American Journal of Plant Sciences
Vol.09 No.08(2018), Article ID:86392,12 pages
10.4236/ajps.2018.98128
Expression of a Bacterial Chitinase (ChiB) Gene Enhances Resistance against Erysiphae polygoni Induced Powdery Mildew Disease in the Transgenic Black Gram (Vigna mungo L.) (cv. T9)
D. K. Das
Post Graduate Department of Biotechnology, T. M. Bhagalpur University, Bhagalpur, India

Copyright © 2018 by author and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: June 21, 2018; Accepted: July 28, 2018; Published: July 31, 2018
ABSTRACT
To enhance the antifungal response of blackgram (Vigna mungo L.), transgenic plants were generated by transferring bacterial chitinase gene with a CaMV 35S promoter. The chopped multiple shoot cells developed on the cotyledonary node were transformed by Particle gun method. Thecalli were raised on the Murashige and Skoog (MS) modified media supplemented with 50 m∙gl−1 kanamycin. The transformation efficiency was 13% approximately. The resultant shoot buds were selected and the antibiotic resistant transgenic plantlets were regenerated. The development of the transgenic plants from the shoot buds took about four to six months. The integration of the transgene was confirmed by PCR, RT-PCR, Southern and western blot analyses. The transgenic plants exhibited higher chitinase activity than the non-transformed ones. The chitinase activity was examined by native polyacrylamide in-gel assay. The transgenic plants showed fungal tolerance as evidenced by the delayed onset of the disease and smaller lesions following an in vitro inoculation of the powdery mildew pathogen (Erysiphae polygoni DC). The transgenic plants adapted well to the greenhouse and did not show any phenotypic alterations.
Keywords:
Particle Gun Bombardment, Organogenesis, Powdery Mildew, Transformation, Erysiphae polygoni DC

1. Introduction
Black gram (Vigna mungo L. Heaper) is an important tropical and tropical annual herbaceous legume cultivated since the ancient times in India. It is one of the main sources of dietary protein for a majority of population in the developing countries of Asia, Africa and Latin America. The productivity of this crop has been greatly limited due to several fungal, viral, bacterial and pest diseases (Singh, 1981) [1] . In vitro culture methods are useful to produce plants with resistant genes to overcome diseases. Most of the varieties that grown in India (e.g. PS 1, Pusa-1, Pusa-2, T9 etc.) are susceptible to many diseases. The most important one among them is powdery mildew caused by Erysiphae polygoni. This disease may be controlled partially by fungicides. But the cost of production may escalate. At the same time, environmental safety concerns may also arise. It would, therefore, be imperative to produce pathogen-resistant varieties of the crop.
2. Materials and Methods
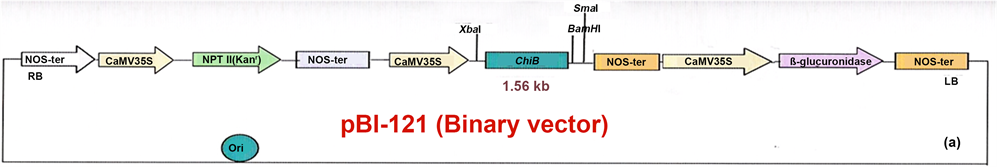
The black-gram seeds were obtained from Bihar Agricultural University Sabour, Bhagalpur, Bihar, India. The multiple shoot explants which were developed on cotyledonary nodes of seeds on MS modified medium supplemented with 50 mg∙l−1 kanamycin under in vitro condition are efficient for transformation and regeneration through organogenesis in black gram. The particle gun-mediated transformation has been demonstrated as quite efficient (1.4% - 22.69%) in several plants i.e. Castor (Ricinus communis L) (Sailaja et al. 2008) [2] , Cowpea (Vigna unguiculata L.) (Ikea et al. 2003) [3] . The gene transfer to blackgram has been achieved till date by Agrobacterium-mediated transformation (Saini and Jaiwal 2007) [4] , but with low efficiency (1% - 4.31%). Particle gun bombardment-mediated transformation of legumes is one of the best options to enhance T.E. (Transformation efficiency). The ballistic device is also overcomes some of the problems associated with the use of Agrobacterium in blackgram. The Bacterial chitinase (ChiB) gene has been expressed in the leaves of tobacco using two photosynthetic gene promoters (Jonathan et al. 1986) [5] . This gene has also been expressed in litchi (Das and Rahman 2010) [6] . The rice chitinase gene (Chi11) has been shown to enhance resistance of bread wheat against herbicide (bialaphos) through biolistics (Chen et al. 1998) [7] . The present work involves the use of Particle