American Journal of Plant Sciences
Vol.5 No.13(2014), Article
ID:46934,10
pages
DOI:10.4236/ajps.2014.513206
Genetic Analysis of Tomato Fruit Ripening at Polypeptide Profiles Level through Quantitative and Multivariate Approaches
Ezequiel Marchionni Basté1,2, Javier H. Pereira da Costa1,2, Gustavo R. Rodríguez1,2*, Roxana Zorzoli2,3, Guillermo R. Pratta1,2
1National Council of Scientific and Technical Researchs (CONICET), Zavalla, Argentina.
2Genetics Cathedra, Faculty of Agronomy Sciences, National University of Rosario, Experimental Field “José F. Villarino”, Zavalla, Argentina
3Council of Researchs of the National University of Rosario (CIUNR), Zavalla, Argentina
Email: *gpratta@unr.edu.ar
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 28 April 2014; revised 25 May 2014; accepted 11 June 2014
ABSTRACT
Multivariate analysis became essential in functional and structural Genomics because of the large quantity of biological data provided by these new research areas. Diallel mating design was widely applied to analyze the heritability of quantitative traits but it was recently used for approaching to the inheritance patterns of other levels of gene expression such as transcript profiles. Investigating the inheritance pattern of total polypeptide profiles with a diallel design remains as a vacancy subject. The objective of the present research was to infer the inheritance of total polypeptides profiles from tomato pericarp tissue at four different ripening stages in a diallel mating design including five recombinant inbred lines (RILs) and their ten second cycle hybrids (SCH). To achieve this objective, a multivariate analysis was applied to identify eventual inheritance patterns through a data mining approach and then univariate analyses were used to verify these patterns. Mainly dominance and also overdominance, though in a minor percentage, contributed to the gene actions involved in their genetic basis. Multivariate analysis was efficient in identifying inheritance patterns of total polypeptide profiles through a data mining approach, and univariate analyses largely verified the identified gene actions.
Keywords:Solanum lycopersicum, Plant Genetic Resources, Proteomics, Cluster Analysis

1. Introduction
Functional and structural Genomics have recently provided scientist with a large amount of biological data causing that multivariate analysis became essential in these new research areas [1] . Also, the accumulation and joint interpretation of transcriptomic, proteomic, metabolomic and phenomic databases have conducted to infer the inheritance patterns of gene expression at those different levels of the genetic information flow [2] -[4] and led to interpret the molecular basis of heterosis, one of the most useful genetic phenomenonin Agronomy [5] .
Tomato (Solanum lycopersicum) fruit ripening is a developmental process with major implications on horticulture production given that commercial quality, including shelf life trait, is defined along it [6] -[8] . Proteome variation and metabolic regulation occurring during tomato ripening were widely reported [9] -[11] . Also, associations among polypeptide profiles from mature green and red ripe periparp tissue and different fruit quality traits were found in various segregating populations [12] -[14] . In many reports [12] -[16] , differential performance of polypeptide bands in F1 hybrids and/or segregating generations were found when compared to the homozygous parents. Other authors [17] -[19] have also widely reported de novo polypeptide bands in different species.
Hence investigating the inheritance pattern of total polypeptide profiles with an appropriate mating design remains as a vacancy subject. Diallel mating design [20] was widely applied to analyze the heritability of quantitative traits but it was recently used for approaching the inheritance patterns of other levels of gene expression such as transcript profiles [21] [22] .
The objective of the present research was to infer the inheritance of total polypeptides profiles from tomato pericarp tissue at four different ripening stages in a diallel mating design including five recombinant inbred lines (RILs) and their ten second cycle hybrids (SCH). To achieve this objective, a multivariate analysis was applied to identify eventual inheritance patterns through a data mining approach and then univariate analyses were used to verify these patterns.
