Journal of Biomaterials and Nanobiotechnology
Vol.4 No.1(2013), Article ID:27379,9 pages DOI:10.4236/jbnb.2013.41013
Antimicrobial Susceptibility, Biofilm Production and Adhesion to HEp-2 Cells of Pseudomonas aeruginosa Strains Isolated from Clinical Samples
![]()
Research Center in Parasitic Diseases and Endemic, University of CEUMA, UniCEUMA, São Luís, Brazil.
Email: *figueiredo.patricia@gmail.com
Received October 7th, 2012; revised November 10th, 2012; accepted December 13th, 2012
Keywords: Pseudomonas aeruginosa; Antimicrobial; Biofilm; Adhesion
ABSTRACT
A hundred Pseudomonas aeruginosa strains from several clinical specimens from five hospitals in São Luís-MA were evaluated for biofilm production, prevalence of the gene algD, adhesion to HEp-2 cells and antimicrobial susceptibility. The most affected clinical specimens and hospital sectors were also evaluated. Most isolates were obtained from the tracheal aspirate (21.0%) and the most affected hospital sector was the ICU (43.0%). The antibiotics with the highest sensitivity rate were amikacin, piperacillin/tazobactam, fluoroquinolones, gentamicin and meropenem and the ones with the highest resistance rate were aztreonam, ceftazidime and cefepime. All samples were sensitive to polymyxin B. In relation to the expression of the gene for ESBL, 50.0% (17/34) of the multiresistant strains showed the enzyme TEM. Most strains showed high hydrophobicity and 96% of the isolates produced biofilm on a polystyrene microplate, 52% were capsule producers, 19% showed mannose-sensitive fimbriae and 39% expressed the gene algD. We observed adhesion to HEp-2 cells and to the coverslip. These factors may be reported in the pathogenesis of this bacterium, what represents a potential risk for colonization of medical devices which favor the establishment of chronic nosocomial infections.
1. Introduction
Pseudomonas aeruginosa is a glucose non-fermenting Gram-negative bacillus, which is widely distributed in nature and in hospital environment, since it has minimum nutritional necessities, being able to survive in several surfaces and in humid places. This bacterium rarely causes severe infections in healthy individuals, nevertheless, it represents a great threat for hospitalized patients [1]. It is an opportunist bacterium, invasive and toxigenic that, over the last few years, has reached an important position among the nosocomial pathogens for causing pneumonia related to mechanical ventilation, bacteremia, endocarditis, meningitis, urinary infections associated to catheters and skin infections, especially in critical ICU patients and in those who are immunocompromised [2].
Nosocomial infections caused by P. aeruginosa are related to high morbimortality rates and among the risk factors for colonization or invasive infection by this pathogen, we can include the prolonged use of antimicrobials, previous hospitalization, severe base disease, operation and immunosuppression [3]. This bacterium shows instrinsic resistance to several antibiotics, combined with the ability of getting new resistance information during treatments, through mutations in porins (decrease in OprD), super expression of efflux pumps (MexAB-OprM) and/or production of hydrolytic enzymes that degrades antimicrobials, such as betalactamase and metallo-β-lactamases [4,5].
The carbapenems have been considered the drugs of choice in the treatment of severe nosocomial infections caused by Gram-negative bacteria. However, the isolation of bacteria is already common in Brazilian hospitals, especially P. aeruginosa, resistant to these antibiotics and in these cases polymyxin is the only therapeutic option [6].
The pathogenesis of the infections caused by P. aeruginosa is multifactorial and involves several virulence factors [7]. Among the factors characterized as adhesins, there are flagella, fimbriae and alginate. The fimbriae or pili promote the adherence of the bacterium to receptors on the surface of the host cell, and the type IV fimbria, especially the adhesin of this bacterium, is responsible for the initial adhesion both in abiotic material and in the surface of epithelial cells, and the flagella, primarily responsible for the motility, may act also as adhesins to the epithelial cells [8].
The alginate, a mucoid exopolysaccharide produced by some strains of P. aeruginosa, in addition to functioning as an adhesin, it also protects the strains from the mucociliary activity, phagocytosis, complement system activity, reduces the antimicrobial action, making its penetration in the bacterium difficult, and it is also associated to biofilm production [9].
Biofilm is a community of bacteria adhered to a biotic and/or abiotic surface, promoting survival advantages for micro-organisms, as resistance to antimicrobials, protection against antiseptics, disinfectants, bacteriophages, host’s immune system, among other [10,11].
The objectives of this study were: to identify the inpatient units and the clinical specimens most affected by P. aeruginosa; to analyze the profile of susceptibility to antimicrobials and the expression of extended-spectrum betalactamase in multiresistant strains by PCR; to evaluate the type of fimbriae, the hydrophobicity and the biofilm production; examine the prevalence of the gene encoding the alginate and check the adhesion capacity of the multiresistant samples in HEp-2 cells.
2. Material and Method
2.1. Bacterial Strains
We evaluated 100 strains of P. aeruginosa isolated from several clinical specimens of inpatients from one private and four public hospitals in São Luís-MA, from March to July, 2010. The distribution of samples per hospital was: HospA (47.0%), HospB (21.0%), HospC (11.0%), HospD (8.0%) e HospE (13.0%). All strains were identified through the automated method Vitek 2 (bioMérieux)Ò
in a local microbiology laboratory. In the tests, the strains P. aeruginosa ATCC 27853 and E. coli ATCC 25922 were used as positive and negative control, respectively.
2.2. Antimicrobial Susceptibility
This test was conducted through the disk diffusion antibiotic sensitivity testing (Kirby & Bauer), with the following antimicrobials (OXOID): amikacin, gentamicin, cefepime, ceftazidime, ciprofloxacin, levofloxacin, aztreonam, imipenem, meropenem, piperacillin/tazobactam and polymyxin B. The reading of the zones of inhibition for each antibiotic followed the criteria established by the Clinical and Laboratory Standards Institute [12] and the results were reported as susceptible, intermediate or resistant. We considered as multiresistant those strains that showed resistance to three of more classes of antibacterials.