gun-mediated transformation to over express bacterial chitinase gene (ChiB) in Blackgram cv. T9. The transgenic fertile plants exhibited higher chitinase activity with increased resistance to powdery mildew disease caused by Erysiphae polygoni DC. The precultured multiple shoots on MS medium supplemented with 2 mg∙l−1 BAP developed on cotyledonary node from in vitro sterilized Seeds of Black gram cv. T9, were chopped into small pieces (explants) and gene construct (binary vector pBI121-ChiB-GUS, with chitinase and GUS genes respectively (Figure 1) under the control of 35S constitutive promoter) was entered into black gram genome through Particle gun-bombardment method (PDS 1000/He of BioRad) using tungsten particles (microcarrier and size is 0.7 - 1 µm) sterilized in ethanol suspension. The de-agglomerate particles were processed in 50 µl aliquots containing ethanol, 5 µl of gene construct DNA,
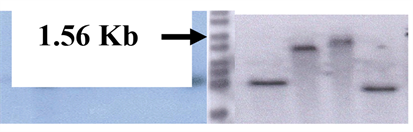
Figure 1. ChiB genes construct in plasmid pBI 121 and its analysis. 1.1 Map of ChiB gene constructs with CaMV 35 S constitutive promoter.
50 µl of CaCl2 (2.5 M) and 20 µl of spermidine (0.1 M). The rest was followed by the protocol of Kikkert et al. (2004) [8] . Shoots were developed on calliin MS modified medium with 50 mg∙l−1 kanamyc in followed the protocol of Das et al. (2002) [9] . For root initiation, elongated shoots were excised and cultured on one?third strength of MS salts supplemented with MS organics, 1 mg∙l−1 IBA and 0.7% agar semi-solid medium. The rooted plantlets (ca. 5 cm) were transferred into autoclaved vermiculite moistened with Hoagland (Hoagland and Arnon 1950) [10] medium in 6-cm plastic pots and covered with a plastic cover to maintain humidity. Two weeks later, the covers were gradually removed over a period of 7 days at high light intensity i.e. 90 µmol∙m−2∙s−1 and temperature range from 28˚C to 37˚C for acclimatization, before the plantlets were finally transferred to soil. Histochemical GUS assays were conducted according to Jefferson et al. (1987) [11] on 2 days after bombarded samples.
To confirm the presence of the Bacterialchitinase gene in the transgenic plants, genomic DNA was isolated from 0.5 g of fresh young black gram leaves as described by (Lodhi et al. 1994) [12] . For the PCR analysis, 200 ng of plant DNA or 4 ng of plasmid DNA was used per 25-µl reaction mixture. The primers were designed to amplify 465 bp fragments of chitinase at 63.6oC (F5’GCTACTGCTTCAAGGAGGAGAAACA3'; R5'CTGGTTGTAGCAATCCAGGTTATCG-3') and 508-bp fragments of npt gene at 52˚C (F-5’AGCTGCGCCGATGGTTTCTACAA3’; R-5’ATCGCCTCGCTCCAGTCAATG 3’). The PCR program profile for both the genes was as follows; initial denaturation at 94˚C for 4 min, followed by 30 cycles of 94˚C for 1 min, 1.5 min at the annealing temperature of each gene and 1 min at 72˚C, with a final extension at 72˚C for 10 min. The amplified products were run on 1% agarose gels and visualized by ethidium bromide staining. In order to confirm the transgene integration and to determine the number of copies of transgene (ChiB) integrated into genomic DNA, Southern blotting (Southern 1975) [13] experiments were performed. Genomic DNA (10 µg) and plasmid (pBI121-ChiB-GUS) as positive control were digested with XbaI or BamHI(New England Biolab), fragments were separated on 1% (w/v) agarose gels at 25 V for 16 hand processed as mentioned earlier Das and Rahman (2012) [14] . Total RNA was prepared from leaf tissues using Trizol Reagent as per the manufacturer’s instructions (Trizol Reagent, Invitrogen life technologies, San Diego, California, USA). To detect the presence of bacterial chitinase mRNA transcripts in the transformants, RT-PCR was carried out by standard procedure as described earlier Das and Rahman (2012) [14] . The chitinase levels in the transgenic black gram were determined using colorimetric enzyme assay, in-gel assay and western blot analysis. Total soluble proteins were extracted from the frozen leaves (placed at −80˚C for a week) of the transformed and non-transformed samples. Leaves were homogenized with a pestle and mortar in liquid nitrogen and the frozen powder was suspended in 5 volumes of 0.1 M sodium citrate buffer (pH 6.0) containing 20 mM sodium ascorbate and polyclar AT. After two rounds of centrifugation at 13,000 rpm for 15 min