Advances in Bioscience and Biotechnology
Vol.3 No.8(2012), Article ID:25835,10 pages DOI:10.4236/abb.2012.38144
Genetic characterization of Burkholderia cepacia strain from Northeast India: A potential bio-control agent
![]()
1Microbial Resources Division, Institute of Bioresources and Sustainable Development, An Autonomous DBT Institute, Government of India, Imphal, India
2Bioinformatics Division, Institute of Bioresources and Sustainable Development, An Autonomous DBT Institute, Government of India, Imphal, India
Email: *sidevi1@yahoo.co.in
Received 12 September 2012; revised 19 October 2012; accepted 22 November 2012
Keywords: Antagonistic Activity; Burkholderia cepacia; Fusarium oxysporum; Rhizoctonia solani; French Bean
ABSTRACT
This paper reports the characterization of naturally occurring rhizobacteria from NE India for the control of wilt and damping-off diseases in French beans. French bean (Phaseolus vulgaris L.) is one of the most important food legumes for direct human consumption which was originated from South and Central America. In NE India wilt and damping-off in beans are serious diseases which are mainly caused by Fusarium spp and Rhizoctonia solani. Out of 625 bacterial isolates obtained from French bean growing fields of Manipur, 15 potential isolates were selected, whose inhibition zone width were more than 15 mm. Strain IBSD-LKBI exhibited strong antagonistic activity and can inhibit both the pathogens. Besides, this strain also produced extracellular hydrolytic enzymes (protease, chitinase, β-1, 3-glucanase), secondary metabolites (Siderophore, HCN). It also produces plant growth promoting hormone (IAA) and can solubilize organic phosphate. Burkholderia cepacia IBSD-LKBI exhibited the highest plant growth promotion and BCA. After 60 days of treatment, plant height, root length, fresh weight and % of disease suppression were significantly (P < 0.05) higher than in control plants. Productions of secondary metabolites were detected using gene specific primer. This bacterium was phenotypically characterized and sequence data of 16S rDNA revealed the closest identity with Burkholderia cepacia (98%). Multiple sequence alignment & phylogram with closely related sequence accessions using MEGA Software revealed this naturally occurring biocontrol bacteria belongs to Burkholderia cepacia complex genomovar I based on the Biolog identification system with 16S rDNA gene sequence analysis. This organism is one of the new reports from NE India for the control of wilt and damping-off diseases in French beans.
1. INTRODUCTION
One of the obstacles in the production of high quality and yield of food crops is the difficulty to control fungal diseases in plant. Every year, plant pathogenic fungi cause damage to crops, worthy of millions of dollars, all over the world despite the extensive use of chemical pesticides. So the demand for alternative biocontrol techniques, without using any synthetic fungicides, is increasing [1]. Also, environmental concerns and development of resistance in target populations have reduced the availability of effective fungicides. Therefore, the interest in so-called microbial biocontrol agents or their product to limit the attack and damage by phytopathogens has been increased and recommended. In North East region of India the yield of French beans are severely affected due to fungal infection by Fusarium oxysporum and R. solani causing charcoal rot, wilt and damping off diseases. Fusarium and R. solani are the most common and destructive plant pathogens and a monotypic soil borne species with a wide host range involving more than 200 species in 75 families [2]. These fungi infect plants from seedling stage to maturity. It is also called summer wilt or dry weather wilt because it often occurs during the summer, when plants are under heat and drought stresses. When severe, the disease reduces yield by killing plants at early reproductive stages. Microsclerotia is a compact mass of hardened mycelium stored with reserve food material that becomes detached and remains dormant until a favourable opportunity for growth occurs.
Continued use of chemical pesticides causes several negative and harmful effects on health and environment and thereby killing all the beneficial microorganisms from the soil. Due to continuous use of chemical pesticides the pathogens become resistant to a wide spectrum of fungicides applied in the soil. The use of microbial biocontrol agents is an alternative approach for reducing the use of chemicals in agriculture because it is ecofriendly in nature and safe to use in agricultural system to increase crop productivity since the action of such bacteria is highly specific and cost effective. It has been suggested that these bacteria might interact more closely with the host plants and act as effective biocontrol agents in sustainable crop production. Several species of bacteria are known to have beneficial effect on plant growth promotion and disease suppression through production of plant growth promoting regulators and secondary metabolite substances. Organisms such as Burkholderia cepacia have shown effective in suppressing many plant diseases and also promote plant growth [3]. Recently, Burkholderia species has been classified into 10 genotypically distinct but phenotypically similar species (genomovars) referring to the Burkholderia cepacia complex (Bcc) [4].
In this study, isolation and characterization of potential indigenous biocontrol bacteria IBSD-LKBI was reported from NE India for the control of two pathogens which caused wilt and damping-off diseases in French bean. The production of different hydrolytic enzymes and production of growth hormone and iron sequestering from soil has been studied. Moreover, this strain played significant functions in promoting growth on beans and this can further lead to the development of a potential bio-inoculants against soil borne fungal pathogens and also to study its ecological role and interactions with plant which can further lead to the development of a potential biocontrol packages/bioinoculants against soil borne fungal pathogens and also to study its ecological role and interaction with plants.
2. MATERIAL AND METHODS
2.1. Sampling
Twenty five French bean field sites were selected from different locations representing the entire Imphal East and Imphal West valley zone of Manipur, which falls in the Indian region of Indo-Burma diversity hotspot of the world and part of other NE region, for the collection of rhizosphere sample. From each sites, 4 - 5 French bean roots along with the rhizosphere soils from both healthy and infected plants were randomly selected and uprooted during the winter seasons of 2006-2007. The collected samples were carried to the laboratory and stored in the freeze (4˚C) prior to isolation of rhizosphere bacteria. The sampling site is given in Figure 1.

