American Journal of Plant Sciences
Vol.4 No.1(2013), Article ID:27635,8 pages DOI:10.4236/ajps.2013.41013
Characterization of a Tos17 Insertion Mutant of Rice Auxin Signal Transcription Factor Gene, OsARF24
![]()
1Faculty of Bioresources and Environmental Sciences, Ishikawa Prefectural University, Nonoichi, Japan; 2Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya, Japan.
Email: *sakamoto@ishikawa-pu.ac.jp
Received October 19th, 2012; revised November 26th, 2012; accepted December 6th, 2012
Keywords: Auxin; Auxin Response Factor (ARF); Mutant; Phyllotaxis; Retrotransposon Tos17; Rice
ABSTRACT
Auxin signaling plays a key role in the regulation of various growth and developmental processes in higher plants. Auxin response factors (ARFs) are transcription factors that regulate the expression of auxin-response genes. The osarf24-1 mutant contains a truncation of domain IV in the C-terminal dimerization domain of a rice ARF protein, OsARF24. This mutant showed auxin-deficient phenotypes and reduced sensitivity to auxin. However, OsARF24 protein contains an SPL-rich repression domain in its middle region and acts as a transcriptional repressor. These results imply that the C-terminal dimerization domain, especially the C-terminal half of domain IV, is essential for the proper regulation of OsARF24 function as a transcriptional repressor in rice.
1. Introduction
Auxins are endogenous phytohormones that play important roles in regulating a wide variety of cellular and developmental processes. Analyses of auxin-insensitive mutants have provided solid evidence to support the models of auxin function proposed by conventional physiological experiments and have also provided new insights and ideas about auxin. Recent molecular genetic studies, mainly on Arabidopsis (Arabidopsis thaliana L.), have made significant progress in elucidating the auxin signaling pathway. The binding of a bioactive auxin such as indole-3-acetic acid (IAA) to members of the TIR1/ AFB family of F-box proteins triggers the degradation of Aux/IAA transcriptional repressors, thereby allowing auxin response factor (ARF) transcription factors, which show either activator or repressor activity, to regulate the expression of auxin-response genes [1-5].
A typical ARF protein contains a conserved N-terminal DNA-binding domain, a non-conserved middle region, and a conserved C-terminal dimerization domain [3,6,7]. The DNA-binding domain of ARF protein binds with specificity to TGTCTC auxin response elements (AuxREs) in promoters of auxin-response genes to regulate their expression [8]. The C-terminal dimerization domain, which is related in amino acid sequence to domains III and IV in Aux/IAA protein, is involved in the homoand hetero-dimerization of ARF proteins and the hetero-dimerization among ARF and Aux/IAA proteins [4,9,10]. The middle region, between the DNA-binding domain and the C-terminal dimerization domain, functions as either an activation domain or a repression domain [3,4,11,12].
The ARF proteins are encoded by a multigene family in plants, and in rice (Oryza sativa L.), 25 OsARF genes have been identified [13]. Among them, 9 OsARF genes encode transcriptional activators and the other 16 OsARF genes encode transcriptional repressors [14]. However, the biological function of most OsARFs is poorly understood. Here, we report the identification and characterization of the osarf24-1 mutant, a line containing a Tos17 retrotransposon insertion in the putative repressor OsARF gene OsARF24. Interestingly, this mutant showed auxindeficient phenotypes and reduced sensitivity to auxin. Because OsARF24 contains an serine proline leucine (SPL)-rich repression domain and indeed acts as a repressor, we hypothesized that the C-terminal dimerization domain, especially the C-terminal half of domain IV, which is truncated in the osarf24-1 mutant, is essential for the regulation of OsARF24 function as a transcriptional repressor.
2. Materials and Methods
2.1. Plant Material
Seeds of wild-type rice (Oryza sativa L. “Nipponbare”)the osarf24-1 mutant, and transformants (described below) were sterilized in 1% NaClO for 30 min and sown on Murashige and Skoog agar medium. Seedlings were grown in a growth chamber at 28˚C under continuous light for 2 weeks. For morphological characterization, seedlings were transplanted and grown in the paddy field at the Experimental Farm of Ishikawa Prefectural University. For gene expression analyses, seedlings were selected for uniformity of growth and adapted to hydroponic culture for 2 days before treatment. IAA treatment (20 mM) was carried out by adding IAA to the culture medium.