2. Material and Methods
Five tomato RILs (Recombinant Inbred Lines ToUNR1, ToUNR8, ToUNR9, ToUNR15, and ToUNR18) derived by selection for fruit weight and shelf life from an inter specific cross S. lycopersicum cv. Caimanta x S. pimpinellifolium LA722 [23] were mated in a complete diallel design without reciprocals. The fifteen genotypes (5 parents and 10 hybrids) were characterized for agronomic traits by [24] . For experiments here reported, two fruits at mature green (MG), breaker (B), red ripe (RR) and ripe on shelves (RS) stages were harvested on three plants per genotypes. MG, B, and RR stages were defined according to [25] and RS stage was assessed according to [15] . Polypeptide extraction from pericarp tissue and SDS-PAGE were made according to [12] . Briefly, pericarp tissue was homogenized with mortar and pestle and the suspended in a SDS buffer: 4% (w/v) SDS, 2% (v/v) 2-mercaptoethanol, 20% (w/v) glycerol, and 2 mm PMSF in 100 mMTris-Cl (pH 8.5). The sample was adjusted to1.5 ml with this extraction buffer and incubated for 10 min at 4C. An equal volume of water saturated phenol was then added. After 10 min with shaking at room temperature, the phases were separated by centrifugation. The phenol phase was recovered and reextracted with an equal volume of extraction buffer. Proteins were precipitated from the phenol phase by addition of 5 vol of 0.1 M ammonium acetate in methanol and incubated at −20˚C overnight. The precipitate was washed three times with the ammonium acetate in methanol and once with acetone. The pellet was dried, solubilized and stored at 4˚C until electrophoresis. Equal amounts of polypeptides (20 ug) were run for 1.5 h at 35 mA constant in a denaturing polyacrylamide gels (12% v/v). Gels were stained with a 0.1% solution of Coomassie Brilliant Blue R-250 and distained with boiling water, scanned and analyzed using the Gelpro Analyzer 3.0 software. Polypeptide bands were evaluated as present or absent in all genotypes at each ripening stage and a binary value of 1 or 0 was respectively assigned.
Polymorphism for presence/absence of total polypeptide bands was evaluated among parents, among hybrids and among all genotypes by stage. Jaccard’s distances were calculated from 1/0 respective matrix and a cluster analysis by UPGMA method was applied to the five parents to find associations among the homozygous genotypes. Then the hybrids were included to investigate associations among homozygous and heterozygous genotypes, considering three possible inheritance patterns for the data mining approach: Unidirectional Dominance (UD): hybrids are equal to just one of their parents either for the presence or the absence of most polypeptide bands, Bidirectional Dominance (BD): hybrids are equal to one of their parents either for the presence or the absence of some polypeptide bands but equal to the other parent for the rest of the bands, and overdominance (OD): hybrids differ from both either from the presence or the absence of polypeptide bands, which represent de novo polymorphism. According to this criterion, UD and BD are properties of parents (though measured through their hybrids, as proposed by 29) while OD is a property of the heterozygous genotypes. Finally, univariate analysis reporting the number of individual bands by stage corresponding to gene action of dominance and over dominance in each cross and by parental RIL was accomplished to verify the inheritance pattern suggested by multivariate analysis.
3. Results
Polypeptide profiles were obtained from each genotype at all ripening stage per triplicate and no differences among repetitions were found according to SDS-PAGE (Figure 1). A total of 21 bands ranging between 102.7 and 27.9 kDa were counted though different numbers were detected according to ripening stages and groups of genotypes (Table 1). Just one band of 84.9 kDa was completely monomorphic. A wide variety of performances
Table 1. Polypeptide profiles of pericarp tissue in groups of parents (RILs), F1 (Hybrids) and all genotypes (RILs + Hybrids) at mature green (MG), breaker (B), red ripe (RR), and ripe on shelves (RS) stages.
kDa: molecular weight of the band; p: polymorphic band; m: monomorphic band; a: absent band; NPB: Number of polymorphic bands; TNB: Total number of bands; % P: percentage of polymorphism (NPB × 100/TNB).