Figure 1. Map of North East India showing sample collection site.
2.2. Climatic Condition
Northeast India has humid sub-tropical climate, ranging from hot to humid summers, severe monsoons with an average rainfall recorded around 10,000 mm and above and mild to chilled winter. Therefore, the variation in weather condition adds on to the exploration of indigenous microbes that which could adapt to changes in various environmental conditions and weather being one of the factors for varied applications.
2.3. Isolation of Bacteria
Ten grams of the root sample along with the rhizosphere soils were transferred to conical flasks with 90 ml of sterile physiological saline water and homogenized by shaking at 120 rpm for 30 mins Suspensions from the collected samples were serially diluted up to 10–5. One ml of the homogenates were spread over plates of King’s B agar medium, and Luria Bertani agar and incubated at 30˚C for 24 - 48 h. Subsequently, the colonies of different colony characteristics and also with microscopically different morphology were selected and purified by dilution streaks. The purified isolates were maintained at –80˚C in 30% glycerol stock for further biochemical analysis.
2.4. Fungal Strains
The fungal pathogens Fusarium oxysporum and Rhizoctonial solani were isolated from naturally infected French bean and identified by standard protocol based on the morphological and microscopical characteristics using Trinocular fluorescent microscope.
3. SCREENING OF ANTAGONISTIC ACTIVITY
The bacterial isolates were tested for their ability to inhibit the growth of R. solani and Fusarium oxysporum using dual plate assay. Fresh cultures of Fusarium oxysporum and R. solani were grown in PDA medium (Potato infusion-200; Dextrose-20; agar-18 (gl–1). A plug from the growing edges (5 mm diameter) of the colony was placed at one site of a fresh PDA plate. The bacterial isolates were grown overnight, spotted along the periphery of 90 mm diameter petri disc equidistant from the mycelial plug. These plates (4 replicates for each test organisms) along with uninoculated control were incubated at 30˚C for about 72 hrs or until the leading edge of the fungus reached the edge of the plate. Antibiosis was assessed by measuring the diameter of the mycelial growth of R. solani and Fusarium oxysporum.
4. DETERMINATION OF PLANT GROWTH PROMOTION MECHANISMS
Plant growth promotion attributes such as production of Indole acitic acid (IAA) like substances and P-solubilization activity of the fifteen selected isolates were determined in broth culture and agar plate using the modified method given [5]. Bacterial cultures were inoculated in LB broth amended with tryptophan (2 mg/ml) and incubated at 30˚C for 2 - 3 days. After centrifugation 2 ml of the supernatant was mixed with 2 drops of orthophosphoric acid along with 4 ml of Solawaski reagents and pink color intensity developed was measured at 530 nm O.D. Level of IAA in culture filtrates was determined from the standard graph obtained from concentration between 0 - 100 ppm IAA (Sigma). The production of P-solubilizing activity was determined in Pikovskaya’s agar which contains tri calcium phosphate as insoluble phosphate and cultures plates were grown in shaking conditions at 30˚C. Siderophore production was determined by the modified method [6] by plate method using Ternary complex chrome azurol S (CAS), Fe3+/Hexadicyl trimethyl ammonium bromide (HDTMA) as an indicator.
5. SCREENING OF DIFFERENT BIOCONTROL MECHANISMS
Different biocontrol mechanisms such as protease, chitinase, ß 1, 3, glucanase, and hydrogen cyanide production were assessed.
Protease activity was determined using the method described by [7]. The bacterial cultures were inoculated in Skim Milk Agar plate and incubated at 28˚C ± 2˚C for 1 - 2 days and clear zone formation was observed.
Chitinase activity was determined using chitin minimal medium. Cells were streaked on chitin minimal medium supplemented with colloidal chitin (1% w/v) and solidified with 1.5% agar [8].
ß 1, 3, glucanase activity was determined by modified method of Katatny et al., 2001 [9]. For plate screening, carboxy methyl cellulose agar medium amended with laminarin were used. Bacterial cultures were streaked at the centre of the plate. Plates were incubated at 25˚C for one day. For gluconolytic activity observations, plates were stained with 0.1% congo red dye for 15 - 20 minutes followed by destaining with 1N NaCl and then with 1N NaOH for 15 minutes for three times.
For quantitative determination of hydrogen cyanide production, bacterial isolates were separately streaked on TSA supplemented with 4.4 g glycine with simultaneous addition of cut filter paper disc soaked with 0.5% picric acid in 1% Na2CO3 and placed in the lid on each petridish by modified method of [10]. In the control tube, no culture was inoculated. These disc were sealed in parafilm and incubated for 4 days at 30˚C. The HCN production was determined by colour change observed in yellow colour turning brown to reddish brown. Scoring was done as weak if the colour changes from yellow to light red; moderate-yellow to brown and strong-yellow to reddish brown.
6. DETECTION FOR GENES INVOLVED IN ANTIBIOTIC PRODUCTION
PCR based detection of Burkholeria cepacia isolates were screened for the presence of biosynthetic loci/gene involved in the production of three known antibiotics with biocontrol activity (2,4, DAPG, pyrrolnitrin, phenazine) using specific PCR amplification from genomic DNA. The production of known antibiotics phenazine, DAPG and pyrrolnitrin (Primer sequences, Table 1) were detected using known gene specific primers. The PCR cycling conditions were as per standard protocol [11]. Amplified products were detected at 1.5% gel.
