Journal of Cancer Therapy
Vol.4 No.2(2013), Article ID:29759,7 pages DOI:10.4236/jct.2013.42072
N-myc Downstream Regulated Gene 1 Increases Differentiation Factors Level in Human Prostate Cancer Cells without Affecting Cell Proliferation and Cell Cycle Profiles
![]()
1Department of Human Biology, Faculty of Natural Sciences, University of Haifa, Haifa, Israel; 2Department of Molecular Genetics, Carmel Medical Center, Haifa, Israel; 3Pediatric Gastroenterology and Nutrition Unit, Carmel Medical Center, Haifa, Israel; 4The Bruce Rappaport Faculty of Medicine, Technion-Israel Institute of Technology, Haifa, Israel.
Email: *ffares@sci.haifa.ac.il
Copyright © 2013 Tina Napso et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received February 6th, 2013; revised March 9th, 2013; accepted March 17th, 2013
Keywords: NDRG1; Human Prostate Cancer; p21; Cytokeratin 8/18
ABSTRACT
N-myc downstream regulated gene 1 (NDRG1), also known as differentiation related gene 1, was previously identified as an up-regulated gene upon cellular differentiation. Even though its sequence along with its expressional pattern in cancer cell lines are evident, the functional aspects concerning cell proliferation, viability, differentiation and cell cycle regulation of NDRG1 remains vague. The aim of the present study was to elucidate the functional role of NDRG1 in human prostate cancer. Our results showed basal levels of NDRG1 expression in PC-3 (poorly differentiated, null p53), DU-145 (moderately differentiated, mutant p53) and LNCaP (well-differentiated, wiled type p53). Nevertheless, NDRG1 sequencing assay disclosed no mutations in the gene. Furthermore, human cDNA of NDRG1 from normal placenta was cloned into a eukaryotic expression vector and transfected into the three cancer cell lines. This resulted in over-expression of NDRG1, which in turn markedly up regulated two differentiation markers of the prostate tissue, p21 and cytokeratin 8/18. Unpredictably, cell cycle progression, cell proliferation and DNA synthesis were unaffected following NDRG1 expression. These results revealed that NDRG1 is functional in prostate cancer cells and able to induce expression of differentiation factors through p53 independent pathway. However, the pathway downstream NDRG1, involving p21 and c8/18, regulating cell cycle progression and DNA synthesis is unfunctional. Loss of sensitivity to p21 cell cycle control may be associated with prostatic cancer behavior. Further studies are required to clarify the intra cellular molecular pathways affecting NDRG1 function in human prostate cancer.
1. Introduction
Prostate cancer emerges as a central problem of male’s public health in recent years, as it develops mainly in elderly men making its incidence to be continuously growing in an era characterized with ageing of the population. The development of prostate cancer is considered to be dependent on a number of gene abnormalities, and in this manner, the molecular biology research in this field is obvious and essential. Comprehending the molecular aspect of prostate cancer will aid in prevention, provide molecular markers for diagnosis, support the development of tumor staging along with classification, and shift towards novel therapeutic targets in the treatment for highly developed and metastasized cancers. In this study, NDRG1, a proliferation related gene, was investigated in order to clarify its functionality in human prostate cancer cells.
N-myc downstream regulated gene 1 (NDRG1), which is mapped to the human chromosome 8q24.2, was initially identified by differential display as up-regulated gene in cellular differentiation [1]. Further studies revealed that NDRG1 is involved in carcinogenesis development, metastatic spreading, atherosclerosis, differentiation and cell survival [2]. This gene encodes a 43 kDa protein that contains three tandem repeats of 10 amino acids (Gly-Thr-Arg-Ser-Arg-Ser-His-Thr-Ser-Glu) at the C-terminal end, and possesses several phosphorylation sites of protein kinase A [3]. NDRG1 is a member of the NDRG family, which belongs to the α/β hydrolase super family with no specific attributable function [4]. This family includes three additional highly conserved proteins: NDRG2, NDRG3 and NDRG4, which are distinct in their expression pattern without any homology to other known proteins [5]. NDRG1 was reported to be up regulated by DNA damaging agents in addition to cellular stress induced by hypoxia, exposure to nickel, homo-cysteine and calcium ionophores, as well as by various differentiation factors e.g. androgens and estrogens [6-9]. ER stressinduced agents, in addition to inhibition of DNA methylation and histon-deacetylation, were also detected as upregulators of NDRG1 gene [10].