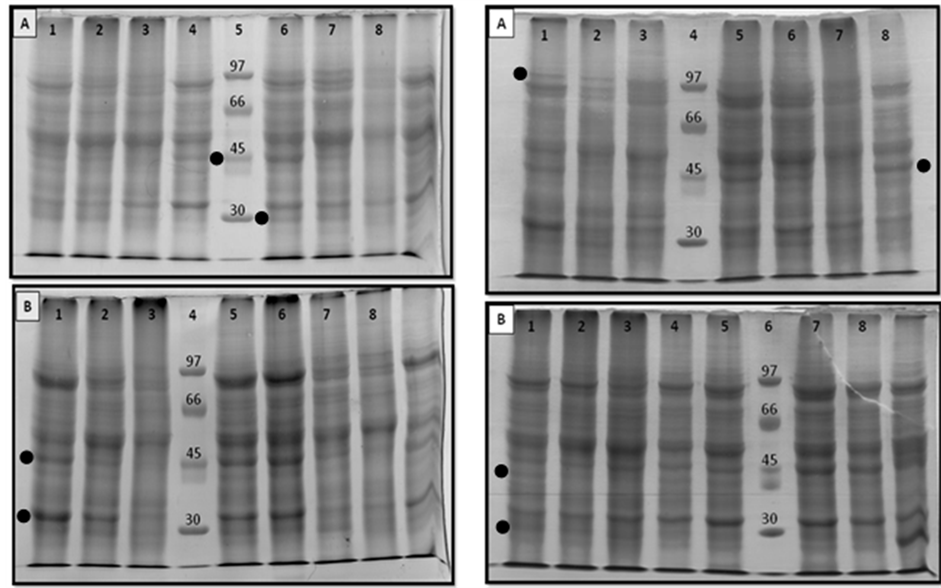 (a) (b)
(a) (b)
Figure 1. Pericarp total polypeptide profiles from different genotypes of the diallel mating design. (a) Gel A: Profiles from H(ToUNR18 × ToUNR8) at Red Ripe, Breaker, Mature Green and Ripe in shelves stages in lanes 1, 2, 3, and 4, respectively. Molecular weight marker (in kDa) is indicated in lanes 5. Profiles from H(ToUNR1 × ToUNR15) at Red Ripe, Breaker and Mature Green stages in lanes 6, 7, and 8, respectively. Gel B: Profiles from H(ToUNR18 × ToUNR9) at Red Ripe, Breaker, Mature Green and Ripe in shelves stages in lanes 1, 2, 3, and 5, respectively. Molecular weight marker (in kDa) is indicated in lane 4. Profiles from H(ToUNR18 × ToUNR1) at Red Ripe, Breaker and Mature Green stages in lanes 6, 7, and 8, respectively; (b) Gel A: Profiles from ToUNR18 at Red Ripe, Breaker and Mature Green stages in lanes 1, 2, and 3, respectively. Molecular weight marker (in kDa) is indicated in lane4. Profiles from H(ToUNR18 × ToUNR15) at Red Ripe, Breaker, Mature Green and Ripe in shelves stages in lanes 5, 6, 7, and 8, respectively. Gel B: Profiles from ToUNR1 at Red Ripe, Breaker and Mature Green stages in lanes1, 2, and 3, respectively. Molecular weight marker (in kDa) is indicated in lane 6. In lanes 4, 5, 7, and 8, profiles at Red in shelves stage from ToUNR8, ToUNR15, ToUNR18 and H(ToUNR18 × ToUNR1), respectively. Points indicate examples of polymorphic polypeptide bands.
was found, for instance the band of 102.7 kDa was monomorphic at B but polymorphic at the other stages while the75.6 kDa was absent in RR but present in MG, B, and RS. The band of 64.1 kDa was just detected at RS in RILs, being a de novo present polypeptide in hybrids at MG, B, and RR. Conversely, the band of 31.1 kDa was a de novo absent polypeptide in hybrids at MG and B, de novo present at RR and undetectable at RS in all genotypes. Overall, the highest percentage of polymorphism was found at MG followed by RR.