7. EVALUATION OF SEED GERMINATION AND DISEASE CONTROL EFFECT
To determine the influence of bacterial strain IBSDLKBI in French bean growth, a pot experiment was conducted during 2008-2009 for which the O.D of the culture broth was adjusted between 0.3 and 0.5 at 600 nm and mixed with an aqueous solution of 1% carboxy-

Table 1. Antibiotic gene specific primer used in this study.
methyl cellulose (CMC) to obtain a cell concentration of 109 cfu/ml. The standardization broths were used for bacterization of seeds. Fifty sterile French bean (Phaseolus mungo L.) seeds of local variety were treated for 30 min with standardized culture broth amended with 1% CMC. Control seeds were treated with mixture of uninoculated LB broth along with 1% CMC. Bacterized seeds were air dried overnight at room temperature. For checking the effectiveness in controlling wilt and damping off diseases in beans inocula of F. oxysporum and R. solani were prepared by incubating mycelial growth in presterilized oat grain for 7 days at 30˚C. Fungal inocula were mixed with soil two days ahead of the seed sowing and the fungal inoculum level was maintained at 105 cfu/g soil. The pot size was of 10 inches diameter containing 2 kg soil. The experiment was conducted using a randomized plot design with three replicates of each treatment. The treatment combinations of the experiment for evaluation of plant growth and biocontrol effect were 1) Control; 2) IBSD-LKBI and for assessing disease suppression experiment the treatments given were 1) IBSDLKBI + Fusarium oxysporum; 2) Fusarium oxysporum infested control plot; 3) IBSD-LKBI + R. solani; 4) R. solani infested soil. Sterilized seeds were planted at a depth of approximately 1 cm from the soil surface and watered with tap water daily. Percentage of disease incidence (%DI) were calculated at 30th day by using the following formula.

The ANOVA approach was used to evaluate the efficiency of biocontrol agent in field. Comparison among treatment and means of appropriate control treatment were made at P = 0.05. The pool data of the two consecutive seasons were subjected to ANOVA.
8. MOLECULAR IDENTIFICATION
8.1. Preparation of Genomic DNA
Genomic DNA was extracted using the modified method [12]. A single colony was inoculated to 5 ml of LB broth (Hi-media, M1245) at 30˚C, overnight. Culture broth of 5OD at A600 nm was centrifuged at 10,000 × g for 5 min (Allegra 22R, Backman-Coulter, USA). The cell pellet was washed twice with sterile distilled water and after removal of the supernatant, the pallet was suspended with 200 µl sterile deionised water (TKA water purification system, Germany) with 10 µl of lysozyme (1 mg·ml–1) GeNei, B3, (Bangalore, India) by vortexing and incubated at 37˚C for 30 min followed by boiling for 5 min and centrifuged at 10,000 × g for 5 min. The supernatant was carefully pipetted out and used for PCR analysis either immediately or stored at –20˚C until required. Purity of the DNA was measured using Nanodrop spectrophotometer by reading the value between 260/280 nm.
8.2. 16S-23S rDNA Gene Sequencing
Genomic DNA of IBSD-LKBI was amplified using the primers: 16SF-R2 (5’-CGCGGGATCCTTGTACA- CACCGCCCGTC-3’) and 23SR-R10 (5’-GGCCGTCGACCCTTTCCCTCACGGTACTG-3’) 0.5 µl (2U) of Taq DNA polymerase (Promega, M1901) and 19.0 µl of autoclaved deionised water (Millipore water purification system). The amplification was performed with a total of 36 PCR cycles in a thermal cycler Biorad, USA) at 94˚C for 10 min. These were then subjected to 36 cycles consisting of 94˚C for 30 sec, annealing at 55˚C for 1 min and elongation at 72˚C for 2 min and amplification product cooled at 4˚C. PCR products were purified and sequenced at Institute of Microbial Technology (IMTECH).
8.3. Nucleotide Sequence Accession Number
The nucleotide sequence of the 16S rDNA gene of IBSD-LKBI strain ishown to have 98% similarity with Burkholderia cepacia and it has been deposited in GenBank under accession number GQ927326.
8.4. Genomovar Identification
The sequences of the genomovar were compiled from NCBI nucleotide database. DNA sequence homology search were performed using the online BLAST search engine with megablast option in GenBank (available at http://www.ncbi.nlm.nih.gov) [13]. Comparative consistency analysis performed using TCOFFEE advance search engine [14]. Phylogenetic molecular evolutionary analysis was conducted using MEGA Version 5.05 [15].
9. RESULTS
9.1. Isolation & Screening of Antagonistic Activity
Out of the total 625 bacterial isolates collected from French bean rhizosphere soil of Imphal East and West different districts of Manipur using King’s B and LB agar, 15 isolates were selected based on their significant reduction in the fungal growth against F. oxysporum and R. solani where their zone width is more than 15 mm diameter after 4 days of incubation on dual plate assay and most potent bacteria were IBSD-LKBI, IBSD-56 and IBSD-38 (Table 2). Among the three bacterial strains IBSD-LKBI exhibited the highest inhibition zone diameter with significant fungal growth inhibition to control the two test fungal pathogens. Therefore, strain IBSDLKBI was selected for further studies.
9.2. Determination of Plant Growth Promotion Assay
The production of plant growth promotion hormone such as Indole acitic acid (IAA) like substances were assessed in culture filtrate supplemented with tryptophan. Level of IAA like substances ranges from 22.1 - 65.2 µg/ml culture filtrate (Table 1) and IBSD-LKBI produced significant level of IAA at the rate of 58.4 µg/ml culture filtrate. Similarly, in addition to this P-solubilization and siderophore production was determined after 24 - 48 hr of incubation of the culture. Out of the 15 selected potential isolates 67% showed P-solubilization activity. In case of siderophore activity formation of bright zone with yellowish fluorescens in dark blue medium were observed. Out of the total 15 isolates only 6 isolates showed siderophore production with highest zone produced by isolate IBSD-P-50 with a zone diameter of 15 mm (Table 2).