The expression alongside the function of NDRG1 is cell type dependent, as it was shown by numerous studies. Decreased levels of NDRG1 were found in colon, breast and prostate cancer tissues compared to their normal cell lines, whereas other studies showed up-regulated NDRG1 during differentiation and growth arrest in colon, esophageal, pancreatic and prostate cancer metastasis, which were further correlated with tumor aggressiveness and poor survival of hepatocellular carcinoma patients [11, 12].
NDRG1’s functional aspects in prostatic tumor progression, local invasion and metastatic spread, remain rather indefinite and controversial. To further elucidate the function of NDRG1 in prostate cancer cells, cDNA of NDRG1 from normal human placenta was cloned and over expressed in three human prostate cancer cell lines with diverse differentiation levels and different genotypic/phenotypic features. Our results indicate that over expression of NDRG1 increases levels of the differentiating factors, p21 and c8/18, without affecting cell cycle, proliferation and DNA synthesis.
2. Materials and Methods
2.1. Cell Culture
The human prostate cancer cell lines; PC-3 (deficient in p53 gene, androgen-independent and poorly differentiated), DU-145 (mutant p53 gene, androgen-independent and moderately differentiated) and LNCaP (wild-type form of p53 gene, androgen-dependent and well differentiated) were used in this study. Cells were maintained in humidified atmosphere of 5% CO2 in air, at 37˚C in RPMI-1640 medium (LNCaP, DU-145) or in F-12 (PC-3) with L-glutamine, supplemented with 10% fetal calf serum (FCS), 100 µg/ml streptomycin and 100 U/ml penicillin. For DU-145 cells, 5 µg/ml of insulin were added.
2.2. NDRG1 Cloning
To create mammalian expression plasmid, total mRNA was isolated from normal human placenta tissue and reverse transcribed to cDNA. cDNA of NDRG1 was amplified by PCR reaction using the forward primer (5’-TTAGGCAGGTACCAGCAGGGACATG-3’), which contained the Kozak sequence and the KpnΙ restriction site, and the reverse primer (5’-GAGGAGGGGGCCACTCGAGAGATCAGAGTC-3’), which included the XhoΙ restriction site. The PCR amplification conducted with initial denaturation step of 95˚C for 2 min and 40 cycles of the following conditions: 95˚C for 30 sec, 58˚C for 1 min and 72˚ for 3 min. Final step of 72˚C for 10 min was performed. The PCR product was cloned into the cloning site of the eukaryotic PCDNA3.1+ expression vector (Invitrogen, San Diego, CA), which contains a CMV promoter. Plasmid insert was fully sequenced to ensure that no mutations existed in the PCR product of the NDRG1 gene. The human prostate cancer cell lines (PC-3, DU145 and LNCaP) were transfected either with PCDNA3.1+/NDRG1 or with an empty vector, using FuGENE 6 (Roche, Mannheim, Germany) according to the manufacturer’s protocol. Cells were analyzed, 72 h post transfection for the expression of NDRG1 by RT-PCR, which was further confirmed by western blot.