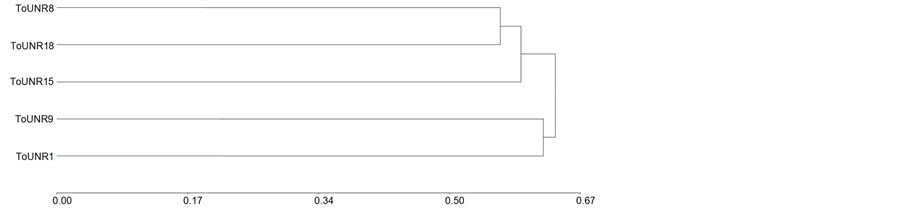
Dendrograms for parent according to all polymorphic polypeptide bands is shown in Figure 2(a), its cophenetic correlation being 0.71. RILs grouped in an unexpected way since ToUNR1, selected for high fruit weight and long shelf life, and ToUNR9, selected for low fruit weight and short shelf life [23] were in the same cluster. Then, ToUNR15, selected for high fruit weight and short shelf life, separated from the two closest RILs: ToUNR8, selected for low weight and long shelf life, and ToUNR18, selected just for its ovate shape, though it presented high fruit weight and long shelf life. Therefore, presence/absence of polypeptide bands appeared to be unrelated to phenotypic traits used as selection criteria. In Figure 2(b), the 15 genotypes of the diallel mating design were clustered, the respective dendrogram having a cophenetic correlation of 0.82. Associations among parents were not modified. Instead, hybrids inserted among them though in a non random way. In fact, ToUNR1 and ToUNR9 continued as a different cluster from the remnant genotypes and no hybrid grouped with them. Partition of the group formed by those remnant genotypes allow separating H(ToUNR15 x ToUNR9) from the rest, and then ToUNR15 and H(ToUNR15 × ToUNR8) clustered apart from the others. Finally, H(ToUNR1 ×
 (a)
(a)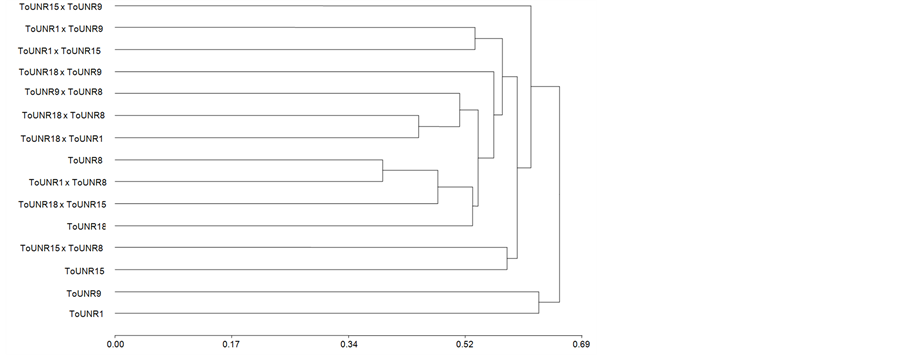 (b)
(b)
Figure 2. Dendrograms of parents (a) and all genotypes (b) of the diallel mating design according to Jaccard’s distances calculated from all polymorphic pericarp polypeptide bands and UPGMA’s method.
ToUNR15) and H(ToUNR1 × ToUNR9) differentiated from ToUNR8 and ToUNR18, which not only remained as the closest RILs but also grouped with most hybrids involving them as at least one of their parents. This arrangement of homozygous and heterozygous genotypes of the diallel design suggested the following inheritance patterns: ToUNR8 and ToUNR18 carried most of polypeptide bands that are dominant, while ToUNR1 and ToUNR9 carried the recessive alternatives. Hence they showed UD, which could explain their differential grouping as parents and the subsequent placing of hybrids around them (no F1 close to the recessive ToUNR1 and ToUNR9, and most F1s close to their dominant parents ToUNR8 and ToUNR18). In contrast ToUNR15 would carry certain polypeptide bands that are dominant in some cases and recessive in others, appearing as a BD inheritance pattern. Finally hybrids H(ToUNR15 × ToUNR9), H(ToUNR1 × ToUNR15), and H(ToUNR1 × ToUNR9) did not group with any of their parents and should present an OD inheritance pattern.