9.3. Extra Cellular Cell Wall Degrading Enzyme Assay
The potential for the production of different fungal cell wall degrading enzymes such as chitinase, protease, ß-1, 3-glucanase activity were assessed for the selected potential strains and evaluated based on different enzymatic activities. All the isolates were inoculated in colloidal chitin supplemented medium for chitinase activity and skim milk agar plates for protease activity and later the plates were incubated at 30˚C for 2 - 3 days and observed for the clear zone formation around the colonies. A total of 10 isolates showed chitin activity ranging from 7 mm to 14.1 mm zone diameter whereas for protease activity the zone diameter ranges from 7.5 - 15.5 mm of which IBSD-LKBI exhibits highest activity of both chitinase and protease activity. In addition to this 9 isolates showed ß-1, 3-glucanase activity. HCN production was observed only in three isolates (Table 2).
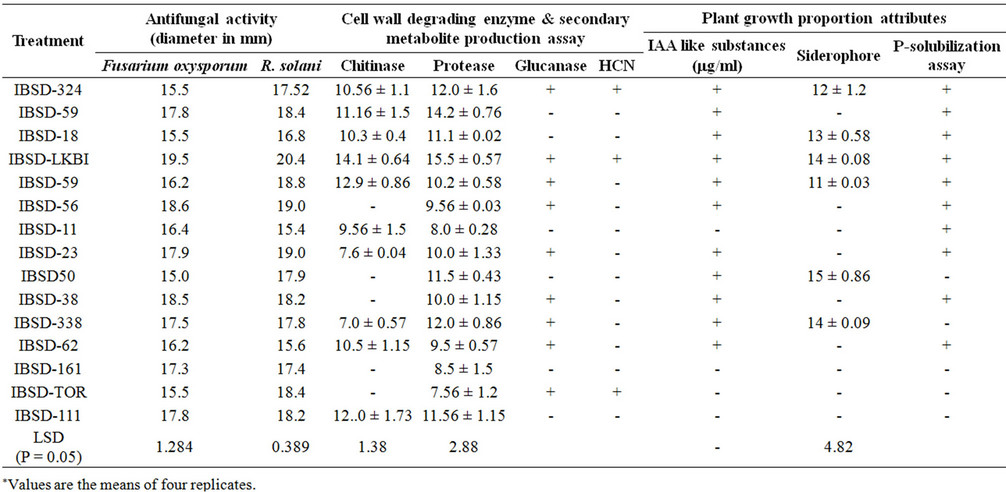
Table 2. Evaluation of different antagonistic and biocontrol potential of the bacterial isolates.
9.4. Biochemical Characteristics of IBSD-LKBI
This bacterial isolates were gram negative, motile, rod shaped bacteria and exhibited positive catalase, oxidase and arginine hydrolysis. It can utilize dextrose, galactose and manose. Based on the above characteristics the isolates could be identified as genus Burkholderia sps.
9.5. Detection of Biosynthetic Genes for Antibiotic Production
During the study on production of genes involved in antibiotic productions such as 2,4 DAPG, pyrolnitrin and phenazine based on gene specific primers, PCR based detection confirms that B. cepacea IBSD-LKBI produces genes responsible for phenazine production (Figure 2).
9.6. Impact Assessment of IBSD-LKBI on Plant Growth Promotion
The effect of plant growth promotion using IBSD-LKBI was assessed in French beans healthy soil during 2008 & 2009. The rate of seed germination percentage was found to significantly increase (88%) as compared to the control plant (68%). After two months of planting the plant height and root length treated with IBSD-LKBI were found to remarkably increase at 40.0 ± 3.0 & 21.0 ± 1.0 respectively and 34.0 ± 2.0 and 14.0 ± 1.5 in case of untreated plants. Whereas the seedling biomass was found to be 27.86 ± 1.2 in case of treated plant against the control untreated plant (16.62 ± 2.0) which is good sign to be used as a strong plant growth promoter in French bean (Table 3).
9.7. Assessment of Biological Control Efficacy
The strain IBSD-LKBI had good control effects on French bean under potconditions in disease infested soils (Table 4). In the Fusarium and R. solani treated plots, % of disease incidence were found to be 65.5% and 68% respectively. As per disease control effect exhibited by IBSD-LKBI treated plant with a control value of 86.9% and 67.4% against the wilt and damping-off diseases in French bean caused by Fusarium oxysporum and R. solani respectively. Evaluation of IBSD-LKBI treated plant showed that its control effects were equivalent to that of inhibition of mycelial growth of the pathogens in vitro conditions. Therefore, these pooled data clearly indicates that indigenous strains IBSD-LKBI could significantly reduce the severity and development of wilting and damping-off disease in French beans and could be used as a potential bio-control agent.
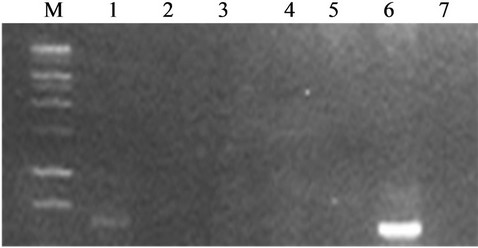
Figure 2. Detection of band for antibiotic using PCR gene specific primer for Phenazine-1-carboxylic acid. Lane M: 100 bp DNA marker, 1: IBSD-38, 2: IBSD-56 3: IBSD-11, 4: IBSD-50, 5: IBSD-TOR, 6: IBSD-LKBI, 7: RFP-18.