at 4˚C, the supernatants were recovered (Yamamoto et al. 2000) [15] . The protein concentrations in the extracts were estimated by the Bradford method (Bradford 1976) [16] . Equal amounts (25 µg) of soluble proteins were resolved on 1D SDS-gels and stained with 0.1% Coomassie brilliant blue R-250 and de-stained in 10% acetic acid overnight. For the western blotting the proteins were transferred into the nitrocellulose membrane and probed with anti-chitinase B antibodies (Generously provided by Dr. M. V. Razam, Deptt. Of Genetics, South Campus, University of Delhi, India) at 1:5000 dilution. The rest procedure was followed as earlier Das and Rahman (2012) [14] . A solubilized, ethylene glycol-chitin (Sigma-Aldrich) was used as a substrate for chitinase activity assay. The colorimetric analysis of chitinase enzyme activity of PCR, Southern and RT-PCR positive transgenic plants were done following the protocol of Stephan and Wolf (1990) [17] with slight modifications. The aliquots of 300 µl of ethylene glycol-chitin (stock 2 mg/ml) were mixed with 100 µl of 200 mM sodium acetate buffer, pH 5.0 and 0.5 ml enzyme solution, and then incubated for 60 min. at 37˚C in the circulating water bath. The rest procedure was done as mentioned by Das and Rahman (2012) [14] . Tolerance potential of the transgenic black gram carrying Bacterial chitinase gene was evaluated against powdery mildew caused by Erysiphaepolygoni. The developing secondary or tertiary leaves were detached from the in vitro grown transgenic plants. There are no reports of endoparasites in black gram plants so far. Two leaves from each transgenic plant were placed adaxial side up onto 0.6% (w/v) agar containing 40 mgl−1 benzodiazoloe in a Petri dish. As control, leaves were taken from non-transformed regenerated plants. Since Erysiphae polygoni is an obligate ectophytic parasite, so it cannot be cultured on an artificial medium (Srivastava 2004) [18] but only on leaf disc culture in water (Morrison 1960) [19] . The spores were collected in the aqueous washing (having 0.01% (v/v) Tween 20) of infected leaves obtained from Horticulture department, Bihar Agricultural University, Sabour, Bhagalpur. The germination and its subsequent growth were measured on the transgenic and control plants. Electron micrography was used for surface ultrastructure study and small (1 - 8 mm) leaves of both transgenic and non-transgenic black gram (Vigna mungo L.) plants were sprayed with pathogenic fungal conidia/spores’ solution and left for 2 - 4 days. For scanning electron microscope (SEM), the specimen was vacuum dried. Fixation of the black gram leaves was performed by incubation in a solution of a buffered chemical fixative, such as 2.5% glutaraldehyde in combination with 2% formaldehyde in 0.1 M phosphate buffer at pH 7.2 containing 0.03 M sucrose for overnight at 4˚C. It was subsequently washed in 0.1 M phosphate buffer with 0.3 M sucrose for 1h. The rest standard procedure was done as mentioned in Das and Rahman (2012) [14] . Electron microscopic studies were done separately for both transformed and untransformed detached leaves of black gram plants. An average of each disease value was taken in triplicates. In each experiment 6 detached leaves were taken in each Petridish. The disease values were rated based on the approximate percentage of leaf necrotic area after 15 - 28 days of inoculation. Since Erysiphae polygoni is an obligate ectophytic parasite, so it can’t be cultured on an artificial medium (Srivastava 2004) [18] but only on leaf disc culture in water (Morrison 1960) [19] . The spores were collected in the aqueous washing (having 0.01% (v/v) Tween 20) of infected leaves obtained from Horticulture department, Bihar Agricultural University, Sabour, Bhagalpur. The spore suspension, 0.5 ml (106 spores/ml) was sprayed on to each Petri dish containing moistened leaf and kept at saturated humidity at 25˚C. The degree of disease severity was scored using a visual assessment scale based on the size and characteristics of necrotic lesions. A 5-point disease rating scale based on the approximate percentage of leaf necrotic area after 15 - 28 days of inoculation (1 = 0%; 2 = 1% - 20%; 2 = 20% - 30%; 3 = 30% - 40%; 4 = 40% - 50%; 5 = 50%) (Yamamoto et al 2000, Jayraj and Punja 2007) [20] was employed.