2.3. Amplification of β-Lactamase Genes by PCR
The detection of the genes blaTEM, blaCTX-M and blaSHV, which encodes the β-lactamase (ESBL) enzymes, was carried out by multiplex PCR, according to what is described by Monstein et al. [13]. The primers used are described in Table 1. The total DNA extraction was carried out by boiling the bacterial cells for 10 minutes. The amplification reactions were performed with 2.5 µL of DNA from each strain, 0.8 µL of specific primers (1 µM), 0.25 µL Taq DNA polymerase (5 u/µL), 5 µL of the buffer solution Green Go Taq Flexi Buffer (5×), 2 µL MgCl2 (2 mM), 0.25 µL dNTPs (20 mM) and 12.2 µL of milli-Q water in a final volume of 25 µL. DNA was amplified in the thermocycler Mycicler (BioRad), using the following protocol: 95˚C for 15 min, 30 cycles at 94˚C for 30 s, 60˚C for 30 s, 72˚C for 2 min and 72˚C for 10 min. The PCR products were detected after electrophoresis in agarose gel 2.5% at 60/80 v for 55 min, then stained with ethidium bromide (20 µg/100mL of water) and viewed under ultraviolet light with the aid of a transilluminator.
Table 1. Primers used in amplification by multiplex PCR for ESBL detection.
 Source: Monstein et al. [13].
Source: Monstein et al. [13].
2.4. Evaluation of Capsule Production
This test was performed by the method of inoculation in Congo Red Agar (CRA), described by Freeman et al. [14], with minor modifications. The CRA was prepared from 37 g∙L−1 of BHI, 50 g∙L−1 saccharose (Difco), 15 g∙L−1 agar base (Difco) and 0.8 g∙L−1 Congo Red (Difco). After overnight growth, the strains were inoculated onto CRA and incubated at 37˚C/24 hours. The black bacterial colonies were considered as capsule producers and the red ones were non-producers.
2.5. Biofilm Induction in Vitro
This test was performed in microtiter plates of polystyrene by the Crystal Violet method, described by Stepanovic et al. [15]. After inoculation in BHI and incubation at 37˚C/24 hours, the samples were diluted (1:40) in sterile BHI and then, 200 µL of each sample in triplicate were added to the wells of the polystyrene plate, with only 200 µL of BHI in triplicate as negative control and the plates were incubated at 37˚C/24 hours. They were subsequently washed (3×) with PBS (pH 7.4), left to dry at room temperature, added crystal violet (200 µL/well) and incubated for 15 minutes at room temperature. Then, each well was washed with sterile distilled water (3×) and left to dry at room temperature. Biofilm formation was evaluated by the reading of the absorbance of each well using a spectrophotometer (BIO-RAD Laboratories PR 2100), at a wavelength of 490 nm. Based on the optical density of the samples (ODi) and on the average of optical density of the negative control (ODc), the samples were classified as strong (4xODc < ODi), moderate (2xODc < ODi ≤ 4xODc), weak (ODc < ODi ≤ 2xODc) or non-producer of biofilm (ODi < ODc).
2.6. Fimbriae Identification
The samples were inoculated in BHI, incubated at 37˚C/24 h and centrifuged three times (360 rpm/15 min), highlighting the supernatant and adding PBS (pH 7.4). The sheep blood was washed three times with PBS (pH 7.4) to prepare a solution of erythrocytes 1% in PBS with and without mannose addition (5.4 g of mannose/1000 mL). On glass slides, 100 µL of the isolates were mixed with 100 µL of the erythrocytes solution with and without mannose. The samples that suffered hemagglutination only in the absence of mannose were considered mannose-sensitive and the one that suffered hemagglutination in presence and absence of mannose, mannoseresistant [16].
2.7. Evaluation of Bacterial Hydrophobicity
After growth in agar BHI for 18 hours at 37˚C, the cultures were suspended in increasing concentrations of ammonium sulfate (0.5 M, 1 M, 1.5 M, 2 M, 2.5 M and 3 M). The formation of clumps within two minutes after suspension indicated positive result [17].
2.8. Search for the Gene algD by PCR
The PCR technique was performed with the oligonucleotides (F-ATGCGA ATCAGCATCTTTGGT and R-CTACCAGCAGATGCCCTCGGC) specific for the gene algD, that encodes GDP mannose 6-dehydrogenase (alginate), amplifying a fragment of 1310 pairs of nitrogenous bases. The total DNA extraction was obtained by boiling the bacterial cells for 10 min. The amplification reactions were performed in a final volume of 25 µL, containing 2 µL of bacterial DNA, 2.5 nM of dNTP, 20 pmol of the primer, 2 mM MgCl2 and 1.0 U of Taq DNA polymerase in reaction buffer 1× (Flexi Buffer). DNA was amplified in the thermocycler Mycicler (BioRad) using the following protocol: 94˚C for 3 min, 30 cycles at 94˚C for 30 s, 55˚C for 1 min, 72˚C for 1 min and 30 s and 72˚C for 5 min. The PCR products were detected after electrophoresis in agarose gel 2.5% at 80 v for 55 min, then stained with ethidium bromide (20 µg/100mL of water) and viewed under ultraviolet light with the aid of a transilluminator [18].
2.9. Test of Adhesion to HEp-2 Cells
Only multiresistant strains were tested. An inoculum with 40 µL of each of these strains cultivated in BHI was added to plates of 24 wells, containing cellular half-confluent monolayer of HEp-2 cells in MEM medium with SFB 2% in absence or presence of D-mannose 2%. After 2 h of incubation at 37˚C in atmosphere of 5% of CO2 (period of infection), the plats were washed with PBS and we added to the medium 1 mL of MEM plus SFB with and without mannose. After 2 h (multiplication period), the plates were washed with PBS and the cells were fixed with methanol for at least 10 minutes. Afterwards, they were stained with May-Grunwald/Giemsa and analyzed according to the adherence pattern on the optical microscope NIKON 19 [19].
2.10. Statistical Analysis
We used the Contingency Coefficient C and the Chisquared distribution. Values of p < 0.05 were considered significant. In data electronic processing, we used the program BioEstat 5.0.
3. Results
The highest percentages of P. aeruginosa isolates were in the tracheal secretion, with 25 samples (25.0%) and in urine, with 19 (19.0%). The places with highest incidence of this bacterium were ICU, with 43 samples (43.0%), followed by the internal medicine ward, with 31 (31.0%) (Table 2).