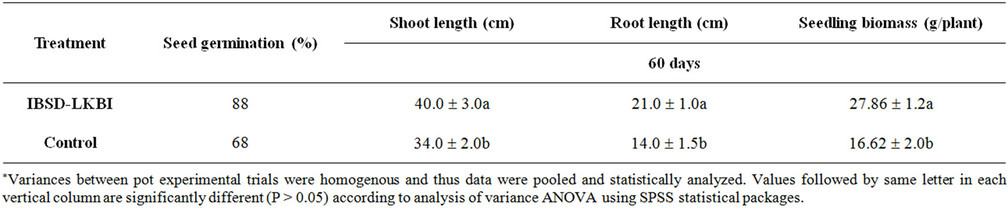
Table 3. Effect of seed bacterization with IBSD-LKBI strain on French bean seedling and plant growth promotion.
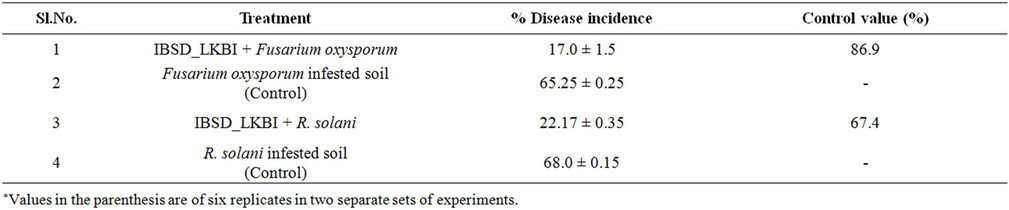
Table 4. Effect of seed bacterization with IBSD-LKBI strain on percentage of disease incidence of French bean seedling in F. oxysporum and R. solani infested soil.
9.8. Comparison of Sequence Identity among the Burkholderia Species Belonging to Different Genomovars
Until now no clear distinction has been established between the ten genomovars of Bcc, but rDNA and recA gene derived primer pairs are available for the identification of B. cepacia genomovar (I), B. multivorans (II), B. cenocepacia (III), B. stabilis (IV), B. vietnamiensis (V), B. dolosa (VI), B. ambifaria (VII), B. anthina (VIII), B. pyrrocinia (IX), B. ubonensis (X), [16-18]. In present study Sixteen (16) sequences of 16S rDNA of Bcc and one (1) recA gene sequence of Burkholderia dolosa (AF323971) were studied for molecular and evolutionary analysis [19-21].
The isolated sequence was compared for consistency analysis [14] of 16S rDNA sequences of isolates from India as well as all representative isolates from other part of the world belonging to various Burkholderia genomovars [6] (Table 5). The Phylogenetic analysis based on 16S rDNA sequence with neighbor joining method at bootstrap values based on 1000 replications establish a clear cut genomovar clusters of Bcc [15] Figure 3. Notably these clusters were further strengthens the consistency analysis for Bcc.
10. DISCUSSION
Antagonistic plant-associated bacteria are an important functional group of beneficial bacteria responsible for the control of soil-borne pathogens (Weller, 1988). B. cepacia was first described by Burkholderia in 1950 isolated from putrefactive onions [22]. Several B. cepacia species are considered to be beneficial in nature and are widespread in nature particularly in plant environment. B. cepacia is a bacterial complex with 10 genomic species which have similar phenotypical traits and different genetic ones and are called genomovars [23,24]. The Burkholderia cepacia complex (Bcc) consists of several species of closely related and extremely versatile gramnegative bacteria found naturally in soil, water, and the rhizosphere of plants. Strains of Bcc have been used in biological control of plant diseases and bioremediation in many cases, while some strains are plant pathogens or opportunistic pathogens of humans with cystic fibrosis. Members of genomovars I, III, VII and IX had been confirmed as having good biocontrol ability, especially effective against Pythium and Rhizoctonia [25,26].
Sergio de los reported Burkholderia cepacia XXVI as a promising biological control agent against the causal agent of anthracnose, C. gloeosporioides, through the production of hydroxamate siderophore [27]. Zaiton established Burkholderia cepacia strain UPM B3, isolated from symptomless palms and its potential as a biocontrol agent against Ganoderma [28]. The ecological versatility of these bacteria is likely due to their unusually large genomes, which are often comprised of several (typically two or three) large replicons, as well as their ability to use a large array of compounds as sole carbon
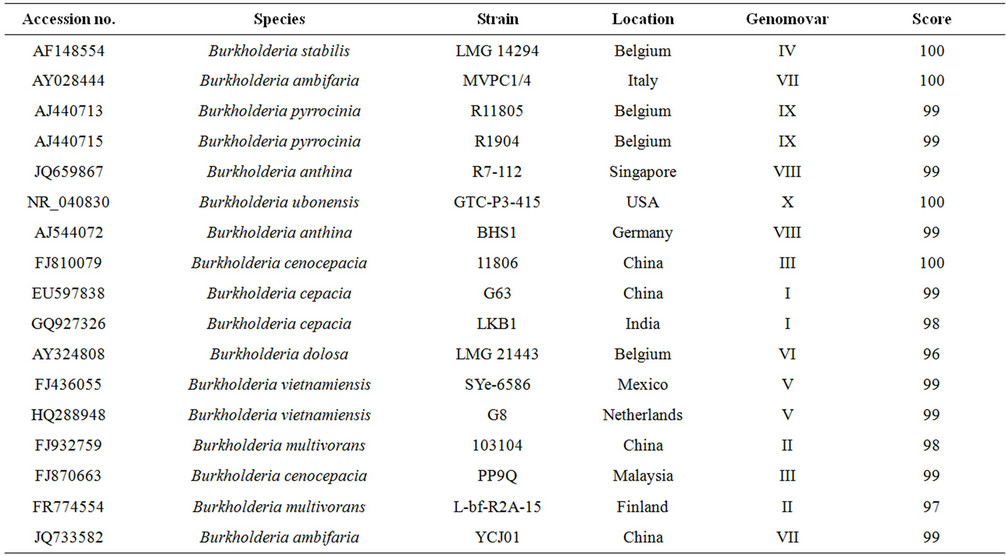
Table 5. The LKB1 isolate from India (GQ927326) is compared at the nucleotide level using TCOFFEE advanced search engine with representative isolates of Burkholderia from India and other part of the world using consistency score.