2.2. Gene Expression Analysis
Total RNA was extracted from whole seedlings of wildtype and mutant rice and from mature leaves of transgenic rice by using an RNeasy Plant Mini Kit (Qiagen, Venlo, Netherlands). Single-strand cDNAs were synthesized by using the Advantage RT-for-PCR Kit (Clontech, Palo Alto, CA, USA). Quantitative RT-PCR was performed with an iCycler iQ real-time PCR system (BioRad Laboratories, Hercules, CA, USA). The primer sequences were 5’-CAGGAAGCTGGTGT GTTGTC-3’ and 5’-CTTGATCAGGCGTGGCTGTG-3’ for OsARF23, 5’-AATGACGCCTGACATCACAC-3’ and 5’-GCTTG ATAAGACTCGATGAGG-3’ for OsARF24, 5’-ACCA AGAGCCGCTCAATGAG-3’ and 5’-ATCACACGTG GGCGAACATC-3’ for OsIAA1, 5’-GATGAACAGGC GGTCGCTGC-3’ and 5’-GGCTC CGGTAGTAGCTTG TG-3’ for OsGH3-1, and 5’-CGCC AGTTTGGTCGCT CTCGATTTCG-3’ and 5’-TCAGGA GCTCCGTGCTC TTCTGGTAC-3’ for Histone H3. These primers specifically amplified the target gene sequences. Expression levels were normalized against the values obtained for Histone H3, which was used as an internal reference gene. For the gene expression experiments, we performed 3 biological repeats.
2.3. Electrophoresis Mobility Shift Assay
Full-length OsARF24 cDNA was inserted in the sense orientation into the pET-32a expression vector (Novagen, Madison, WI, USA) to generate a thioredoxin fusion protein when expressed in BL21(DE3) E. coli cells (Stratagene, La Jolla, CA, USA). The recombinant protein was purified by using Talon Metal Affinity Resin (Clontech Co., Palo Alto, CA). OsIAA1 promoter fragments containing WT or MT AuxRE were amplified by PCR with rice genomic DNA. The primer sequences were 5’-GGTTGAAATTGGAACGATGTG-3’ and 5’-G GAACTTTCATCTACTACTAC-3’ for OsIAA1 AuxRE (WT), and 5’-TTTGGATTCTCCATTATGAGAAAATC AAAACATGGTTTTTT-3’ and 5’-TTAATAAAAAAC CATGTTTTGATTTTCTCATAATGGAGAATCC-3’ for generating the AuxRE mutation (MT). The amplified fragments were cloned into pBluescript II SK (Stratagene) and their identities were confirmed by sequence analysis. The PCR-amplified fragments were excised with restriction endonucleases, purified by 10% PAGE, and labeled with biotin using a Biotin 3’ End DNA Labeling Kit (Pierce, Rockford, IL, USA). The electrophoresis mobility shift assay was performed by using a LightShift Chemiluminescent EMSA Kit (Pierce).
2.4. Production of Transgenic Rice
The entire OsARF24 coding region was inserted between the rice Actin promoter and the nopaline synthase polyadenylation signal of the hygromycin-resistant binary vector pAct-Hm2. This vector was modified from pBIH1 [15] to contain a rice Actin promoter. The resulting construct was introduced into Agrobacterium tumefaciens strain EHA105, and Agrobacterium-mediated transformation of rice (O. sativa L. “Nipponbare”) was performed as described [16]. Transgenic plants were selected on Murashige and Skoog agar medium containing 50 mg·L-1 hygromycin, and then grown in a greenhouse at 28˚C under ambient light conditions.