Table 2 summarizes the percentage of dominant and over dominant bands per cross, and detailed analysis are presented as Supplementary Materials I-IV. As suggested by the cluster analysis, dominance and over dominance were found for genetic determination of polypeptide polymorphism, the former being higher than the latter (70.3 vs. 29.7 in average, respectively). H(ToUNR1 × ToUNR8) had the greatest percentage of polypeptide bands determined by dominant gene actions (92.0%, Table 2 and Supplementary Material IV), most of them contributed by ToUNR8. Accordingly, this hybrid and its male parent were the closest genotypes in the dendrogram (Figure 2(b)). Conversely, one of the more divergent hybrid, H(ToUNR1 × ToUNR9), had the highest number and percentage of over dominant gene actions (48.8%, Table 2). Interestingly, this hybrid was obtained by crossing the two RILs that had the lowest number of dominant polypeptide bands (Supplementary Material III footnote). In respect to this subject, and as also suggested by cluster analysis, ToUNR8 and ToUNR18 contributed with the largest amount of dominant bands (54 and 53, respectively), followed by ToUNR15 (50, see Supplementary Material III footnote). Though the numbers of dominant bands is quite similar, performance of these RILs was very different in two ways. First, ToUNR8 and ToUNR18 had more dominant gene actions in all crosses, while ToUNR15 could be considered as recessive in crosses with both of
Table 2. Number of total polymorphic bands (NTB) and percentage of polymorphism (% P) in each cross segregated by the gene action underlying each polypeptide: Dominance and Overdominance, and in Total.
them and dominant in crosses with ToUNR1 and ToUNR9. This result confirms the trends to UD and BD previously mentioned. Secondly, while ToUNR8 and ToUNR18 showed dominance gene actions for the presence of the band (69% and 78%, respectively), ToUNR15 showed equal number of dominance for either band presence (57% of the dominance gene actions) or absence (43%). These results were calculated from the total of data that are presented as examples in Supplementary Material I.
4. Discussion
Polypeptide profiles from pericarp tissue at four different ripening stages were obtained, polymorphism at the earliest stage being higher than at the others. A greater polymorphism at MG was also found by [14] [16] . Additionally, [26] reported a wider diversity for free amino acid content in fruits at MG compared to RR stage. Examples of presence/absence of total bands in all genotypes by stage, and polymorphism by cross, are presented as Supplementary Materials I-II.
Cluster from the polypeptides profiles allowed to infer their inheritance patterns. Therefore results from this data mining through multivariate analysis were verified by univariate analysis, evaluating the gene action of individual polypeptide bands by each cross at all ripening stages. This finding explained the relatively unexpected position of most hybrids involving ToUNR15 as a parent in the cluster analysis since though they did not present an outstanding percentage of over dominance bands, most dominance gene actions correspond to absence of the bands, which are not computed in Jaccard’s distance as a cause of similarity. Though presence of the band has been proposed to be dominant, [2] have reported all possible kinds on gene actions (dominance of higher parent, dominance of lower parent, and positive and negative over dominance) for transcript profiles in a quantitative study of gene expression.
Non additive gene actions were reported by other authors in determining gene expression [2] [27] [28] but this is the first report on the application of a diallel mating design to the presence/absence of polypeptide bands. In this sense, significant and positive General Combining Abilities could be assigned to ToUNR8 and ToUNR18, while significant and negative ones could be assigned to ToUNR1 and ToUNR9 [29] . Also, significant and positive Specific Combining Ability could be assigned to H(ToUNR1 × ToUNR9) and significant and negative one to H(ToUNR15 × ToUNR9). Adaptations of AMOVA to diallel analysis for this dichotomy molecular variable could be accomplished to estimate the statistical significance of these observations. Furthermore, plant material used in this research comprises Second Cycle Hybrids [30] , which represent different arrangements of selected alleles contributed by just two homozygous original parents. [31] and [32] reported de novo AFLP and microsatellite variation, respectively, occurring in rice RILs respecting their original parents. Hence the great diversity here found for polypeptide expression could be due to recombination, genome reorganization and eventual mutations occurring during the selfing process along with selection was practiced.