The percentage response of disease rating scale was different in different black gram plant leaves. The statistical significance on the disease value was calculated by one-way ANOVA followed by Tukey’s multiple comparison tests. All data analyses were performed using the Graph Pad software (Graph Pad in Stat. Software Inc. San Diego, CA 92130, USA).
3. Results
In this study multiple transformed shoots were developed by Particle gun bombardment method showing GUS positive may confirm integration of gene into black gram genome. The transgene was examined by PCR analysis using gene specific primers that generated a 465 bp fragment and npt (neomycin phosphotransferase) gene was also detected by PCR with gene specific primers that gave rise to a 508 bp fragment (Figure 2). The transformation process induced cellular necrosis to some extent, but recovered soon following special treatments (Vidal et al. 2003) [21] . Large numbers of shoot buds in the MS liquid medium were formed. A few of these shoot buds on calli elongated and the rest necrosed and perished on subsequent transfer to semisolid medium. Necrosis could be due to localization of high concentration of antibiotics and less formation of escapes in the semi solid medium. The transformation efficiency was approx. 13% that may be attributed to strong physical force of bombardment (Kikkert et al. 2004) [8] . The shoot buds rooted well and developed in robust form as sturdy rooted plantlets. Land transfer of the plantlets was 90% successful without somaclonal abnormalities (Figures 3(a)-(k)). All nine transformants (B-Chi1, 2, 4,
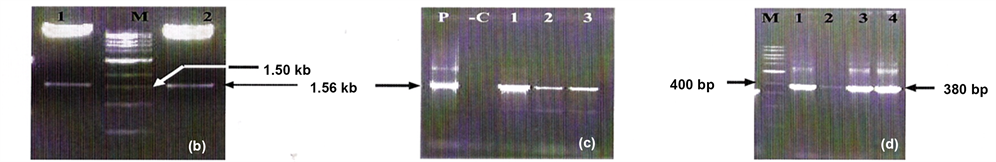
Figure 2. (b) Recombinant clone is digested by Xba I and SmaI; LaneM: 1 kb DNA ladder, Lanes1 and 2: showing 1.56 kb ChiB fragment. (c) Colony PCR of recombinant clone using gene specified primers. Lane P: recombinant plasmid as a positive control, Lane C: negative control without the recombinant plasmid, Lanes 1, 2 and 3 showing amplicons of 1.56 kb ChiB gene. (d) Colony PCRof recombinant clone using GUS gene specific primers, Lane M: 100 bp DNA ladder, Lanes 1, 2, 3 and 4 showing GUS specific amplicon of 380 bp.
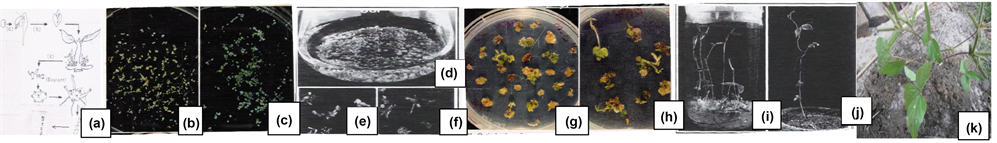
Figure 3. Production of transgenic black gram plants harboring the bacterial chitinase gene (ChiB) by the biolistics (a) Development of multiple shoots on cotyledonary node, excision of multiple shoots and chopped into small pieces (samples) (b) & (c) GUS positive samples (d), (e) & (f) Development of shoot buds on calliin liquid culture. (g) & (h) Shoots were selected on growing calli in semi-solid culture supplemented with kanamycin. (i) Full-grown in vitro plantlet. (j) & (k) Transformed plantlets in vermiculite &field soil bearing pods with seeds.