3. Results and Discussion
3.1. Characterization of osarf24-1 Mutant Rice
osarf24-1 is a mutant of OsARF24 caused by insertion of the Tos17 retrotransposon. osarf24-1 showed a reduction in plant height (the height of osarf24-1 was 91% that of the wild-type, n = 10, P < 0.001; Figure 1(a) and Table 1) and a reduction in the leaf angle of flag leaves from the wild-type value of 22.3˚ to 13.0˚ (n = 10, P < 0.001). This mutant also showed the narrow leaf phenotype typical of auxin-deficient or auxin-insensitive rice mutants (Figure 1(b)) [17,18], and the ratio of blade width to blade length of the flag leaves was reduced (0.031, versus 0.050 in wild-type, n = 10, P < 0.001). Wild-type rice leaves form in a distichous alternate phyllotactic manner, and successive leaves develop on opposite sides of the shoot apical meristem with 180˚ of divergence (Figure 1(c), left plant). However, disordered phyllotaxis was found in the osarf24-1 mutant. In these seedlings, leaf divergence was not 180˚, but instead twisted gradually (Figure 1(c), right plant). Because similar twisted leaf development was also observed in 2,4-D-treated wild-type rice and constitutively active Aux/IAA repressor transgenic rice [19], an adequate auxin signal is important for normal rice leaf development. These phenotypes of osarf24-1 suggest that the osarf24-1 mutant has some defects in auxin response.
Sequence analysis revealed that osarf24-1 had an insertion of Tos17 in exon 13 (Figure 1(d)). In osarf24-1,

Figure 1. Phenotype of the osarf24-1 mutant. (a) Comparison of gross morphology between wild-type (left) and osarf24-1 mutant (right) rice. Bar, 20 cm. (b) Leaf morphology of wild-type (left) and osarf24-1 mutant (right) rice. Bar, 1 cm. (c) Phyllotaxis of wild-type (left) and osarf24-1 mutant (right) rice. Numbers indicate leaf number from top to bottom. (d) The upper panel indicates the genomic structure of the OsARF24 gene and the insertion site of Tos17. White and black boxes indicate non-coding and protein-coding regions, respectively. Lines between the boxes represent introns. In osarf24-1, Tos17 was inserted into exon 13 of OsARF24. The lower panel indicates the modular structure of the OsARF24 protein and the insertion site of Tos17. DBD, MR, and CTD indicate the DNA-binding domain, the middle region (which acts as repression domain), and the C-terminal dimerization domain, respectively. In osarf24-1, Tos17 was inserted into the part of the gene encoding the C-terminal dimerization of the OsARF24 protein at the 733 th leucine residue. (e) Sequence alignment of C-terminal dimerization domain of OsARF24 in wild-type (OsARF24) and osarf24-1 mutant (osarf24-1) rice. Lowercase letters indicate amino acid changes caused by the mutation. Bars above the sequences indicate domain III and domain IV, respectively.
Table 1. Phenotype of the osarf24-1 mutant.

aEach column represents mean ± s.d. of 10 independent plants.
Tos17 was inserted into the part of the gene encoding the C-terminal dimerization domain of the OsARF24 protein at the 773th leucine residue (Figure 1(d)). This insertion altered the amino acid sequence in the C-terminal dimerization domain of the OsARF24 protein; specifically, the C-terminal half of domain IV, which has a similar amino acid sequence to Aux/IAA proteins, was changed and truncated (Figure 1(e)). Therefore, osarf24- 1 is considered to be a truncation mutant of the C-terminal dimerization domain in OsARF24 protein.