5. Conclusion
Inheritance patterns of total polypeptide profiles from tomato pericarp tissue at four different ripening stages were inferred from a diallel mating design including five recombinant inbred lines (RILs) and their ten second cycle hybrids (SCH). Main dominance, either unidirectional or bidirectional, and also overdominance, though in a minor percentage, contributed to the gene actions involved in their genetic basis. Multivariate analysis was efficient in identifying these inheritance patterns through a data mining approach, and univariate analyses largely verified the identified gene actions.
References
- Jombart, T., Pontier, D. and Dufour, A.B. (2009) Genetic Markers in the Playground of Multivariate Analysis. Heredity, 102, 330-341. http://dx.doi.org/10.1038/hdy.2008.130
- Swanson-Wagner, R.A., Jia, I., De Cook, R., Borsuk, L.A., Nettleton, D. and Schnable, P.S. (2006) All Possible Modes of Gene Action Are Observed in a Global Comparison of Gene Expression in a Maize F1 Hybrid and Its Inbred Parents. Proceedings of the National Academy of Sciences of the United States of America, 103, 6805-6810. http://dx.doi.org/10.1073/pnas.0510430103
- Hoecker, N., Keller, B., Muthreich, N., Chollet, D., Descombes, P., Piepho, H.P. and Hochholdinger, F. (2008) Comparison of Maize (Zea mays L.) F1-Hybrid and Parental Inbred Line Primary Root Transcriptomes Suggests Organ-Specific Patterns of Nonadditive Gene Expression and Conserved Expression Trends. Genetics, 179, 1275-1283.http://dx.doi.org/10.1534/genetics.108.088278
- Lisec, J., Römisch-Margl, L., Nikoloski, Z., Piepho, H.P., Giavalisco, P., Selbig, J., Gierl, A. and Willmitzer, L. (2011) Corn Hybrids Display Lower Metabolite Variability and Complex Metabolite Inheritance Patterns. The Plant Journal, 68, 326-336. http://dx.doi.org/10.1111/j.1365-313X.2011.04689.x
- Hochholdinger, F. and Hoecker, N. (2007) Towards the Molecular Basis of Heterosis. Trends in Plant Sciences, 12, 427-432. http://dx.doi.org/10.1016/j.tplants.2007.08.005
- Akihiro, T., Koike, S., Tani, R., Tominaga, T., Watanabe, S., Iijima, Y., Aoki, K., Shibata, D., Ashihara, H., Matsukura, C., Akama, K., Fujimura, T. and Ezura, H. (2008) Biochemical Mechanism on GABA Accumulation during Fruit Development in Tomato. Plant and Cell Physiology, 49, 1378-1389. http://dx.doi.org/10.1093/pcp/pcn113
- Yin, Y.G., Tominaga, T., Iijima, Y., Aoki, K., Shibata, D., Ashihara, H., Nishimura, S., Ezura, H. and Matsukura, C. (2010) Metabolic Alterations in Organic Acids and γ-Aminobutyric Acid in Developing Tomato (Solanum lycopersicum L.) Fruits. Plant and Cell Physiology, 51, 1300-1314. http://dx.doi.org/10.1093/pcp/pcq090
- Oms-Oliu, G., Hertog, M.L.A.T.M., Van de Poel, B., Ampofo-Asiama, J., Geeraerd, A.H. and Nicolaï, B.M. (2011) Metabolic Characterization of Tomato Fruit during Preharvest Development, Ripening, and Postharvest Shelf-Life. Postharvest Biology and Technology, 62, 7-16. http://dx.doi.org/10.1016/j.postharvbio.2011.04.010