2.3. Western Blot
Proteins were extracted using RIPA buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA) containing protease inhibitors (30 μg/ml aprotinin, 2.5 μg/ml leupeptin, 10 μg/ml pepstatin and 1 mM phenylmethyl fluoride). Samples were electrophoresed on nondenaturing 10% sodium dodecyl sulfate-polyacrylamide gels. Proteins were then transferred to a 0.2 µm pore size nitrocellulose membrane (Sigma, St. Louis, MO) at 250 mA for 2.5 h. The nitrocellulose membrane was incubated in 5% nonfat dry milk for 1.5 h at room temperature, followed by overnight incubation with primary antibodies; anti-NDRG1, anti-p21 and anti-c8/18 goat polyclonal antibodies obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The next day, three consecutive washes in PBS containing 0.1% tween-20 (10 min/wash). The membrane was then incubated with secondary antibody conjugated to horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA). Finally, the nitrocellulose paper was reacted with enhanced chemiluminescent substrate (Biological Industries, Beit-Haemek, Israel) for 5 min, and exposed to X-ray film. The western blotting results were quantified using a densitometer, and actin level (as standard protein occurring naturally in these cells) was taken as a reference.
2.4. Cell Viability
Cell viability was determined by XTT cell proliferation assay kit (Biological Industries, Beit-Haemek, Israel) according to manufacturer’s protocol. The absorbance of each sample was measured with a spectrophotometer (Tecan, Spectra (Shell), Austria) at 450 nm wavelength.
2.5. DNA Synthesis
BrdU (5-bromo-2’-deoxyuridine) kit from Roche (Mannheim, Germany) was used according to manufacturer’s protocol. In brief, cells were plated onto 96-well plates (5 × 104 cells/ml) and transfected 24 h later. Cells were incubated with 10 µM BrdU, for 4 h and BrdU was detected with monoclonal anti-BrdU-POD and Fab fragments using spectrophotometer at a wavelength of 405 nm.
2.6. FACS Analysis
Cells were trypsinized 72 h post transfection and were washed by PBS along with fixation with 70% ETOH for 1 h. Cells were incubated with 0.1% NP-40 for 5 min at 4˚C and washed again with PBS followed by incubation with 100 µg/ml RNase and 50 µg/ml PI (Sigma, St. Louis, MO) for 20 min. Finally, cells were analyzed using FACS Calibur flow (Becton Dickinson, NJ, USA).
2.7. Statistical Analysis
Each experiment was repeated at least three times, and results were expressed as a mean ± S.D. Statistical analysis of data presented were performed using SPSS software, significance was determined at P < 0.05. For analysis of protein expression, two-tailed student t-test was used, while two-tailed Man-Whitney analysis was performed for cell viability and DNA synthesis experiments.
3. Results
3.1. NDRG1 Shows Basal Expression Levels in Human Prostate Cancer Cells
Untransfected cells, which were tested for NDRG1 expression in both mRNA and protein levels showed basal expression of NDRG1 in the three cell lines tested; PC-3, DU-145 and LNCaP (Figure 1). Keeping in mind that NDRG1 gene induced differentiation in colon cancer cell lines and was up regulated in differentiated conditions, a mutation in the gene was suspected, enabling NDRG1 to be expressed in the cells without fulfilling its functions. Consequently, cDNA from the cell lines were fully sequenced. However, no mutations were detected in the coding sequence of the gene (data not shown). To further investigate the functional role of NDRG1 in the prostatic cancer cells, the NDRG1 gene from normal human placenta biopsy, was cloned into a eukaryotic expression fected to the human prostate cancer cell lines. Gene ex-


Figure 1. Over-expression of NDRG1 in the prostate cancer cells. pcDNA3.1+/NDRG1 was transfected into the PC-3, DU145 and LNCaP cells and mRNA expression levels of the gene were detected by RT-PCR (a) and protein levels by western blotting (b). Cells which transfected with PCDNA/ NDRG1 marked with (+) and control cells that were transfected only with PCDNA3.1 marked with (−). For protein amount equalization, mouse anti-human β-actin monoclonal antibody was used. The figure shown is representative of three experiments. Bands detected were subtracted from background noise and density values calculated as a control from the proper β-actin and as a percent of control. Statistical significance determined by a two-tailed Student’s t-test. Significance was determined as follows: *P < 0.05.
vector PCDNA3.1 under a CMV promoter, and transpression following transfection was confirmed by RTPCR (Figure 1(a)) and further substantiated by western blot analysis (Figure 1(b)). NDRG1 was mostly overexpressed in DU-145 cells (4 folds of control), followed by PC-3 cells (2 folds of control). The over expression in LNCaP was mild but statistically significant. Western blot results revealed that NDRG1 expression was partially phosphorylated as reported earlier [3].