3.2. The osarf24-1 Mutant Is Less Sensitive to Auxin
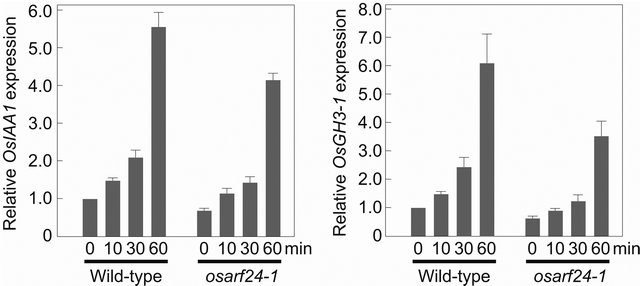
The treatment of rice seedlings with IAA induced an increase in the expression of auxin-response genes OsIAA1 and OsGH3-1 [20,21]. After 10 min, IAA treatment of wild-type seedlings increased the expression of both OsIAA1 and OsGH3-1 to 1.5 times the levels in untreated seedlings (Figure 2). The expression levels gradually increased until 60 min after IAA treatment to 5.5 and 5.9 times those in untreated seedlings, respectively. In osarf24-1 seedlings, the steady-state levels of OsIAA1 and OsGH3-1 mRNA were 71% and 66%, respectively, of those in the wild-type seedlings (Figure 2). The kinetics of the increases in OsIAA1 and OsGH3-1 mRNA levels of osarf24-1 after IAA treatment were similar to those of wild-type rice. At 10 min after IAA treatment, the levels of OsIAA1 and OsGH3-1 mRNA in osarf24-1 seedlings were 1.0 and 0.9 times, respectively, those in the untreated wild-type control, and at 60 min after the treatment, had increased to 3.9 and 3.5 times, respectively. The levels of both OsIAA1 and OsGH3-1 mRNA in IAA-treated osarf24-1 were lower than those in the wild-type. However, the ratios of the relative expression levels of OsIAA1 and OsGH3-1 mRNA in IAA-treated osarf24-1 to untreated osarf24-1 were approximately 1.4 after 10 min and 5.3-5.4 after 60 min, demonstrating that osarf24-1 still has some ability to respond to IAA. Although multiple OsARF proteins are considered to regulate the expression of OsIAA1 and OsGH3-1, our results suggest that the osarf24-1 mutant is less sensitive to auxin than wild-type rice. Because the C-terminal dimerization domain in ARF proteins is considered to function in hetero-dimerization among ARF and Aux/IAA proteins [4,9,10], it is possible that the OsARF24 repressor protein with a C-terminal dimerization domain truncation in the osarf24-1 mutant cannot interact with Aux/ IAA proteins in rice, and therefore that this mutant
 (a) (b)
(a) (b)
Figure 2. Effect of IAA treatment on the expression of OsIAA1 and OsGH3-1 in wild-type and osarf24-1 mutant rice seedlings. Expression levels were normalized against the values obtained for Histone H3. The value obtained from wild-type plants without IAA treatment was arbitrarily set at 1.0. Each column represents mean ± s.d. of 3 biological repeats.
OsARF24 protein represses auxin-response genes constitutively.
3.3. Molecular Characterization of OsARF24
Among the 25 ARF genes in rice, the deduced amino acid sequence of OsARF24 is most closely related (71.4% identity) to OsARF23 (previously designated as OsARF1 [22]), and phylogenetic analysis grouped OsARF24 and OsARF23 with an Arabidopsis ARF, AtARF2 [13]. The predicted open reading frames (ORFs) of OsARF23 and OsARF24 encode proteins of 836 and 840 amino acids, respectively. The similarity of the deduced amino acid sequences of OsARF23 and OsARF24 is 71.4%, and the sequences are most closely related (55.1% and 52.7% similarity, respectively) to Arabidopsis AtARF2/HSS (At5g62010, 853 amino acids) [23]. The structures of OsARF23 and OsARF24 are similar to that of AtARF2/ HSS throughout their lengths: three domains found in repressor ARFs-a DNA-binding domain, an SPL-rich repression domain, and a C-terminal dimerization domain that is related in amino acid sequence to domains III and IV in Aux/IAA proteins-are highly conserved. Because the SPL-rich repression domain is a characteristic of repressor ARF proteins [24], both OsARF23 and OsARF24 are considered to function as transcriptional repressors [13,14].