- Carrari, F. and Fernie, A.R. (2006) Metabolic Regulation Underlying Tomato Fruit Development. Journal of Experimental Botany, 57, 1883-1897. http://dx.doi.org/10.1093/jxb/erj020
- Faurobert, M., Mihr, C., Bertin, N., Pawlowski, T., Negroni, L., Sommerer, N. and Causse, M. (2007) Major Proteome Variations Associated with Cherry Tomato Pericarp Development and Ripening. Plant Physiology, 143, 1327-1346.http://dx.doi.org/10.1104/pp.106.092817
- Kahlau, S. and Bock, R. (2008) Plastid Transcriptomics and Translatomics of Tomato Fruit Development and Chloroplast-to-Chromoplast Differentiation: Chromoplast Gene Expression Largely Serves the Production of a Single Protein. The Plant Cell, 20, 856-874. http://dx.doi.org/10.1105/tpc.107.055202
- Rodríguez, G.R., Sequin, L., Pratta, G.R., Zorzoli, R. and Picardi, L.A. (2008) Protein Profiling in F1 and F2 Generations of Two Tomato Genotypes Differing in Ripening Time. Biologia Plantarum, 52, 548-522.http://dx.doi.org/10.1007/s10535-008-0107-3
- Rodríguez, G.R., da Costa J.H.P., Tomat, D.D., Pratta, G.R., Zorzoli, R. and Picardi, L.A. (2011) Pericarp Total Protein Profiles as Molecular Markers of Tomato Fruit Quality Traits in Two Segregating Populations. Scientia Horticulturae, 130, 60-66. http://dx.doi.org/10.1016/j.scienta.2011.06.004
- Gallo, M., Rodríguez, G.R., Zorzoli, R. and Pratta, G.R. (2011) Ligamiento entre caracteres cuantitativos de calidad de fruto y perfiles polipeptídicos del pericarpio en dos estados de madurez en tomate. Revista de la Facultad de Ciencias Agrarias Universidad Nacional de Cuyo, 43, 145-156.
- Gallo, M., Rodríguez, G.R., Zorzoli, R., Picardi, L.A. and Pratta, G.R. (2010) Proteómica de la madurez del tomate. Revista de la Facultad de Ciencias Agrarias Universidad Nacional de Cuyo, 42, 119-133.
- Liberatti, D.R., Rodríguez, G.R., Zorzoli, R. and Pratta, G.R. (2013) Tomato Second Cycle Hybrids Differ from Their Parents at Three Levels of Genetic Variation. International Journal of Plant Breeding, 7, 1-6.
- Mahmoud, A.A., Sukumar, S. and Krishnan, H.B. (2008) Interspecific Rice Hybrid of Oryza sativa × Oryza nivara Reveals a Significant Increase in Seed Protein Content. Journal of Agricultural and Food Chemistry, 56, 476-482. http://dx.doi.org/10.1021/jf071776n
- Hoecker, N., Lamkemeyer, T., Sarholz, B., Paschold, A., Fladerer, C., Madlung, J., Wurster, K., Stahl, M., Piepho, H.P., Nordheim, A. and Hochholdinger, F. (2008) Analysis of Nonadditive Protein Accumulation in Young Primary Roots of a Maize (Zea mays L.) F1-Hybrid Compared to Its Parental Inbred Lines. Proteomics, 8, 3882-3894. http://dx.doi.org/10.1002/pmic.200800023
- Cheema, N.M., Malik, M.A., Qadir, G. and Rabbani, M.A. (2010) Characterization of Castor Bean Genotypes under Various Environments Using SDS-PAGE of Total Seed Storage Proteins. Pakistan Journal of Botany, 42, 1797-1805.
- Griffing, B. (1956) Concept of General and Specific Combining Ability in Relation to Diallel Crossing Systems. Australian Journal of Biological Sciences, 9, 463-493.