The strains showed most sensitivity to amikacin (75.0%), piperacillin/tazobactam (69.0%), fluoroquinolones (68.0%), gentamicin (64.0%) and meropenem (62.0%). The antibiotics with the highest resistance rates were aztreonam (48.0%), imipenem (43.0%), ceftazidime (42.0%), cefepime (41.0%) and meropenem (38.0%). All the isolates were susceptible to polymyxin B.
From all the isolates, 34.0% (34/100) showed a multiresistance profile. From these, 50.0% (17/34) were sensitive only to polymyxin B. Our results also showed that 53.0% (18/34) of the multiresistant strains were from the ICU. These strains were also submitted to the multiplex PCR technique for detecting ESBLS, and 50.0% (17/34) of the samples expressed the gene TEM, there was no expression of the genes SHV and CTX-M.
In the evaluation of bacterial hydrophobicity, 50.0% - 53.0% of the isolates had clumps formation in ammonium sulfate concentrations equal or above 2.5 M. In the identification of the isolates which produced fimbriae, 19.0% were producers of fimbriae mannose-sensitive, and in the others, the fimbriae were considered mannoseresistant.
The evaluation of the capability of P. aeruginosa in producing capsule as a presumptive test for biofilm pro-
Table 2. Distribution of P. aeruginosa samples according to clinical specimens and hospital sectors.

Diverse secretions: Purulent sec., ear sec., biliary sec., drain sec., oropharynx sec., tendon sec., abdominal sec and peritoneal sec. *Other: Liquor, bronchial washing, pleural fluid, sputum, ascitic fluid, oral cavity plate, nasal swab, blood. **Other: kidney transplant, nephrology ward, orthopedic ward, urgency and oncology. n = number of isolates.
duction by the method CRA, showed that 52.0% of the analyzed samples were capsule producers. In relation to biofilm formation on polystyrene microplates, from the 86 (86.0%) strains that adhered to polystyrene, 19 (22.1%) were strongly adhered, 41 (47.7%) were moderate and 26 (30.2%) were weakly adhered.
From the 100 strains of P. aeruginosa tested, 39% showed presence of the gene algD which participates in the synthesis of alginate, the main component of the matrix produced by bacteria associated to biofilm. It was possible to observe a fragment of approximately 1310 pb by electrophoresis in agarose gel.
About the capacity of adhesion of multiresistant strains to HEp-2 cells, 26.5% (9/34) showed an aggregative adhesion pattern, 2.9% (1/34) local pattern and 20.6% (7/34) adhesion without a well defined pattern (Figure 1), and