3.4. Expression of OsARF24 in Wild-Type Rice Plants
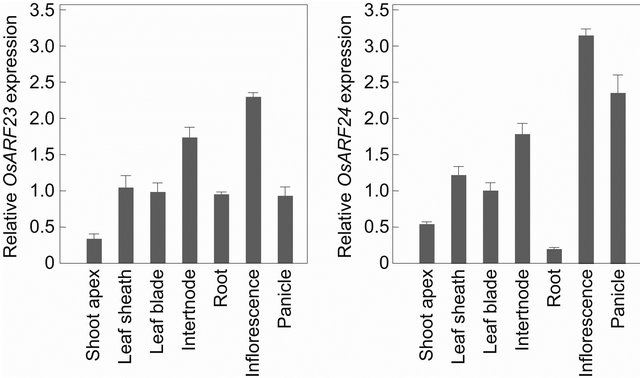
Quantitative reverse-transcription PCR analysis revealed that OsARF24 and OsARF23 were expressed at different levels in all the organs of wild-type rice that we tested, including the vegetative shoot apices, leaf sheaths, leaf blades, elongating internodes, roots, inflorescences (immature panicles), and panicles at flowering time (Figure 3). Both genes were expressed at the highest level in inflorescences. OsARF24 was also preferentially expressed in panicles at flowering time, elongating internodes, leaf sheaths, and leaf blades, and at low levels in vegetative shoot apices and roots. OsARF23 was also preferentially expressed in elongating internodes, leaf sheaths, leaf blades, roots, and panicles at flowering time, and at a low level in vegetative shoot apices.
Previous observations indicate that the expression levels of some ARF genes were not affected by IAA treatment, and those of the others were either increased or decreased [10,13]. Thus, we examined the effect of exogenously applied auxin on the expression of OsARF23 and OsARF24. IAA treatment increased the expression of both OsARF23 and OsARF24 to 1.5 times that in untreated plants within 10 min (Figure 4). However, the kinetics of the increases in OsARF23 and OsARF24 mRNA levels after IAA treatment were different. The
 (a) (b)
(a) (b)
Figure 3. Relative mRNA levels of OsARF23 and OsARF24 in various organs of wild-type rice. Expression levels were normalized against the values obtained for Histone H3. Each column represents mean ± s.d. of 3 biological repeats.

Figure 4. Effect of IAA treatment on the expression of OsARF23 and OsARF24 in wild-type rice seedlings. Expression levels were normalized against the values obtained for Histone H3. The values obtained without IAA treatment were arbitrarily set at 1.0. Each column represents mean ± s.d. of 3 biological repeats.
expression level of OsARF23 gradually increased until 60 min after IAA treatment to 2.5 times that in untreated plants, whereas the expression level of OsARF24 reached a maximum within 10 min and was maintained at that level until 60 min after IAA treatment. These results suggest that the expression of OsARF23 and OsARF24 is regulated by different mechanisms.
3.5. OsARF24 Binds to AuxRE in the OsIAA1 Promoter
The AuxRE has been identified in the promoters of some early auxin-response genes, and ARFs bind the AuxRE to regulate the transcription of these genes [24]. To examine whether the recombinant OsARF24 protein interacts with the AuxRE in the OsIAA1 promoter, we performed electrophoresis mobility shift assays (Figure 5). OsARF24 bound to a 377-bp OsIAA1 fragment containing the intact AuxRE (WT). The amount of retarded complex was reduced by the addition of increasing concentrations of unlabeled WT fragment as a competitor (“WT comp”, in Figure 5). The binding of OsARF24 with the WT fragment was not affected by addition of the unlabeled OsIAA1 fragment containing the mutated AuxRE sequence as a competitor (“MT comp”, in Figure 5). These results demonstrate that OsARF24 can bind to the promoter sequence of OsIAA1, and that this interaction depends only on the presence of an intact AuxRE sequence in the promoter fragment.