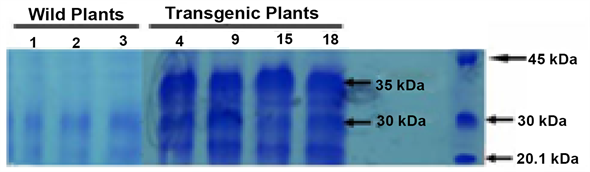
5, 9, 10, 14, 15, and 18) were positive for the 508-bp npt band (Figure 4(a)) but only five (B-Chi 1, 4, 9, 15 and 18) were found to possess 465-bp Bacterial chitinasegene (Figure 4(b))). There was no amplification in the untransformed plant and only five plants were used for further analysis. The foreign genes and their copy number pattern in the nuclear genome of the PCR positive transgenic lines were shown by Southern hybridization. The XbaI and BamHI fragments were released as the ChiB gene cassette (~1.56 kb). The blot probed with 32P-dCTP labeled ChiB cDNA in all the five transgenic lines showed ~1.56 kb band as expected. The genomic DNA was also digested with SacII, the lone restriction site on the T-DNA region, probed with 32P-dCTP labeled npt gene fragment. Single band appeared suggesting single copy integration in all five lines (Figure 5(a) and B).Molecular analyses demonstrated successful integration of T-DNA into the plant genomic DNA. RT-PCR analysis should the expression of ChiB gene. A 465-bp amplified fragment of the ChiB transcript of the Bacterial chitinase gene was clearly observed. No amplification was observed in the RNA samples isolated from the un-transformed plant. In-gel assay analysis of proteins of the transformed and untransformed plants resolved on the SDS-PAGE showed a number of chitinase isoforms (Figure (6)). Untransformed plants extract displayed chitinase isoforms of molecular weights at 21 and 30 kDa. But an additional isoform of 35 kDa was seen only in the transformed lines, as expected. Western blot analysis of the representative lines employing polyclonal antibodies raised against bacterial chitinase showed presence of a single prominent band
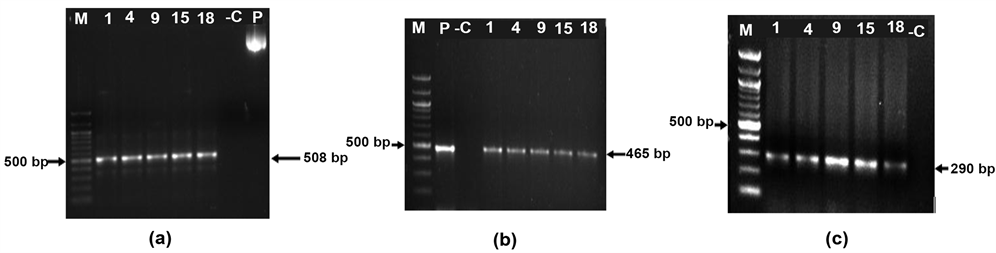
Figure 4. PCR analysis of plant lines for transgenes (a) npt gene), (b) ChiB gene). 200 ng of plant genomic DNA was used for each reaction. Numbers indicate plant lines (1, 4, 9, 15 and 18), P-Plasmid positive control; -C-untransformed plant negative control; M-1 Kb marker; Arrow indicates amplicon size. (c) RT-PCR analysis of plant lines for transgenes. 400 ng of total RNA was used for each reaction. Numbers indicate plant lines (1, 4, 9, 15 and 18); -C-untransformed plant negative control; M-100 bp marker; Arrow indicates amplicon size.
Figure 5. Southern Blot analysis of transgenic black gram cv T9.Genomic DNA (5 µg) and plasmid digested with XbaI (a) or BamHI (b) were probed with PCR-generated fragments of chitinase (ChiB) and npt gene. (Lanes-C-untransformed plant, P-plasmid-pBI121-ChiB, 4, 9, 15, 18-different transgenic lines, M-1 Kb marker).