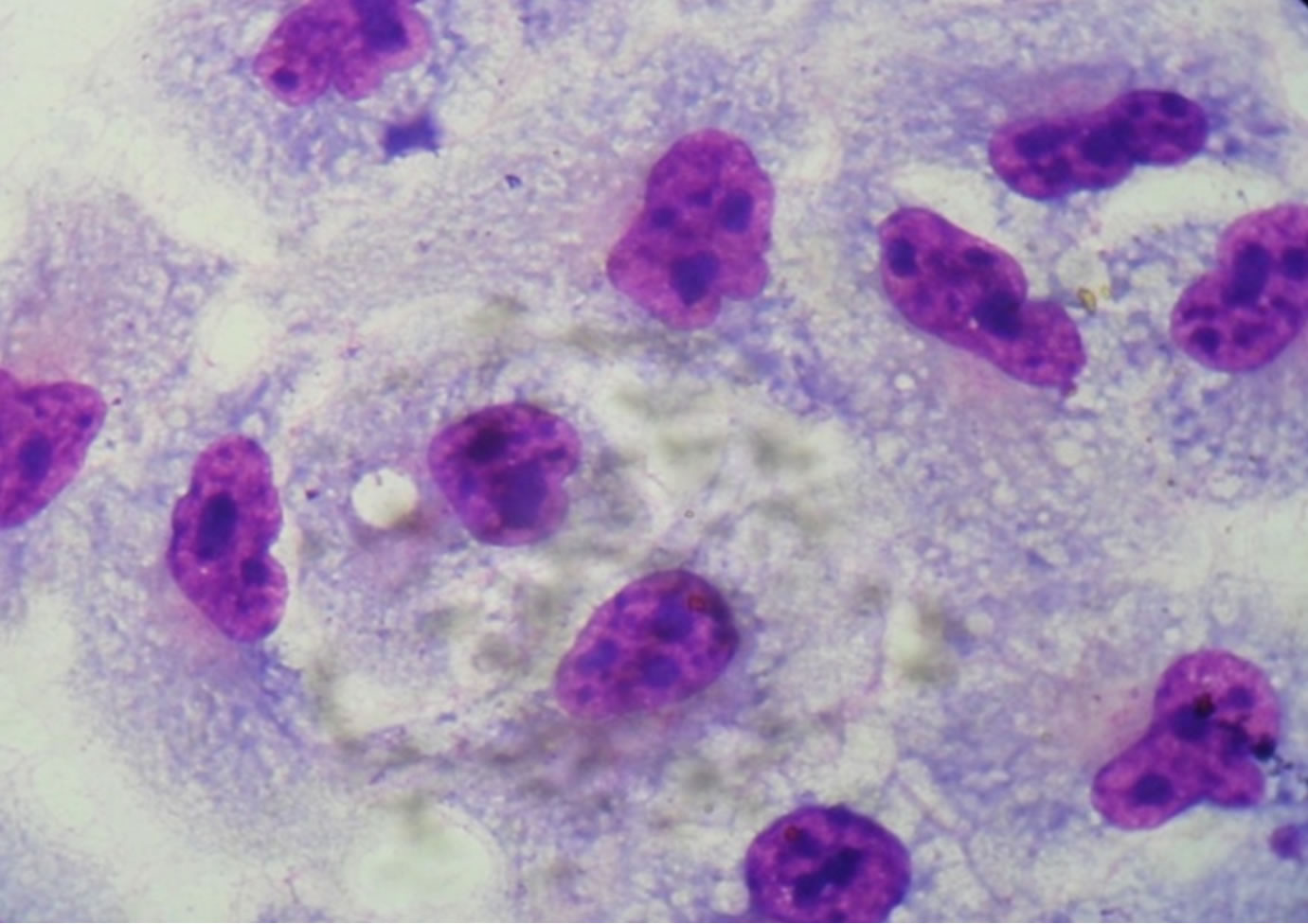 (a)
(a)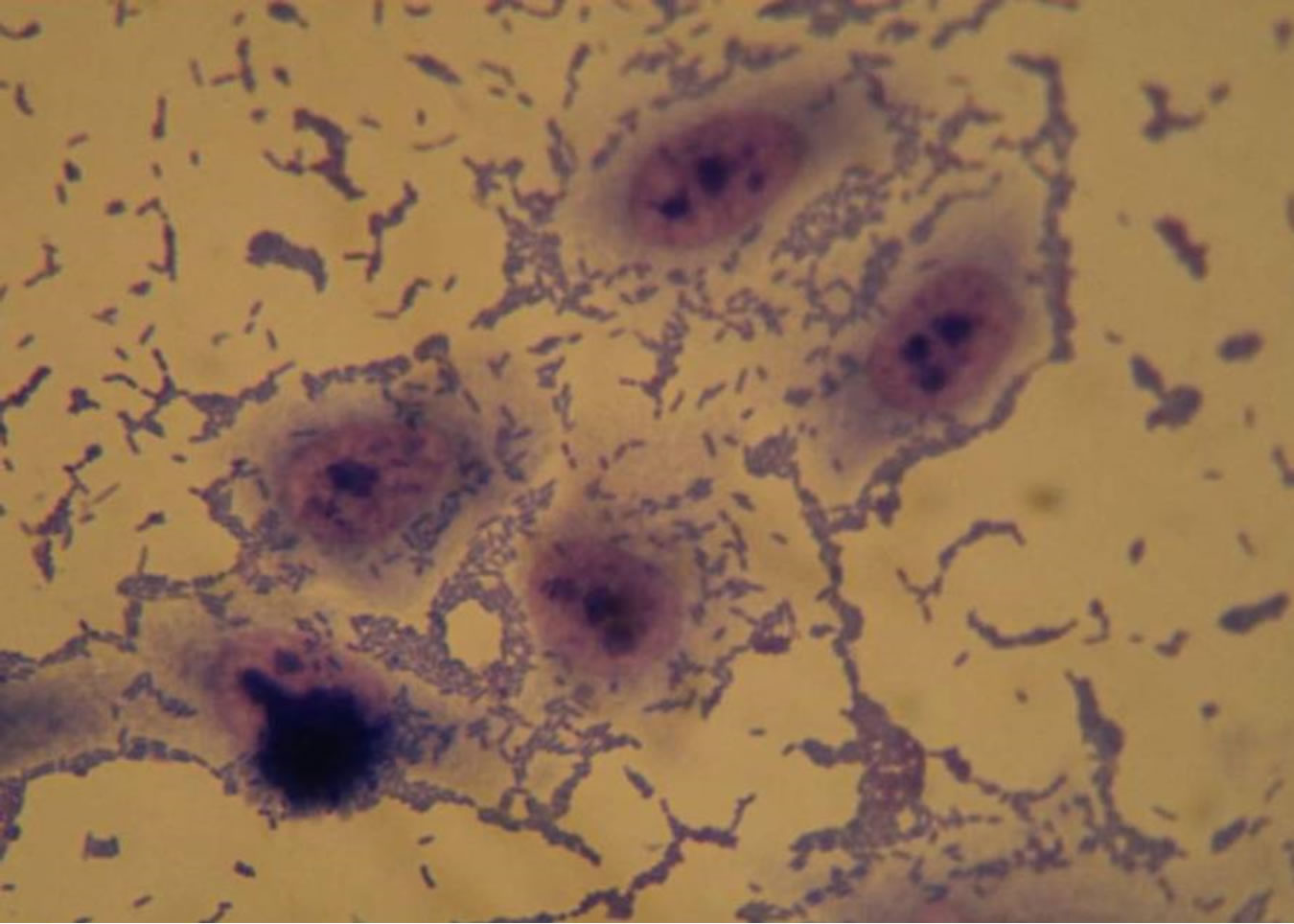 (b)
(b) (c)
(c)
Figure 1. Optical microscopy of adhesion on HEp-2 cells of P. aeruginosa strains (1000×). (a) HEp-2 cells alone; (b) Strain Pa31 with aggregative adhesion pattern; (c) Strain Pa87 without a well defined adhesion pattern.
41% (14/34) of the samples adhered more to plastic (coverslips) than to cells.
4. Discussion
The infections caused by P. aeruginosa are associated to significant morbimortality rates, due to this pathogen capability of adapting to the environmental conditions, developing resistance to antibiotics and producing a variety of virulence factors [20].
In this study, most samples were isolated from tracheal secretion and urine of inpatients and the ICU was the hospital sector with the highest isolation of this bacterium. Studies report that the high frequency of isolation of this pathogen from respiratory tract samples is related to mechanical ventilation, usually causing pneumonia [21]. A study by Lucchetti et al. [22] showed that P. aeruginosa was the main isolated agent causing infections in the urinary tract, and according to epidemiologic data, 35.0% to 45.0% of all acquired nosocomial infections are urinary and 80.0% are related to catheter use. Similar results were found by Torres et al. [23], who found the highest percentual of Pseudomonas spp isolates in the tracheal tract, followed by urine, and the place with the highest incidence was the ICU.
The high incidence of this bacterium in the ICU is probably due to the fact that P. aeruginosa is an opportunist pathogen that causes bacteremia in immunocompromised patients, burn victims, patients with urinary infections related to catheters use and nosocomial pneumonia, related to mechanical ventilation, especially in this unit [23-25].
Monitoring the susceptibility of P. aeruginosa hospital samples to antimicrobials is a helpful tool for choosing the right scheme to treat infections caused by this agent [26]. The analyzed strains showed high sensitivity rates to amikacin, piperacillin/tazobactam, fluoroquinolones, gentamicin and meropenem, what makes them good choices for treating infections by this pathogen. Figueiredo et al. [25] found high sensitivity for amikacin, piperacillin/tazobactam, meropenem and imipenem.