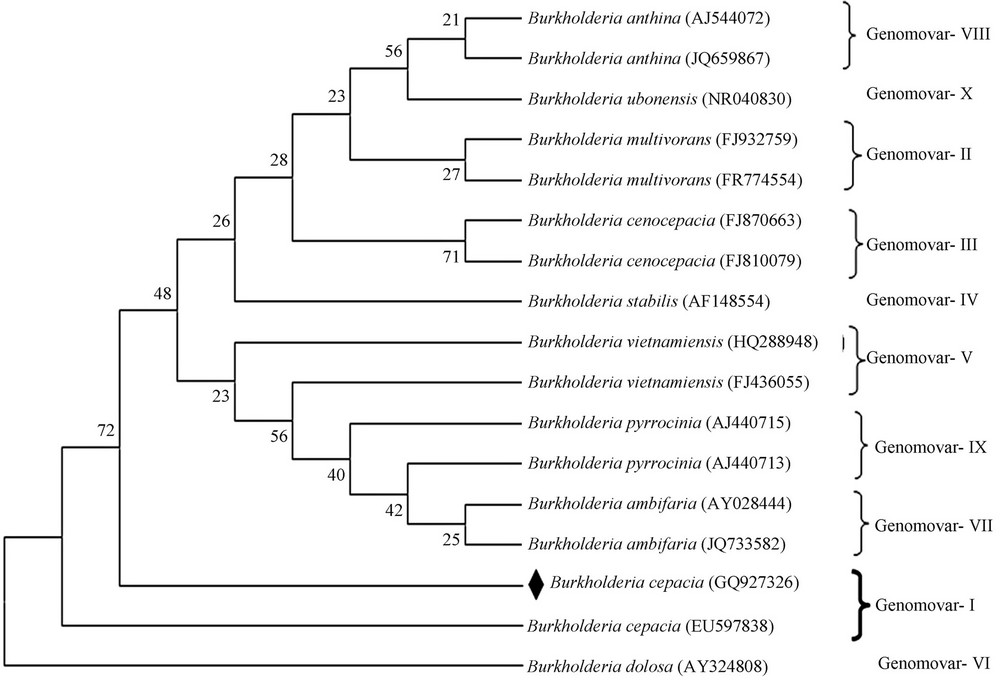
Figure 3. Phylogenetic tree based on 16S rRNA (except AY324808 genomic DNA) sequences, representing the relatedness among the Burkholderia genomovars. Distance and clustering was performed by using the software package MEGA 5.05. Boot strap values based on 1000 replication are listed at the branching point. The sequence of present study dotted in black.
sources. B. cepacia has been reported to promote growth of maize, enhance crop yield [29], and degrade diverse pesticides [30].
In the present study, several indigenous bacterial strains were isolated from Imphal East and West districts of French bean growing area and characterized by phenotypic and genotypic approach and investigated for their ability to inhibit the growth of Fusarium oxysporum and Rhizoctonia solani. To the best of our knowledge, indigenous native isolates performs best in terms of its specificity in a particular environment where it survive [31]. The use of indigenous bacterial isolates such as Pseudomonas and Burkholderia species has not been reported so far from NE India for the control of soil borne diseases in french beans.
To find new biocontrol agents antagonistic to the soilborne fungus Fusarium oxusporum and R. solani, 625 rhizobacterial isolates were evaluated using a combination of different screening steps. As a result of the first screening, isolates were found to produce detectable inhibition zones against the two test fungus on agar plate assay. The production of clear zones in dual plate-culture screens is due to the production of cell wall degrading enzymes and toxic metabolites.
Because of the discrepancies, in vitro and in vivo studies, the assay should be combined with other investigations as was done here. In the present study, we found examples for a good correlation between in vitro and in vivo results e.g. IBSD-LKBI. The identification of an antagonistic bacterium is another important step in the assessment of the BCA.
Production of cell wall degrading enzymes such as glucanase, chitinase, protease, and production of secondary metabolites (Siderophore and HCN) are common mechanisms that bacteria use to inhibit fungal pathogens [5,32,33]. In this study, IBSD-LKBI found to produce different cell wall degrading extracellular enzymes such as protease, glucanase, chitinase and stipulated its role in biological control. The result also supported with the earlier studies [34,35]. This biocontrol strain also produces phenazine when assessed with gene specific primers (Figure 2).
IBSD-LKBI isolates also shows a strong plant growth promotion activity as supported by the production of plant growth hormone such as IAA. Several soil microorganisms have the ability to solubilise insoluble phosphate and maintain soil health and quality. IBSD-LKBI showed the capacity to solubilize inorganic phosphate. In addition to this it can produce Fe+ chelating siderophore which are a good sign of PGP activity.
To select bacterial isolates with highest antagonistic properties for approaches in biocontrol application, a multistep screening for biocontrol activity against Fusarium oxysporum and R. solani and PGR combined with its genotypic and phenotypic characterization was performed. All of these data were integrated into an assessment scheme to identify the best antagonist. Based on this assessment scheme, IBSD-LKBI strain could be considered good biocontrol candidates which can control wilting and damping-off diseases in French bean pathosystems. B. cepacia (Pseudomonas cepacia formerly) has been reported to prevent Pythium diseases of cucumber and peas and R. solani stem rot of poinsettia [26].