3.2. Up-Regulation of NDRG1 Alters Expression Levels of Differential Factors.
Two differentiation related proteins; c8/18 and p21 were analyzed for the effect of NDRG1 on the differentiation and growth pattern of cancer cells. Cytokeratin 8/18 (c8/ 18) plays an important role in differentiation, tissue specialization and functions to preserve the overall structure of epithelial cells. Alternatively, p21 is an inhibitor of the activity of cyclin-CDK2 complexes and consequently operates as a cell cycle progression regulator. Alterations in expression levels of these proteins, 72 h post transfection, were tested by western blot. The NDRG1 over-expression markedly up regulated the two differential markers, p21 (Figure 2(a)) and c8/18 (Figure 2(b)), compared to the controls. The highest relative levels (transaction vs. control) of p21 were detected in PC-3 and LNCaP cell lines, while the highest relative expression of c8/18 was evidenced in PC-3 cell line. Both, p21 and c8/18 were less over-expressed in DU-145 cell line.
3.3. NDRG1 Fails to Influence Cell Proliferation and DNA Synthesis Rates
Cell proliferation (Figure 3), DNA synthesis (Figure 4) and cell cycle analysis (Figure 5) were conducted 72 h post transfection. Since p21 and c8/18 showed remarkable increase following transfection, a reduction in proliferation and a tendency toward differentiation were expected. Surprisingly, the results indicated that over-expression of NDRG1 had no effect on cell viability and on DNA synthesis levels. Furthermore, over-expression of NDRG1 failed to influence the cell cycle phases of the treated cells.
4. Discussion
NDRG1 was identified previously as a gene that was down regulated in tumors of colon, pancreas, esophagus, breast and prostate [11-13]. Its expression is induced by differentiation stimuli in cancer cells, and its up regulation in colon cancer cells induced morphological and molecular changes that were consistent with colon cancer cell line differentiation [10]. Moreover, studies reported
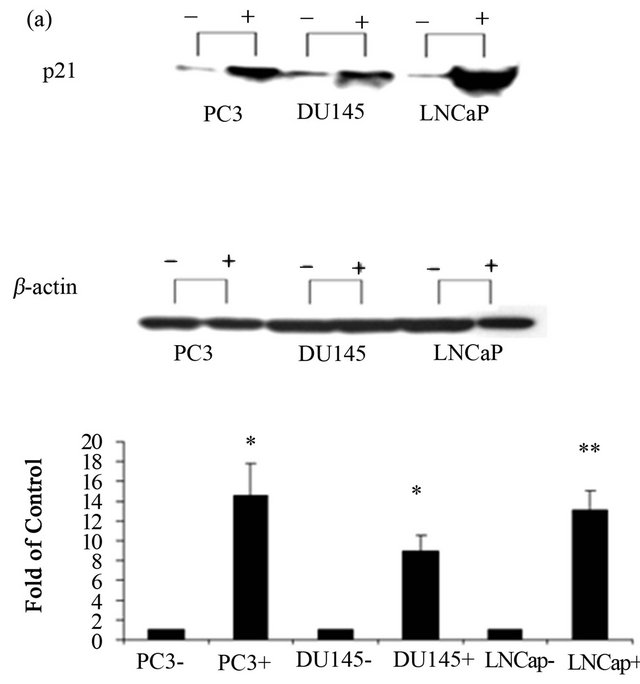

Figure 2. Effect of NDRG1 on the expression level of differentiation markers. 60 µg of protein were extracted from transfected cells and were analyzed via western blot. p21, and c8/18 expression levels were detected. Similar amounts of protein were present in all lanes as it was detected with the house keeping gene β-actin. Figure shown is representative of three experiments. Results were analyzed using Quantity-One software. Statistical significance determined by a two-tailed Student’s t-test. Significance was determined as follows: *P < 0.05.