In agreement to literature data, all strains were susceptible to polymyxin B. Nevertheless, it is indicated for specific situations, due to its toxicity. There are reports about P. aeruginosa strains with reduced sensitivity to this drug [5,25].
The isolates showed high resistance to aztreonam, ceftazidime and cefepime, however, in the study by SantosFilho et al. [27], gentamicin, cefpirome and aztreonam were the most resistant. The abusive use of 3rd and 4th generation cephalosporins, especially ceftazidime (3rd generation), seems to be the main cause of this resistance. Resistance to aztreonam may be due to a superexpression of the MexA-MexB-OprM efflux pump, which also contributes to the expulsion of cephalosporin and betalactamase inhibitors [28].
In relation to carbapenems, the samples were more resistant to imipenem than to meropenem. Variations in the resistance rates between these antibiotics have been previously describe [29]. Rodríguez-Martínez et al. [29] observed higher resistance rates to imipenem when compared to meropenem, corroborating our results [30], in a study in Brazil, described higher resistance to meropenem. This susceptibility difference among carbapenems is explained by several resistance mechanisms, such as loss of proteins of external membrane OprD, that causes resistance to imipenem and not to meropenem; superexpression of efflux systems; and carbapenemase production [29,31].
Our results also showed that 34.0% of the strains had a multiresistance profile, where 50.0% were susceptible only to polymyxin B. The emergence of P. aeruginosa multiresistant to drugs has been reported as an increasing problem in hospitals worldwide [32,33].
A remarkable feature in infections by P. aeruginosa acquired in the ICU is multiresistance, what has been confirmed in this study, in which 53% of the multiresistant strains were from the ICU, suggesting that it might be indiscriminate use of antimicrobials in the treatment of nosocomial infections.
Currently, the combination of different antibiotics is studied in vitro and in vivo, with the intention of providing new therapeutic alternatives for infections caused by multiresistant bacteria [6].
The difficulty in detecting ESBLs is higher in P. aeruginosa strains, possibly due to the higher impermeability of the external membrane when compared to the family Enterobacteriaceae and also to the presence of the enzymes AmpC that may disguise the presence of ESBLs. Therefore, detecting these enzymes in P. aeruginosa strains is a great challenge for the clinical laboratory, since the phenotypic test of disk approximation does not have satisfactory sensitivity and specificity when used for this pathogen. Thus, it is probable that its prevalence in hospitals is underestimated [34].
The dissemination of the genes for these enzymes may play an important role in antimicrobial resistance and the presence of P. aeruginosa producing ESBL can limit the antibiotics choice for treating severe infections by this pathogen. That is why it is important to use molecular methods to detect ESBLs in multiresistant samples of P. aeruginosa [35].
In this study, 50.0% (17/34) of the multiresistant strains have expressed the enzyme TEM, what corroborates studies performed in France, where four variants of TEM (TEM-4, TEM-21, TEM-24 and TEM-42) were described in P. aeruginosa [35].
Thus, knowing about the occurrence of ESBLs in P. aeruginosa samples is essential so that the internist can choose an appropriate antimicrobial therapy, since the use of broad-spectrum cephalosporin and monobactams in the treatment of infections caused by ESBL-producers may result in therapeutical failure [36].
Bacterial adhesion may be divided into primary and secondary stages. The primary is reversible and determined by physico-chemical variables, as hydrophobic interactions, that determine adhesion between the two surfaces, the bacterial cell and the surface of interest. In secondary adhesion, a molecular mediation happens between specific adhesins and the surface and the microorganism consolidates the adhesion through producing an exopolysaccharide complex and/or connecting specific receptors in the fimbriae to the material surface. In the end of this stage, adhesion is irreversible [37,38]. Therefore, adhesion depends basically on factors that involve features from the micro-organism, the surface and environmental conditions.
In our study, most strains of P. aeruginosa showed clumps formation in concentrations of 2.5 M and 3 M of ammonium sulfate, that is, our isolates had high hydrophobicity. Studies show that the higher the hydrophobicity, the better the microbial adhesion, both in cell surface and in adhesion substrate [38]. Thus, this data suggests that P. aeruginosa has a strong adhesion capacity and, consequently, a higher tendency to biofilm formation, since it has high hydrophobicity (p = 0.02; C = 0.35).
Fimbriae are structural components of the surface of P. aeruginosa and also ease adhesion [40]. These structures mediate the connection between the pathogen and the surfaces of the host’s cells, starting colonization and formation of a film of microcolonies which will mature, forming biofilm, critical for the infection establishment [40]. In this study, we identified mannose-sensitive and mannose-resistant fimbriae.
There are several factors related to biofilm formation, the main are: physico-chemical features of the material on which it is adhered and expression of virulence factors by the micro-organisms, as exopolymeric capsule production and fimbrial and non-fimbrial adhesins synthesis [41].
One of the factors related to the micro-organism that may influence the biofilm formation is the presence of capsule, as recently found by Jain and Agarwal [42] and corroborated by this study.
Adhesion is the first step for biofilm formation. In this model, the assays performed with the crystal violet method provided information about the adhesion of the microorganism tested to an abiotic surface, as P. aeruginosa produced biofilm on a microtiter polystyrene plate. Therefore, this strong capacity of adhering to non-biological material showed by P. aeruginosa isolates, represents a great problem in the treatment of patients that need a catheter or other medical device in which this micro-organism can produce biofilm, leading to chronic infections. In addition to his, biofilm protects the bacterium from the host’s immune system and antibiotics action, being more resistant to drugs because, according to Trautner and Dauroiche [40], the juxtaposition of cells in the biofilm eases the transfer of resistance genes from one bacterium to another through plasmids.
In our study, there was no association between the presence of fimbriae and/or capsule with the production or degree of biofilm (p = 0.4; c2 = 12.5), suggesting that other kinds of fimbriae or adhesins may be present, since 63% of P. aeruginosa isolates that were strong biofilm producers and 46% of the moderate producers did not show any of the studied fimbriae (non-demonstrated data). In addition, not all the isolates that were biofilmproducers produced capsule, suggesting that, besides the capsule and fimbriae investigated here, other components of this bacterium can also ease adhesion and biofilm formation with type IV fimbria, flagellum, alginate and LPS [40].