Rhizosphere plant associated bacteria are able to interact more closely with the host plant and fungal pathogens living in soil and plant tissue [36]. Our findings confirm that they could be efficient BCAs. More efficient methods on characterization and identification of BCC are now available for precise evolutionary relationship. Notably, these methods enable the study of fastidious or pathogenic species without employing tricky or expensive microbiological methods. Consistency and distance values prove B. cepacia (IBSD-LKBI) relates to the genomovar I phylogenetic tree of BCC genomovar relatedness based on 16S r DNA are shown in Figure 3. The rhizosphere associated B. cepacia IBSD-LKBI is an interesting candidate for biological control. In further experiments, these selected BCAs will be evaluated in field trial to select the best candidate for commercialization. B. cepacia strains reported in this study as a strong BCA is required to study their ecological role in soil and may be used as a biocontrol agent in sustainable agricultural practices.
11. ACKNOWLEDGEMENTS
We are thankful to Department of Biotechnology for giving financial assistance in this work.
REFERENCES
- Copping, G.L.. and Menn, J.J. (2000) Biopesticides: A review of their action, applications and efficacy. Pest Management Sciences, 65, 651-676. doi:10.1002/1526-4998(200008)56:8<651::AID-PS201>3.0.CO;2-U
- Parmeter, J.R. (1970) R. solani. Biology of pathology. University of California Press, Berkeley Los Angles & London.
- Govindarajan, M., Balandreau, J., Kwon, S.W., Weon, H.Y. and Lakshminarasimhan, C. (2008) Effects of the inoculation of Burkholderia vietnamensis and related endophytic diazotrophic bacteria on grain yield of rice. Microbial Ecology, 55, 21-37. doi:10.1007/s00248-007-9247-9
- Payne, G.W., Vandamme, P., Morgan, S.H., LiPuma, J.J., Coenye, T., Weightman, A.J., Jones, T.H. and Mahenthiralingam, E. (2005) Development of a recA gene-based identification approach for the entire Burkholderia genus. Applied and Environmental Microbiology, 71, 3917-3927. doi:10.1128/AEM.71.7.3917-3927.2005
- Indira, S., Talukdar, N.C., Chandradev Sharma, K., Jeyaram, K and Rohinikumar. M. (2011) Screening of rhizobacteria for their plant growth promotion ability and antagonism against damping off and root rot diseases of broad bean (Vicia faba L.). Indian Journal of Microbiology, 51, 14-21. doi:10.1007/s12088-011-0069-6
- Samad, A., Ajaykumar, P,V., Gupta, M.K., Shukla, A.K., Darokar, M.P., Somkuwar, B.G and Alam, M. (2008) Natural infection of periwrinkle (Catharanthus roseus) with Cucumber mosaic virus, Subgroup IB. Australian. Plant Disease Notes, 3, 30-34. doi:10.1071/DN08013
- Berg, G., Roskot, N., Steidle, A., Ebert, L., Zock, A. and Smalla, K. (2002) Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Applied and Environmental Microbiology, 68, 3328-3338. doi:10.1128/AEM.68.7.3328-3338.2002
- Chernin, L.S., D.E. Fuente, L, Sovolev, V., Haran, S., Vorgias, C.E., Oppenheim, A.B. and Chet. (1997) Molecular cloning, structural analysis, and expression in Escherichia coli of a chitinase gene from Enterobacter agglomerans. Applied and Environmental Microbiology, 63, 834-839.
- Katatny, El., M.H., Gudelj, M., Robra, KH., Elnaghy, M.A. and Gubitz, G.M. (2001) Characterization of a chitinase and an endo-ß-1,3-glucanase from Trichoderma harzianum Rifai T24 involved in control of the phytopathogen Sclerotium rolfsii. Applied and Microbial Biotechnology, 56, 137-143.
- Millar, R.L. and Higgins, V.J. (1970) Association of cyanide with infection of birds foot trefoil by Stemphyllium loti. Phytopathology, 60, 104-110. doi:10.1094/Phyto-60-104
- Raajmakers, J.M., Weller, D.M. and Thomashow, L.S. (1997) Frequency of antibiotic-production Pseudomonas spps. in natural environments. Applied and Environmental Microbiology, 63, 881-887.
- Zhang, Y.C., Ronimus, R.S., Turner, N. Zhang, Y. and Morgan, H.W. (2002) Enumeration of thermophillic Bacillus species in composts and identification with a Random Amplified Polymorphic DNA (RAPD) protocol system. Applied Microbiology, 25, 618-626. doi:10.1078/07232020260517760
- Zhang, Z., Schwartz, S., Wagner, L. and Miller, W. (2000) A greedy algorithm for aligning DNA sequences. Journal of Computational Biology, 7, 203-214. doi:10.1089/10665270050081478
- Poirot, O., O’Toole, E. and Notredame, C. (2003) Tcoffee@igs; A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acid Research, 13, 3503-3506. doi:10.1093/nar/gkg522
- Kumar, S., Tamura, K., Jakobsen, I.B. and Nei, M. (2001) MEGA2: Molecular evolutionary genetics analysis software. Arizona State University, Tempe.