that NDRG1 suppressed invasion of bladder cancer cells in vitro, liver metastasis in nude mice and tumor metastasis of prostate cancer in vivo [11]. On the other hand, over-expression of NDRG1 was proved to be an indicator of poor prognosis in hepatocellular carcinoma [14]. Although many are known regarding the genomic events which regulate NDRG1 expression, little is known about the non-genomic and protein level regulators of this gene.

Figure 3. Cell viability following over expression of NDRG1 in PC-3, DU145 and LNCaP. Cell viability detected using the XTT method. Data presented are average of three experiments, each conducted in five replicates (mean ± SD) and are expressed as percentages of the respective control. Statistical significance determined by a two-tailed MannWhitney test in which significance was determined to be P < 0.05.
Since cell differentiation is a pivotal function of NDRGI, two differentiation factors: p21 and c8/18, representatives of cell cycle control and cytoskeleton network in human prostate cancer, respectively, were explored in this study.
Previous studies showed that NDRG1 was identified as a gene that is rapidly induced by p53 and it is necessary but not sufficient for apoptosis initiated by p53 [15]. To date, the exact biological role of NDRG1 remains unclear. In order to clarify the role of NDRG1 in prostate cancer cells we transfected the cDNA of NDRG1 into three different cell lines of human prostate cancer with diverse differentiation levels, androgen receptor presence and p53 status. Prior to transfection, basal levels of NDRG1 were tested in the cells. The results revealed that all three cell lines expressed basal levels of NDRG1, matching in this regard, the report of Urlix [9] who showed that highest basal expression of the mRNA of the NDRG1 was detected in the PC-3 cell line which is considered to be a poorly differentiated one. Since the differentiation gene is expressed in such undifferentiated

Figure 4. DNA synthesis following over expression of NDRG1. PC-3, DU145 and LNCaP cells were transfected and 72 h post transfection DNA synthesis was measured using the BRDU kit. Data presented are average of three experiments, each conducted in three replicates (mean ± SD), and are expressed as percentages of the respective control. Statistical significance determined by a two-tailed MannWhitney test in which significance was determined to be P < 0.05.
prostatic cell line, a deactivation mutation was suspected to exist, however no mutations were detected in the coding sequence of the gene. Contrary to some publications [16], we concluded that no correlation exists between the basal expression of the gene and the differentiation degree of the cells lines.
To further clarify this enigma we over expressed the gene, which was cloned from a normal human placenta, that was previously reported to express the wild-type form of the NDRG1 gene. We hypothesized that over-expression of NDRG1 would increase the expression of differentiation factors and induce cell differentition resulting in cell cycle arrest. Indeed, both p21 and c8/18 proteins, showed clear up regulation in all three cell lines. Increased expression of p21 protein is known to prevent cell-cycle progression by inhibiting the activity of CDKs and therefore regulating the progression through the G1/S checkpoint of the cell cycle. Cytokeratin 8/18 (c8/18) was reported as an up regulated protein during differentiation of the prostate cells [17]. The present results revealed that p21 was up regulated extremely in the LNCaP cells, which expressed the transfected NDRG1 in
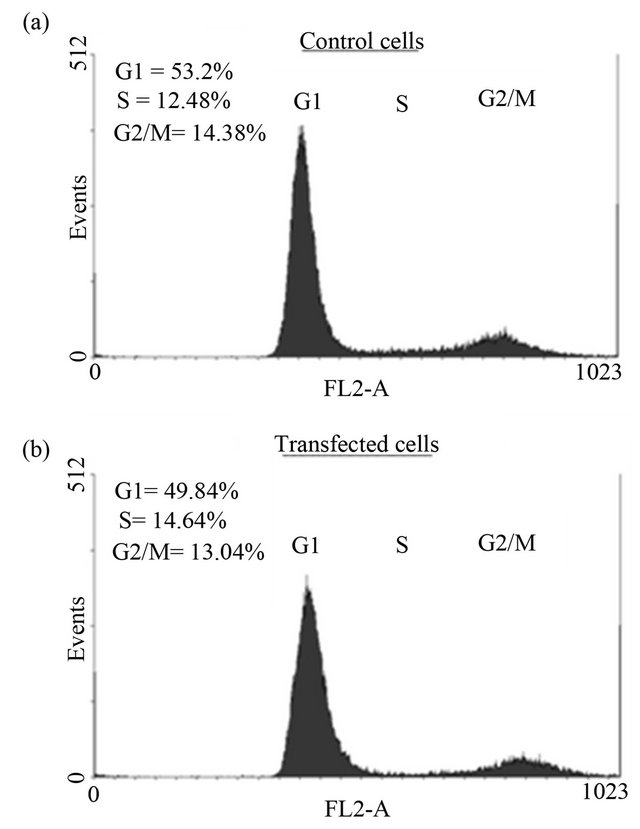
Figure 5. The effect of NDRG1 on cell cycle progression of prostate cancer cells. Cells were transfected in T-25 flasks and fixated with Ethanol 70% after 72 h of tansfection. Cells then were prepared for flow-cytometry analysis as described in the methods. The histograms demonstrated are of the PC-3 cells and are representative figures of four experiments each conducted in duplicates.