The genetic cluster algACD is responsible for the synthesis of alginate, which is a component of the surface of P. aeruginosa that eases adhesion. The expression of algD, that encodes the enzyme GDP mannose dehydrogenase, is frequently used as an indicator of the expression of the other genes responsible for biosynthesis of alginate, which is a primary compound of the organic polymer matrix, that involves biofilm in P. aeruginosa and is considered by some authors as responsible for the antimicrobial resistance, observed in biofilm [11,40].
In our study, 39.0% of the isolates expressed the gene algD. In another study, from 202 strains analyzed, 91.1% expressed this gene [20]. Despite alginate is a mucoid exopolysaccharide that forms a prominent capsule in the bacterial surface, favoring adhesion and later biofilm formation, the statistical analysis of data did not show correlation between biofilm formation and the expression of gene algD (p = 0.13; c2 = 2.19), suggesting that other genes governing alginate synthesis (algC, algR, algP, algB, algU) may be present.
Studying the adherence capacity, 34 multiresistant strains of P. aeruginosa were analyzed and they showed adhesion to HEp-2 cells and glass slides. Three adhesion patterns were observed: aggregative, local and without a well defined pattern.
Adhesion to HEp-2 cells suggests that P. aeruginosa recognizes receptors in epithelial cells and therefore can adhere to other epithelial cells, what has been confirmed by Di Martino et al. [43] when demonstrating the adhesion of this bacterium to pneumocytes (lineage A549), where they observed diffuse, local and aggregative patterns, suggesting that the presence of this bacterium in the respiratory tract cells contributes for the pulmonary pathogenesis of this micro-organism, especially in patients carrying cystic fibrosis, in whom it causes a chronic pulmonary infection.
The strains analyzed in the adhesion test were from nosocomial infections and in most cases, patients were using medical devices, once 53.0% of them were in the ICU. Adhesion to plastic (coverslips) of 41.0% (14/34) of the samples analyzed in this study confirms the literature about the capacity of this bacterium in adhering to inert surfaces as catheters, tubes, respiratory equipments, implants and other medical devices, what represents an important step in colonization of patients who are immunocompromised or using these devices [13].
Data found in this study suggest that antimicrobial susceptibility profiles should be monitored, supporting the therapeutics and avoiding treatment failures. They also indicate the great capacity of P. aeruginosa being hydrophobic and forming biofilm on abiotic surface, what may induce colonization of medical devices, with risk to the hospitalized patient. Besides, samples adhered to HEp-2 cells, and these characteristics confirm that P. aeruginosa is one of the most important nosocomial pathogens, therefore there is interest on elucidating the adaptation and pathogenicity mechanisms of this microorganism.
5. Acknowledgements
We thank Fundação de Amparo à Pesquisa e ao Desenvolvimento Científico e Tecnológico do Maranhão (FAPEMA), for financial suports (ATB-04138/10; Edital FAPEMA n 026/2010 Bancada; APP-Universal-00442/11).
REFERENCES
- J. M. Blatt, “Mecanismo de Resistência e Detecção das Betalactamases de Espectro Ampliado,” Newslab, Vol. 40, 2000, pp. 86-96.
- R. A. Bonomo and D. Szabo, “Mechanisms of Multidrug Resistance in Acinetobacter species and Pseudomonas aeruginosa,” Clinical Infectious Diseases, Vol. 43, No. 2, 2006, pp. 49-56. doi:10.1086/504477
- P. A. Bradford, “Extended-Spectrum Betalactamases in the 21st Century: Characterization, Epidemiology and Detection of This Important Resistance Threat,” Clinical Microbiology Reviews, Vol. 14, No. 4, 2001, pp. 933-951. doi:10.1128/CMR.14.4.933-951.2001
- S. Bratu, J. Quale, S. Cebular and R. Heddurshetti, “Multidrug-Resistant Pseudomonas aeruginosa in Brooklyn, New York: Molecular Epidemiology and in Vitro Activity of Polymyxin B,” European Journal of Clinical Microbiology and Infectious Diseases, Vol. 24, No. 3, 2005, pp. 196-201. doi:10.1007/s10096-005-1294-x
- S. Clegg and D. C. Old, “Fimbriae of Escherichia coli K-12 Strain AW405 and Related Bacteria,” Journal of Bacteriology, Vol. 137, No. 2, 1979, pp. 1008-1012.
- Clinical Laboratory and Standards Institute, “Performance Standards for Antimicrobial. Susceptibility Testing,” 20th Informational Supplement, CLSI Document M100-S20. WAYNE, CLSI, 2010.
- A. L. Clutterbuck, A. C. Cochrane, J. Dolman and S. L. Percival, “Evaluating Antibiotics for Use in Medicine Using a Poloxamer Biofilm Model,” Annals of Clinical Microbiology and Antimicrobials, Vol. 15, No. 6, 2007, pp. 1-10.
- Di P. Martino, H. Gagniere, H. Berry and L. Bret, “Antibiotic Resistance and Virulence Properties of Pseudomonas aeruginosa Strains from Mechanically Ventilated Patients with Pneumonia in Intensive Care Units: Comparison with Imipenem Resistant Extra-Respiratory Tract Isolates from Uninfected Patients,” Microbes and Infection, Vol. 4, No. 6, 2002, pp. 613-620. doi:10.1016/S1286-4579(02)01579-4
- D. Q. Figueiredo, L. F. S. Castro, K. N. Santos, L. Teixeira and S. S. B. Mondino, “Detection of Metallo-Betalactamases in Hospital Strains of Pseudomonas aeruginosa and Acinetobacter baumanni,” Brazilian Journal of Pathology and Laboratory Medicine, Vol. 45, No. 3, 2009, pp. 177-184.
- J. Flach, C. Karnopp and G. Corção, “Biofilm Formation from Milk in Contact with Raw Material: Virulence Factors Involved,” Act Scient Veterinariae, Vol. 33, No. 3, 2005, pp. 291-296.