Figure 6. Biochemical analysis of transformed and untransformed black gram plants. Detection of chitinolytic activity of chitinase after glycol chitin SDS-PAGE electrophoresis of untransformed and transformed black gram plants. (Lanes-1, 2 and 3 is untransformed plants showing two chitinase isoforms of 21 and 30 kDa. Lanes-4, 9, 15, and 18 are individual transformed lines showing three isoforms of chitinsaei e. 21 kDa and 30 kDa like normal untransformed plants and 35 kDa of unique size translated from bacterial chitinase (ChiB) gene. M-Rainbow marker was used as protein molecular weight standard.
corresponding to the size of 35 kDa indicating its robust expression (Figure 7). Higher chitinase activity in all the transgenic plants (chi4, chi9, chi15 and chi18) than in the nontransgenic ones (Table 1) was quite obvious. Lines 4 and 15 showed approximately two and three fold more increase in the enzyme activity than the nontransgenic ones, while lines 9 and 18 showed approximately one
Figure 7. Chitinase enzyme assay of different transgenic lines of black gram. Inset shows Western blots analysis showing 35 kDa bands.
Table 1. Tolerant potential of transgenic black gram plants to Erysiphae polygoni and number of days required for onset of disease and the complete leaf necrosis on detached leaves from Bacterial chitinase transgenic lines.
and half fold increase in the activity (Figure 7). This is correlated well with the degree of resistance to the pathogens. The statistical correlation coefficient between chitinase activity and disease rating scale is −0.1726. Thus the chitinase activity of the transgenic plants increases with corresponding reduction in the disease rating scale. Previous reports in the transgenic canola (Broglie et al. 1991), cotton (Emani et al. 2003) [22] adequately support the present observations. The detached leaves of the transgenic plants were tested for resistance to the obligate foliar ectoparasitic fungus, Erysiphae polygoni. Both Chi-4 and Chi-15 lines showed disease rating scores of 2.0 and 1.3 as an average score of three experiments, respectively, as compared to a score of 4.7 for the non-transformant (Table 1). These results indicated that the two transformants exhibited partial resistance to Erysiphae polygoni because there was delay in the spread of lesion areas of the disease. The degree of disease symptoms correlated well with the level of chitinase enzyme activity. The number of days required for the onset of necrosis was also studied (Table 1). Corresponding to the results on disease index, both Chi-4 and Chi-15 lines took longer period for the necrosis to develop and completely cover the whole leaf area (Reddy et al. 1987) [23] . However, with longer durations all leaves succumbed to the disease. Besides the detached leaves, pathogenicity of Erysiphae polygoni on leaves of both transgenic and non transgenic plants was also studied in the intact plants. The number of days required for the onset and complete chlorosis in each leaf in comparison to control (Figures 8(a)-(c)) were recorded. The diseased leaves were digitally photographed. A portion of leaves of both non transgenic and transgenic plants were electron micro-graphed which showed that pathogenic fungal spores could easily germinate, ramify mycelia and also invade the leaf surface cells of the non transgenic regenerated black gram plants (Figures 8(d)-(e)). These leaves developed powdery mildew disease. In the transgenic plants, pathogenic spores germinated well but mycelial growth was stunted leading to suppression of the disease.
4. Discussion
It was found that the spores were able to germinate but unable to develop mycelia and produce disease symptoms on the leaves of the transgenic plant. The
Figure 8. Evaluation of pathogenicity of Erysiphae polygoni both on transgenic and nontransgenic detached leaves of black gram plants (a) Transgenic black gram leaf showing spores couldn’t grow and no development of powdery mildew disease; (b) Untransformed black gram leaf showing growth of spores &development of disease; (c) Transformed black gram with high yield (d) Non transformed black gram with very low yield (e) Electron micrographs of untransformed (a) and transformed (b) black gram leaves’ showing the growth of mycelium of pathogen.
spores, however, germinated very fast, developed mycelia and symptoms of disease (as white powdery patches) on the leaves of the non-transformed plant by 8 - 18 days. Other green parts later became dull colored. These patches gradually increased in size and covered both surfaces of leaves in 20 - 69 days. In severe infection, foliage became yellow causing premature defoliation. These symptoms were significantly resisted in the transgenic lines. In vitro inoculation method was followed using detached leaves under controlled conditions to evaluate the resistance against the powdery mildew disease in the blackgram transgenic plants. Significant resistance by the transgenic plant leaves (at least till the 18th day) as compared to the controls was observed. Beyond this period resistance afforded by the transgene started waning. It may be due to non-availability of necessary ingredients to the detached leaves for survival and resistance from the mother plant. However, for 18 days these leaves resisted against the pathogen entirely on their own.