the lowest amount. Apparently, LNCaP cells are more sensitive to the NDRG1 influence compared to the other transfected cell lines. This cell line differs from the PC-3 and DU145 cells in the expression of the androgen receptor and in the activity of p53. While LNCaP cells express the androgen receptor and the wild-type p53, PC-3 and DU145 cell lines are androgen receptor negative and p53 un or less functional, respecttively. It can be concluded that p53 contributes to the p21 and c8/18 induction by NDRG1, however, this process can be activated independently of p53 as it was proven in PC-3 and DU145 cells.
Most recent interactive data reinforce the role of NDRG1 in differentiation and cell adhesion [10]. NDRG1 was found to interact directly with two key control proteins: E-cadherin and β-catenin, cell adhesion molecule and anchorage to the actine cytoskeleton, respectively. Moreover, the loss of E-cadherin or disruption of cadherin-catenin interaction is known to potentiate metastasis spread in prostate cancer [11]. Combining the NDRG1 interactome [10] and the present data, it is evident that NDRG1 is involved in the cytoskeleton network (β-catenin, c8/18), differentiation and cell specialization (p21) and epithelial integrity (E-cadherin) in the human prostatic cell lines investigated.
Differentiation level of cells is characterized by decreased proliferation and by cell cycle arrest. After establishing the fact that NDRG1 up regulated p21 and C8/18, two classical features of differentiation state, the proliferation rate and cell cycle progression of the cells were examined, in order to reinforce our assumption that over expression of NDRG1 would cause cell differentiation. Surprisingly, the viability of the cells after transfection and the proliferation and DNA synthesis rate of the cells revealed unexpected results. Evidently, there was an up regulation of the differentiation markers of the prostate tissue, nonetheless, this did not influence the growth arrest of the cells nor the proliferation rate or the DNA synthesis rate of the transfected cells. Apparently, the human prostate cancer cells PC-3, DU145 and LNCaP lost their sensitivity to the inhibitory effect of the p21, with regard to cell proliferation and cell cycle progresssion. Earlier studies also reported the loss of function of p21 gene in these cell lines of the prostate [18]. These studies tested the functional activity of hypoxia-induced factor-1 (HIF-1), which is a known inducer of the NDRG1, in prostate cell lines. Transient transfection of the HIF-1 into these cells induced expression of p21. However, neither expression of p21 nor hypoxia caused growth arrest of the cells. The loss of p21 ability to inhibit tumor growth was reported also by Fizazi et al., [19], claiming that the cause of this phenomenon is not understood. The assumption that it can be related to genetic alteration of the p21 was disqualified because mutations in this gene are rare in the human neoplasm, including prostate cancer. An alternative explanation can immerge from the recent discovery of several mutations in the CHEK2 and its downstream target gene TP53, in the three prostatic cell lines presently investigated [17]. It is possible that mutated CHEK2 and TP53 in LNCaP, PC-3 and DU145 cell lines or some other undiscovered mutations are at the origin of the p21 loss of cell cycle inhibitory effect. More recent studies substantiated the loss of sensitivity of LNCaP toward p21 induction [18], while others reported an inhibitory effect of induced p21 on cell cycle in prostatic cancer cell lines, without manipulating NDRG1 expression [19]. Most probably, prostatic cell lines sensitivity to the inhibitory effect of p21 on cell cycle is p21- pathway dependent. Over expressed NDRG1 induced p21 incapable to regulate while induction of p21 by different pathways maintained its inhibitory function of the cell cycle.