- D. J. Freeman, F. R. Falkiner and C. T. Keane, “New Method for Detecting Slime Production by Coagulase Negative Staphylococci,” Journal of Clinical Pathology, Vol. 42, No. 8, 1989, pp. 872-874. doi:10.1136/jcp.42.8.872
- A. C. Galles, R. N. Jones, J. Turnidge, R. Rennie and R. Ramphal, “Characterization of Pseudomonas aeruginosa Isolates: Occurrence Rates, Antimicrobial Susceptibility Patterns, and Molecular Typing in the Global Sentry Antimicrobial Surveillance Program, 1997-1999,” Clinical Infectious Diseases, Vol. 32, No. 2, 2001, pp. S146-S155.
- D. O. Garcia, “Proteases (Caseinase and Elastase), Hemolysins, Adhesion and Susceptibility to Antimicrobials of Stenotrophomonas maltophilia Isolates Obtained from Clinical Specimens,” Brazilian Journal of Microbiology, Vol. 33, No. 2, 2002, pp. 157-162. doi:10.1590/S1517-83822002000200012
- V. Hemalatha, S. Uma and K. Vijaylakshmi, “Detection of Metallo Betalactamase Producing Pseudomonas aeruginosa in Hospitalized Patients,” Indian Journal of Medical Research, Vol. 122, 2005, pp. 148-152.
- A. Jain and A. Agarwal, “Biofilm Production, a Marker of Pathogenic Potential of Colonizing and Commensal Staphylococci,” Journal of Microbiological Methods, Vol. 76, No. 1, 2009, pp. 88-92. doi:10.1016/j.mimet.2008.09.017
- J. A. Karlowsky, D. C. Draghi, M. E. Jones, C. Thornsberry, I. R. Friedland and D. F. Sahm, “Surveillance for Antimicrobial Susceptibility among Clinical Isolates of Pseudomonas aeruginosa and Acinetobacter baumannii from Hospitalized Patients in the United States, 1998 to 2001,” Antimicrobial Agents and Chemotherapy, Vol. 47, No. 5, 2003, pp. 1681-1688. doi:10.1128/AAC.47.5.1681-1688.2003
- E. Kipnis, T. Sawa and J. Wiener-Kronish, “Targeting Mechanisms of Pseudomonas aeruginosa Pathogenesis,” Médecine et Maladies Infectieuses, Vol. 36, No. 2, 2006, pp. 78-91. doi:10.1016/j.medmal.2005.10.007
- M. Klausen, G. Morten, I. Kreft and T. Tolken-Nielsen, “Dymanics of Development and Dispersal in Sessile Microbial Communities: Examples from Pseudomonas aeruginosa and Pseudomonas putida Model Biofilms,” FEMS Microbiology Letters, Vol. 261, No. 1, 2006, pp. 1-11. doi:10.1111/j.1574-6968.2006.00280.x
- P. Lanotte, S. Watt, L. Mereghetti, N. Dartiguelongue, A. Rastegar-Lari, A. Goudeau and R. Quentin, “Genetic Features of Pseudomonas aeruginosa Isolates from Cystic Fibrosis Patients Compared with Those of Isolates from Other Origins,” Journal of Medical Microbiology, Vol. 53, No. 1, 2004, pp. 73-81. doi:10.1099/jmm.0.05324-0
- G. Lucchetti, A. J. Silva, S. M. Y. Ueda, M. C. D. Perez and L. M. J. Mimica, “Infecções Do Trato Urinário: Aná- lise da Freqüência e Do Perfil de Sensibilidade Dos Agentes Causadores de Infecções Do Trato Urinário Em Pacientes Com Cateterização Vesical Crônica,” Journal Brasileiro de Patologia e Medicina Laboratorial, Vol. 41, No. 6, 2005, pp. 383-389. doi:10.1590/S1676-24442005000600003
- A. R. Marra, C. A. Pereira, A. C. Gales, L. C. Menezes, R. G. Cal, J. M. de Souza, M. B. Edmond, C. Faro and S. B. Wey, “Bloodstream Infections with Metallo-β-Lactamase Producing Pseudomonas aeruginosa: Epidemiology, Microbiology, and Clinical Outcomes,” Antimicrobial and Agents Chemotherapy, Vol. 50, No. 1, 2006, pp. 388-390. doi:10.1128/AAC.50.1.388-390.2006
- E. A. Menezes, F. V. V. Macedo, F. A. Cunha, M. S. S. Andrade and M. V. P. Rocha, “Perfil de Infecção e Resistência Aos Antimicrobianos de Bacilos Gram-Negativos Não Fermentadores Isolados No Laboratório de Patologia Clínica Dr. Edilson Gurgel da Santa Casa de Misericórdia de Fortaleza-CE,” Revista Brasileira de Análises Clínicas, Vol. 36, No. 4, 2004, pp. 209-212.
- E. A. Menezes, K. M. Sá, F. A. Cunha, M. R. F. Ângelo, I. R. N. Oliveira and M. N. C. Salviano, “Frequência e Percentual de Suscetibilidade de Bactérias Isoladas em Pacientes Atendidos Na Unidade de Terapia Intensiva do Hospital Geral de Fortaleza,” Journal Brasileiro de Patologia e Medicina Laboratorial, Vol. 43, No. 3, 2007, pp. 149-155. doi:10.1590/S1676-24442007000300003
- I. Mitov, T. Strateva and B. Markova, “Prevalence of Virulence Genes among Bulgarian Nosocomial and Cystic Fibrosis Isolates of Pseudomonas aeruginosa,” Brazilian Journal of Microbiology, Vol. 41, No. 3, 2010, pp. 588-595. doi:10.1590/S1517-83822010000300008
- C. S. Mitsugui, M. C. B. Tognim, F. E. Carrara-Marrone and L. B. Garcia, “Efeito Antimicrobiano in Vitro da Associação de Polimixina B e Ceftzidima em Amostras Clínicas de Pseudomonas aeruginosa,” Rev Ciênc Cuid Saúde, Vol. 7, 2008, pp. 76-81.