- Zwarts, L., Magwire, M.M., Carbone, M.A., Verstevena, M., Herteleer, L., Anholt, R.R.H., Callaertsa, P. and Mackay, T.F.C. (2011) Complex Genetic Architecture of Drosophila Aggressive Behavior. Proceedings of the National Academy of Sciences of the United States of America, 108, 17070-17075. http://dx.doi.org/10.1073/pnas.1113877108
- Yang, D.G., Ye, C.Y., Ma, X.F., Zhu, Z.H., Zhou, X.J., Wang, H.F., Meng, Q.Q., Pei, X.Y., Yu, S.X. and Zhu, J. (2012) A New Approach to Dissecting Complex Traits by Combining Quantitative Trait Transcript (QTT) Mapping and Diallel Cross Analysis. Chinese Science Bulletin, 57, 2695-2700. http://dx.doi.org/10.1007/s11434-012-5196-x
- Rodríguez, G.R., Pratta, G.R., Zorzoli, R. and Picardi, L.A. (2006) Recombinant Lines Obtained from an Interspecific Cross between Lycopersicon Species Selected by Fruit Weight and Fruit Shelf Life. Journal of the American Society for Horticultural Science, 131, 651-656.
- MarchionniBasté, E., Liberatti, D.R., Mahuad, S.L., Rodríguez, G.R., Pratta, G.R., Zorzoli, R. and Picardi, L.A. (2010) Diallel Analysis for Fruit Traits among Tomato Recombinant Inbred Lines Derived from an Interspecific Cross Solanum lycopersicum × S. pimpinellifolium. Journal of Applied Horticulture, 12, 21-25.
- Giovannoni, J.J. (2004) Genetic Regulation of Fruit Development and Ripening. The Plant Cell, 16, S170-S180. http://dx.doi.org/10.1105/tpc.019158
- Pratta, G., Rodríguez, G.R., Zorzoli, R., Picardi, L.A. and Valle, E.M. (2011) Biodiversity in a Tomato Germplasm for Free Amino Acids and Pigment Content of Ripening Fruits. American Journal of Plant Sciences, 2, 255-261. http://dx.doi.org/10.4236/ajps.2011.22027
- Rapp, R.A., Udall, J.A. and Wendel, J.F. (2009) Genomic Expression Dominance in Allopolyploids. BMC Biology, 7, 18. http://dx.doi.org/10.1186/1741-7007-7-18
- Marcon, C., Schützenmeister, A., Schütz, W., Madlung, J., Piepho, H.P. and Hochholdinger, F. (2010) Nonadditive Protein Accumulation Patterns in Maize (Zea mays L.) Hybrids during Embryo Development. Journal of Proteome Research, 9, 6511-6522. http://dx.doi.org/10.1021/pr100718d
- Vianna, J.M.S. (2000) The Parametric Restrictions of the Griffing Diallel Analysis Model: Combining Ability Analysis. Genetics and Molecular Biology, 23, 877-881. http://dx.doi.org/10.1590/S1415-47572000000400029
- Hill, J., Lethenborg, P., Li, P.W., Rahman, M.H., Sørensen, H. and Sørensen, J.C. (2003) Inheritance of Progoitrin and Total Aliphatic Glucosinolates in Oilseed Rape (Brassica napus L). Euphytica, 134, 179-187. http://dx.doi.org/10.1023/B:EUPH.0000003857.57573.2f
- Wang, Y.M., Dong, Z.Y., Zhang, Z.J., Lin, X.Y., Shen, Y., Zhou, D. and Liu, B. (2005) Extensive de Novo Genomic Variation in Rice Induced by Introgression from Wild Rice (Zizania latifolia Griseb.). Genetics, 170, 1945-1956. http://dx.doi.org/10.1534/genetics.105.040964
- Dong, Z.Y., Wang, H.Y., Dong, Y.Z., Wang, Y.M., Liu, W., Miao, G.J., Lin, X.Y., Wang, D.Q. and Liu, B. (2013) Extensive Microsatellite Variation in Rice Induced by Introgression from Wild Rice (Zizania latifolia Griseb.). PLoS ONE, 8, Article ID: e62317. http://dx.doi.org/10.1371/journal.pone.0062317
NOTES

*Corresponding author.