5. Conclusion
Briefly, the transformation of multiple shoots derived from the cotyledonary nodes of Vigna mungo L.cv. T9 were regenerated into rooted plants. Particle gun method enhanced the T.E. of the plant tissue. Successful integration of the bacterial chitinase gene into the black gram genome showed significant resistance to powdery mildew disease not only on the rooted plant leaves but also on the detached leaves. These findings suggest that the Bacterial chitinase ChiB gene could be utilized as a genetic source of disease control for breeding and improving crop plants.
Acknowledgements
Author is grateful to Dr. M. K. Reddy, Plant Molecular Biology Group and Dr. V. S. Reddy, Plant Transformation group, International Centre for Genetic Engineering and Biotechnology, New Delhi for providing Bacterial chitinase and antibody and providing facility for use of Particle gun bombardment, Mr. S.C.B. Sharma, Advanced Instrumentation Research Facility, JNU, New Delhi for providing Electron micrography, and Prof. N. K. Sah, Head, Deptt. of Botany, T. N. B. College, T. M. B. U. Bhagalpur for manuscript updating.
Cite this paper
Das, D.K. (2018) Expression of a Bacterial Chitinase (ChiB) Gene Enhances Resistance against Erysiphae polygoni Induced Powdery Mildew Disease in the Transgenic Black Gram (Vigna mungo L.) (cv. T9). American Journal of Plant Sciences, 9, 1759-1770. https://doi.org/10.4236/ajps.2018.98128
References
- 1. Singh, D.P. (1981) Breeding for Resistance to Disease in Green Gram and Blackgram. Theoretical and Applied Genetics, 59, 1-10.
- 2. Sailaja, M., Tarekeswari, M. and Sujatha, M. (2008) Stable Genetic Transformation of Castor (Ricinuscommunis L.) via Particle Gun—Mediated Gene Transfer Using Embryo Axes from Mature Seeds. Plant Cell Reports, 27, 1509-1519. https://doi.org/10.1007/s00299-008-0580-3
- 3. Ikea, J., Ingelbrecht, I., Uwalfo, A. and Thottapilly, G. (2003) Stable Gene Transformation in Cowpea (Vignaunguiculata L. walp). African Journal of Biotechnology, 2, 211-218.
- 4. Saini, R. and Jaiwal, P.K. (2007) Agrobacterium tumefaciens-Mediated Transformation of Black Gram: An Assessment of Factors Influencing the Efficiency of uidA Gene Transfer. Biologia Plantarum, 51, 69-74. https://doi.org/10.1007/s10535-007-0014-z
- 5. Jones, J.D.G., Dean, C., Gidoni, D., Gilbert, D., Bond-Nutter, D., Lee, R., Bedbrook, J. and Dunsmuir, P. (1986) Expression of Bacterial Chitinase Protein in Tobacco Leaves Using Two Photosynthetic Gene Promoters. Molecular and General Genetics, 212, 536-542. https://doi.org/10.1007/BF00330861
- 6. Das, D.K. and Rahman, A. (2010) Expression of a Bacterial Chitinase (ChiB) Gene Enhances Antifungal Potential in Transgenic Litchi chinensis Sonn. (cv. Bedana). Current Trends of Biotechnology and Pharmacy, 4, 820-833.
- 7. Chen, W.B., Xu, X., Liang, G.H., Muthukrishnan, S., Chen, P.D., Liu, D.J. and Gill, B.S. (1998) Introduction and Constitutive Expression of a Rice Chitinase Gene in Bread Wheat Using Biolistic Bombardment and bar Gene as a Selectable Marker. Theoretical and Applied Genetics, 97, 1296-1306. https://doi.org/10.1007/s001220051022
- 8. Kikkert, J.R., Vidal, J.R. and Reisch, B.I. (2004) Stable Transformation of Plant Cells by Particle Bombardment/Biolistics. Methods in Molecular Biology, 286, 61-78. https://doi.org/10.1385/1-59259-827-7:061
- 9. Das, D.K., Bhomkar, P., Shiva Prakash, N. and Sarin, N.B. (2002) Improved Method of Regeneration of Black Gram (Vigna mungo L.) through Liquid Culture. In Vitro Cellular & Developmental Biology—Plant, 38, 456-459. https://doi.org/10.1079/IVP2002336
- 10. Hoagland, M. and Arnon, D. (1950) The Water Culture Method for Growing Plants without Soil. California Agricultural Experiment Station, 347, 1950.