- H. J. Monstein, A. Ostholm-Balkhed, M. V. Nilsson, M. Nilsson, K. Dornbusch and L. E. Nilsson, “Multiplex PCR Amplification Assay for the Detection of blaSHV, blaTEM and blaCTX-M Genes in Enterobacteriaceae,” Acta Pathologica, Microbiologica et Immunologica Scandinavica, Vol. 115, No. 12, 2007, pp. 1400-1408. doi:10.1111/j.1600-0463.2007.00722.x
- S. Navon-Venezia, R. Bem-Ami and Y. Carmeli, “Update on Pseudomonas aeruginosa and Acinetobacter baumannii Infections in the Healthcare Setting,” Current Opinion in Infectious Diseases, Vol. 18, No. 4, 2005, pp. 306-313. doi:10.1097/01.qco.0000171920.44809.f0
- R. C. Picão and A. C. Gales, “β-Lactamases de Espectro Ampliado (ESBL) em Pseudomonas aeruginosa: Pesadelo ou Só Imaginação?” Prática hospitalar. Ano IX, No 49, 2007.
- S. U. Picoli, “Metallo-ß-Lactamase and Pseudomonas aeruginosa,” Revista Brasileira de Análises Clínicas, Vol. 40, No. 4, 2008, pp. 273-277.
- L. B. Rodrigues, L. R. Santos, N. N. Rizzo, V. Z. Taglieti, V. P. Oliveira, G. Trenhago, S. C. Rodegheri, R. M. Tglieti, E. L. Dickel and V. P. Nascimento, “Avaliação da Hidrofobicidade e da Formação de Biofilme em Poliestireno por Salmonella Heidelberg Isoladas de Abatedouro Avícola,” Acta Scientiae Veterinarie, Vol. 37, No. 3, 2009, pp. 225-230.
- J. M. Rodríguez-Martínez, P. Laurent and P. Nordmann, “Molecular Epidemiology and Mechanisms of Carbapenem Resistance in Pseudomonas aeruginosa,” Antimicrobial Agents and Chemotherapy, Vol. 53, No. 11, 2009, pp. 4783-4788. doi:10.1128/AAC.00574-09
- L. Santos-Filho, I. B. Santos, A. M. L. Assis and D. E. Xavier, “Determinação da Produção de Metalo-β-Lactamases em Amostras de Pseudomonas aeruginosa Isoladas em João Pessoa, Paraíba,” Journal Brasileiro de Patologia e Medicina Laboratorial, Vol. 38, No. 4, 2002, pp. 79-84. doi:10.1590/S1676-24442002000400007
- I. C. A. Scaletsky, M. L. M. Silva and L. R. Trabulsi, “Distinctive Patterns of Adherence of Enteropathogenic Escherichia coli to HeLa Cells,” Infection and Immunity, Vol. 45, No. 2, 1984, pp. 534-536.
- S. Sharma, R. Kaur, V. Yadav, K. Harjai and K. Joshi, “Contribution for Exotoxin A of Pseudomonas aeruginosa in Acute and Chronic Experimental Renal Infection,” Journal of Infectious Diseases, Vol. 57, No. 3, 2004, pp. 119-120.
- F. M. Silva, H. S. Sader, R. N. Jones and A. C. Gales, “Antimicrobial Susceptibility Profile of Pseudomonas aeruginosa Causing Bloodstream Infections in Brazilian Medical Centers: Results of the SENTRY Antimicrobial Surveillance Program (2001-2006),” Institute for Defence Studies and Analyses, poster 521, 2007.
- S. Stepanovic, D. Vukovic, V. Hola, G. Bonaventura, S. Djukic, I. C. Irkovic and F. Ruzicka, “Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci,” Acta Pathologica, Microbiologica et Immunologica Scandinavica, Vol. 115, No. 8, 2007, pp. 891-899. doi:10.1111/j.1600-0463.2007.apm_630.x
- J. C. N. Torres, E. A. Menezes, M. R. F. Ângelo, I. R. N. Oliveira, M. N. C. Salviano, D. E. Xavier and L. S Filho, “Cepas de Pseudomonas aeruginosa Produtoras de Metalo- β-Lactamases Isoladas No Hospital Geral de Fortaleza,” Journal Brasileiro de Patologia e Medicina Laboratorial, Vol. 42, No. 5, 2006, pp. 313-319. doi:10.1590/S1676-24442006000500003
- B. W. Trautner and R. O. Darouiche, “Role of Biofilm in Catheter-Associated Urinary Tract Infection,” American Journal of Infection Control, Vol. 32, No. 3, 2004, pp. 177-183. doi:10.1016/j.ajic.2003.08.005
- G. F. Weldhagen, L. Poirel and P. Nordmann, “Ambler Class A Extended-Spectrum Beta-Lactamase in Pseudomonas aeruginosa: Novel Development and Clinical Impact,” Antimicrobial Agents and Chemotherapy, Vol. 47, No. 8, 2003, pp. 2385-2391. doi:10.1128/AAC.47.8.2385-2392.2003
- S. Vesterlund, J. Paltta, M. Karp and A. C. Ouwerhand, “Measurement of Bacterial Adhesion in Vitro Evaluation of Different Methods,” Journal of Microbiological Methods, Vol. 60, No. 2, 2005, pp. 225-233. doi:10.1016/j.mimet.2004.09.013
- A. P. Zavascki, A. L. Barth, J. F. Fernandes, A. L. D. Moro, A. L. S. Gonçalves and L. Z. Goldani, “Reappraisal of Pseudomonas aeruginosa Hospital-Acquired Pneumonia Mortality in the Era of Metallo-β-Lactamase-Mediated Multidrug Resistance: A Prospective Observational Study,” Critical Care, Vol. 10, No. 4, 2006, p. R114. doi:10.1186/cc5006
- A. P. Zavascki, L. Z. Goldani, J. Li and R. L. Nation, “Polimyxin B for the Treatment of Multidrug-Resistant Pathogens: A Critical Review,” Journal of Antimicrobial Chemotherapy, Vol. 60, No. 6, 2007, pp. 1206-1215. doi:10.1093/jac/dkm357
- H. Schmidt, E. Schloricke, R. Fislage, H. A. Schulze and R. Guthoff, “Effect of Surface Modifications of Intraocular Lenses on the Adherence of Staphylococcus epidermidis,” Zentralblatt für Bakteriologie, Vol. 287, No. 1-2, 1998, pp. 135-145. doi:10.1016/S0934-8840(98)80159-X
NOTES
*Corresponding author.

