefert and R. Panstruga, “Molecular Phylogeny and Evolution of the Plant-Specific Seven- Transmembrane MLO Family,” Journal of Molecular Evolution, Vol. 56, No. 1, 2003, pp. 77-88. doi:10.1007/s00239-002-2382-5
- L. P. Martinez-Castilla and E. R. Alvarez-Buylla, “Adaptive Evolution in the Arabidopsis MADS Box Gene Family Inferred from Its Complete Resolved Phylogeny,” Proceeding of the National Academy Science of United State of America, Vol. 100, No. 23, 2003, pp. 13407- 13412.
- D. O. Niu-Liu, L. Zhang and M. R. Foolad, “Sequence Comparison and Characterization of DNA Fragments Amplified by Resistance Gene Primers in Tomato,” Acta Horticulturae, Vol. 625, 2003, pp. 49-58.
- F. Ch. Trognitz and B. R. Trognitz, “Survey of Resistance Gene Analogs in Solanum caripense, a Relative of Potato and Tomato, and Update on R Gene Genealogy,” Molecular Genetics and Genomics, Vol. 274, No. 6, 2005, pp. 595-605. doi:10.1007/s00438-005-0038-z