- 11. Jefferson, R.A. (1987) Assaying Chimeric Genes in Plants. The GUS Gene Fusion System. Plant Molecular Biology Reporter, 5, 387-405. https://doi.org/10.1007/BF02667740
- 12. Lodhi, M., Ye, G.N., Weeden, N.F. and Reisch, B.I. (1994) A Simple and Efficient Method for DNA Extraction from Grapevine Cultivars and Vitis Species. Plant Molecular Biology Reporter, 12, 6-13. https://doi.org/10.1007/BF02668658
- 13. Southern, E.M. (1975) Detection of Specific Sequences among DNA Fragments Separated by Gel Electrophoresis. Journal of Molecular Biology, 98, 503-517. https://doi.org/10.1016/S0022-2836(75)80083-0
- 14. Das, D.K. and Rahman, A. (2012) Expression of a Rice Chitinase Gene Enhances Antifungal Response in Transgenic Litchi (cvBedana). Plant Cell Tissue and Organ Culture, 109, 315-325. https://doi.org/10.1007/s11240-011-0097-2
- 15. Yamamoto, T., Iketani, H., Ieki, H., Nishizawa, Y., Notsuka, K., Hibi, T., Hayashi, T. and Matsuta, N. (2000) Transgenic Grapevine Plants Expressing a Rice Chitinase with Enhanced Resistance to Fungal Pathogens. Plant Cell Reports, 19, 639-646. https://doi.org/10.1007/s002999900174
- 16. Bradford, M.M. (1976) A Rapid and Sensitive Method for the Quantization of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Analytical Biochemistry, 72, 248-254. https://doi.org/10.1016/0003-2697(76)90527-3
- 17. Stephan, J. and Wolf, G.A. (1990) Dye-Labelled Substrate for the Assay and Detection of Chitinase and Lysozyme Activity. Journal of Microbiological Methods, 12, 197-205. https://doi.org/10.1016/0167-7012(90)90031-Z
- 18. Srivastava, S. (2004) Fruit and Vegetable Diseases. Disease Management of Fruits and Vegetables, 1, 307-355.
- 19. Morrison, R.M. (1960) Studies of Clonal Isolates of Erysiphae cichoracearum on Leaf Disc Culture. Mycologia, 52, 388-393. https://doi.org/10.2307/3755954
- 20. Jayraj, J., Anand, A. and Muthukrishnan, S. (2004) Pathogenesis-Related Proteins and Their Roles in Resistance to Fungal Pathogen. In: Punja, Z.K., Ed., Fungal Disease Resistance in Plants-Biochemistry, Molecular Biology and Genetic Engineering, Food Products Press (Haworth Press), New York, 139-178.
- 21. Vidal, J.R., Kikkert, J.R., Wallace, P.G. and Reisch, B.I. (2003) High-Efficiency Biolistic Co-Transformation and Regeneration of “Chardonnay” (Vitis vinifera L.) Containing nptII and Antimicrobial Peptide Genes. Plant Cell Reports, 22, 252-260. https://doi.org/10.1007/s00299-003-0682-x
- 22. Emani, C., Garcia, J.M., Lopata-Finch, E., Pozo, M.J., Uribe, P., Kim, D.J., Sunikumar, G., Cook, D.R., Kenerley, C.M. and Rathore, K.S. (2003) Enhanced Fungal Resistance in Transgenic Cotton Expressing an Endochitinase Gene from Trichoderma virens. Plant Biotechnology, 1, 321-326. https://doi.org/10.1046/j.1467-7652.2003.00029.x
- 23. Reddy, K.S., Pawar, S.E. and Bhatia, C.R. (1987) Screening for Powdery Mildew (Erysiphae polygoni DC) Resistance in Mungbean (Vigna radiata (L.) Wilzek) Using Excised Leaves. Proceedings: Plant Sciences, 97, 365-369.
Abbreviations
NPT II: Neomycin phosphotransferase gene,
ChiB: Bacterial chitinase gene,
MS: Murashige and Skoog (1962),
NAA: α-naphthalene acetic acid,
IBA: Indole3-butyric acid,
GUS: β-glucuronidase gene,
RT-PCR: Reverse transcriptase polymerase chain reaction,
CaMV 35S: Cauliflower mosaic virus constitutive promoter,
NOS: Nopaline synthase,
BAP: 6-benzyl amino purine,
T.E: Transformation efficiency