The results of the present study indicated that over expressed NDRG1 augment p21 and c8/18 expression in a p53 and androgen receptor independent pathways, without affecting cell cycle progression, proliferation rate and DNA synthesis. Future studies are needed to explore the exact molecular mechanism of the NDRG1 in prostate cancer cells.
REFERENCES
- N. van Belzen, W. N. Dinjens, M. P. Diesveld, N. A. Groen, A. C. van der Made, Y. Nozawa, et al., “A Novel Gene Which Is Up-Regulated during Colon Epithelial Cell Differentiation and Down-Regulated in Colorectal Neoplasms,” Laboratory Investigation: A Journal of Technical Methods and Pathology, Vol. 77, No. 1, 1997, pp. 85-92.
- N. van Belzen, W. N. Dinjens, B. H. Eussen and F. T. Bosman, “Expression of Differentiation-Related Genes in Colorectal Cancer: Possible Implications for Prognosis,” Histology and Histopathology, Vol. 13, No. 4, 1998, pp. 1233-1242.
- K. L. Agarwala, K. Kokame, H. Kato and T. Miyata, “Phosphorylation of RTP, an ER Stress-Responsive Cytoplasmic Protein,” Biochemical and Biophysical Research Communications, Vol. 272, No. 3, 2000, pp. 641- 647. doi:10.1006/bbrc.2000.2833
- R. H. Zhou, K. Kokame, Y. Tsukamoto, C. Yutani, H. Kato and T. Miyata, “Characterization of the Human NDRG Gene Family: A Newly Identified Member, NDRG4, Is Specifically Expressed in Brain and Heart,” Genomics, Vol. 73, No. 1, 2001, pp. 86-97. doi:10.1006/geno.2000.6496
- T. Okuda and H. Kondoh, “Identification of New Genes ndr2 and ndr3 Which Are Related to Ndr1/RTP/Drg1 but Show Distinct Tissue Specificity and Response to Nmyc,” Biochemical and Biophysical Research Communications, Vol. 266, No. 1, 1999, pp. 208-215. doi:10.1006/bbrc.1999.1780
- D. Zhou, K. Salnikow and M. Costa, “Cap43, a Novel Gene Specifically Induced by Ni2+ Compounds,” Cancer Research, Vol. 58, No. 10, 1998, pp. 2182-2189.