- Vandamme, P., Holmes. B., Vancanneyt, M., Coenye, T., Hoste, B., Coopman, R., Revets. H., Lauwers, S., Gillis, M., Kersters, K. and Govan, J.R. (1997) Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderai multivorans sp. nov. International Journal of Systemic Bacteriolog, 47, 1188-1200. doi:10.1099/00207713-47-4-1188
- Coenye, T., Vandamme, P., Govan, J.R. and Lipuma, J.J. (2001c) Taxonomy and identification of the Burkholderia cepacia complex. Journal of Clinical Microbiology, 39, 3427-3436. doi:10.1128/JCM.39.10.3427-3436.2001
- Perin, L., Martinez-Aguilar, L., Castro-Gonzalez, P., Estrada-de, LSP., Cabellos-Avelar, T., Guedes, H.V., Reis V.M. and Caballero-Mellado, J. (2006) Diazotrophic Burkholderia species associated with field grown maize and sugercane. Applied and Environmental Microbiology, 72, 3103-3110. doi:10.1128/AEM.72.5.3103-3110.2006
- Mahenthiralingam, E. and Vandamme, P. (2005) Taxonomy and pathogenesis of the Burkholderia cepacia complex. Chronic Respiratory Disease, 2, 209-217. doi:10.1191/1479972305cd053ra
- Spreet, D.P. (2001) Understanding Burkholderia cepacia: Epidemiology, genomovars, and virulent. Infections in Medicine, 18, 49-56.
- Weisburg, W.G., Barns, S.M., Pelletier, D.A. and Lane, D.J. (1992) 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology, 173, 697-703.
- Allan, N.D., Kooi, C., Sokol, P.A. and Beveridge, T.J. (2003) Putative virulence factors are released in association with membrane vesicles from Burkholderia cepacia. Canadian Journal of Microbiology, 49, 613-624. doi:10.1139/w03-078
- Cardona, S.T., Wopperer, J., Eberl, L. and Valvano, M.A. (2005) Diverse pathogenecity of Burkholderia cepacia complex strains in the Caenorhabditis elegans host model. FEMS Microbiology, 14, 277-286.
- Chiarini, L., Bevivino, A., Dalmastri, C., Tabacchioni, S. and Visca, P. (2006) Burkholderia cepacia complex species: Health hazards and biotechnological potential. Trends in Microbiology, 14, 277-286. doi:10.1016/j.tim.2006.04.006
- Milus, E.A. and Rothrock, C.S. (1997) Efficacy of bacterial seed treatments for controlling Pythium root rot of winter wheat. Plant Disease, 81, 180-184. doi:10.1094/PDIS.1997.81.2.180
- Cartwright, D.K. and Benson, O.M. (1995) Comparison of Pseudomonas species and application techniques for biocontrol of Rhizoctonia stem rot of poinsettia. Plant Disease, 79, 309-313. doi:10.1094/PD-79-0309
- de los Santos-Villalobos, S., Barrera-Galicia, G.C., MirandaSalcedo, M.A. and Peña-Cabriales J.J. (2012) Burkholderia cepacia XXVI siderophore with biocontrol capacity against Colletotrichumgloeosporioides. World Journal of Microbiology and Biotechnology, 28, 2615-2623. doi:10.1007/s11274-012-1071-9
- Zaiton, S., Sariah, M. and Zainal Abidin, M.A. (2008) Effect of endophytic bacteria on growth and suppression of Ganoderma infection in oil palm. International Journal of Agriculture and Biology, 10, 127-132.
- Bevivino, A., Sarrocco, S., Dalmastri, C., Tabacchion, I.S., Cantale, C. and Chiarini, L. (1998) Characterization of a free-living maize-rhizosphere population of Burkholderia cepacia: Effect of seed treatment on disease suppression and growth promotion of maize. FEMS Microbiology Ecology, 27, 225-237. doi:10.1111/j.1574-6941.1998.tb00539.x
- McLoughlin, T.I., Quinn, J.P., Bettermann, A. and Bookland, R. (1992) Pseudomonas cepacia suppression of sunflower wilt fungus and role of antifungal compounds in controlling the disease. Applied and Environmental Microbiology, 58, 1760-1763.
- Larralde, C.P., Santiago-Mena, M.R., Sifuentes-Rincon A.M., Rodrigue-Luna, I.C., Rodriguez-Perez, M.A., Shiraj, K. and Narvaez-Zapata, J.A. (2008) Biological potential and polyphasic characterization of novel native Trichoderma strains against Macrophomina phaseolina from Sorgum and common bean. Applied Microbiology and Biotechnology, 80, 167-177. doi:10.1007/s00253-008-1532-0
- Raaijmakers, J.M., Vlami, M. and de Souza, J.T. (2002) Antibiotics production by bacterial biocontrol agents. Antonie Van Leeuvenhoek International Journal, 81, 537- 547. doi:10.1023/A:1020501420831
- Kim, P.I. and Chung, K.C. (2004) Production of an antifungl protein for control of Colletotrichum lagenarium by Bacillus amyloliquefaciens MET0908. FEMS Microbiology Letters, 234, 177-183.
- Ogawa, K., Yoshida, N., Kariya, K., Ohnishi, C. and Ikeda, R. (2002) Purification and characterization of a novel chitinase from Burkholderia cepacia strain KH2 isolated from the bed log of Lentinus edodes, Shiitake mushroom. The Journal of General and Applied Microbiology, 48, 25-92. doi:10.2323/jgam.48.25
- Fridlender, M., Inbar, J. and Chet, I. (1993) Biological control of soilborne plant pathogens by a ß-1,3 glucanase-producing Pseudomonas cepacia. Soil Biology and Biochemistry, 25, 1121-1221. doi:10.1016/0038-0717(93)90217-Y
- Bal, A.S. and Chanway, C.P. (2000) Isolation and identication of endophytic bacteria from lodgepole pine and western red cedar. Auburn University website. http://www.ag.auburn.edu/argentina/pdfmanuscripts/bal.pdf
NOTES
#Corresponding author.