- K. Kokame, H. Kato and T. Miyata, “Homocysteine-Respondent Genes in Vascular Endothelial Cells Identified by Differential Display Analysis. GRP78/BiP and Novel Genes,” The Journal of Biological Chemistry, Vol. 271, No. 47, 1996, pp. 29659-29665. doi:10.1074/jbc.271.47.29659
- H. Park, M. A. Adams, P. Lachat, F. Bosman, S. C. Pang and C. H. Graham, “Hypoxia Induces the Expression of a 43-kDa Protein (PROXY-1) in Normal and Malignant Cells,” Biochemical and Biophysical Research Communications, Vol. 276, No. 1, 2000, pp. 321-328. doi:10.1006/bbrc.2000.3475
- W. Ulrix, J. V. Swinnen, W. Heyns and G. Verhoeven, “The Differentiation-Related Gene 1, Drg1, Is Markedly Upregulated by Androgens in LNCaP Prostatic Adenocarcinoma Cells,” FEBS Letters, Vol. 455, No. 1-2, 1999, pp. 23-26. doi:10.1016/S0014-5793(99)00845-5
- R. J. Guan, H. L. Ford, Y. Fu, Y. Li, L. M. Shaw and A. B. Pardee, “Drg-1 as a Differentiation-Related, Putative Metastatic Suppressor Gene in Human Colon Cancer,” Cancer Research, Vol. 60, No. 3, 2000 , pp. 749-755.
- S. Bandyopadhyay, S. K. Pai, S. C. Gross, S. Hirota, S. Hosobe, K. Miura, et al., “The Drg-1 Gene Suppresses Tumor Metastasis in Prostate Cancer,” Cancer Research, Vol. 63, No. 8, 2003, pp. 1731-1736.
- S. Bandyopadhyay, S. K. Pai, S. Hirota, S. Hosobe, Y. Takano, K. Saito, et al., “Role of the Putative Tumor Metastasis Suppressor Gene Drg-1 in Breast Cancer Progression,” Oncogene, Vol. 23, No. 33, 2004, pp. 5675- 5681. doi:10.1038/sj.onc.1207734
- N. van Belzen, W. N. Dinjens, M. P. Diesveld, N. A. Groen, A. C. van der Made, Y. Nozawa, et al., “A Novel Gene Which Is Up-Regulated during Colon Epithelial Cell Differentiation and down-regulated in Colorectal Neoplasms,” Laboratory Investigation, Vol. 77, No. 1, 1997, pp. 85-92.
- T. Okuda, Y. Higashi, K. Kokame, C. Tanaka, H. Kondoh and T. Miyata, “Ndrg1-Deficient Mice Exhibit a Progressive Demyelinating Disorder of Peripheral Nerves,” Molecular and Cellular Biology, Vol. 24, No. 9, 2004, pp. 3949-3956. doi:10.1128/MCB.24.9.3949-3956.2004
- S. Stein, E. K. Thomas, B. Herzog, M. D. Westfall, J. V. Rocheleau, R. S. Jackson 2nd, et al., “NDRG1 Is Necessary for p53-Dependent Apoptosis,” The Journal of Biological Chemistry, Vol. 279, No. 47, 2004, pp. 48930- 48940. doi:10.1074/jbc.M400386200
- R. J. Guan, H. L. Ford, Y. Fu, Y. Li, L. M. Shaw and A. B. Pardee, “Drg-1 as a Differentiation-Related, Putative Metastatic Suppressor Gene in Human Colon Cancer,” Cancer Research, Vol. 60, No. 3, 2000, pp. 749-755.
- E. R. Sherwood, G. Theyer, G. Steiner, L. A. Berg, J. M. Kozlowski and C. Lee, “Differential Expression of Specific Cytokeratin Polypeptides in the Basal and Luminal Epithelia of the Human Prostate,” The Prostate, Vol. 18, No. 4, 1991, pp. 303-314. doi:10.1002/pros.2990180404
- K. Salnikow, M. V. Blagosklonny, H. Ryan, R. Johnson and M. Costa, “Carcinogenic Nickel Induces Genes Involved with Hypoxic Stress,” Cancer Research, Vol. 60, No. 1, 2000, pp. 38-41.
- K. Fizazi, L. A. Martinez, C. R. Sikes, D. A. Johnston, L. C. Stephens, T. J. McDonnell, et al., “The Association of p21((WAF-1/CIP1)) with Progression to Androgen-Independent Prostate Cancer,” Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, Vol. 8, No. 3, 2002, pp. 775-781.
NOTES
*Corresponding author.