3.6. Overexpression of OsARF24 in Transgenic Rice
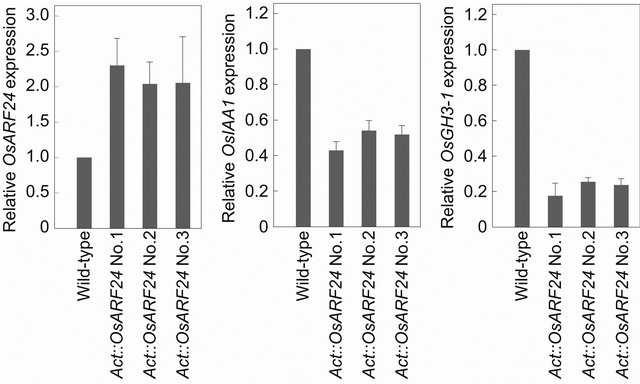
To assess the activity of the OsARF24 gene product in vivo, we fused the full-length OsARF24 cDNA to the rice Actin promoter in the sense orientation (Act::OsARF24) and introduced the construct into wild-type rice by Agrobacterium-mediated gene transfer (Figure 6). In this experiment, we selected 3 transgenic lines in which the OsARF24 expression level was doubled relative to wild-type (Figure 7). The Act::OsARF24 transformants showed a slight reduction in plant height (the average plant height of 5 plants from each of 3 different transgenic lines was 85% that of wild-type; Figure 6(a)). These transformants also showed the narrow leaf phenotype typical of auxin-deficient or auxin-insensitive rice mutants (Figure 6(b)), and the ratio of blade width to blade length of the flag leaves was reduced to 0.034 (the average of 5 flag leaves from each of 3 different transgenic lines), versus 0.050 in wild-type. Disordered phyllotaxis was also found in Act::OsARF24 transgenic seedlings (Figure 6(c)). These results suggest that the Act:: OsARF24 transformants have some defects in auxin response, as was the case for the osarf24-1 mutant.
We also examined the expression of auxin-response genes in the Act::OsARF24 transformants. The steadystate levels of OsIAA1 and OsGH3-1 mRNA in the leaves of Act::OsARF24 transformants decreased to about 50% and 20%, respectively, of those in the leaves of wild-type plants (Figure 7), confirming that OsARF24, which has an SPL-rich middle region, acts as a transcriptional repressor of auxin signaling in rice.
3.7. The C-Terminal Dimerization Domain Is Essential for the Regulation of OsARF24 Function
As previously mentioned, OsARF24 protein, which contains the SPL-rich repression domain, is considered to be
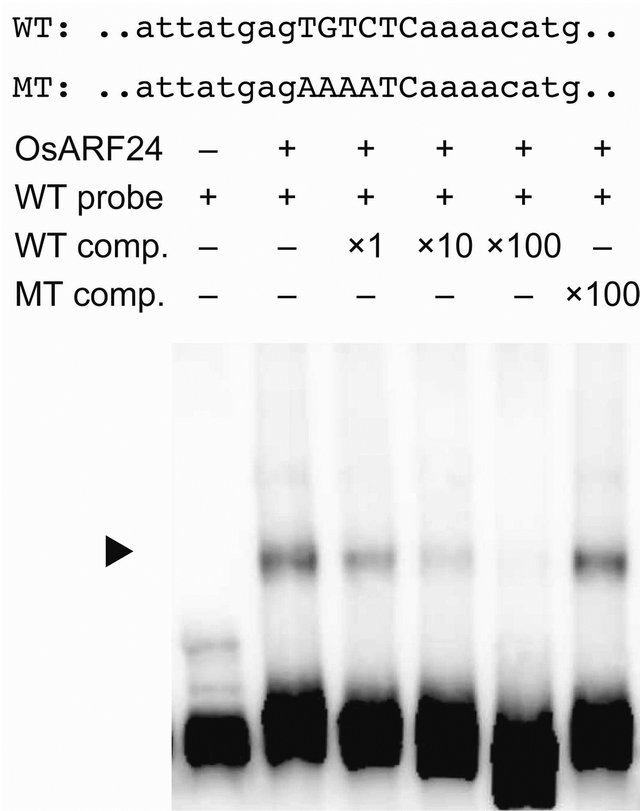
Figure 5. OsARF24 binds to the AuxRE in OsIAA1. Upper panel represents the core sequence of the AuxRE found in OsIAA1 (WT) and a mutated AuxRE sequence (MT). A labeled WT fragment was used as the probe. Binding affinity was estimated using non-labeled WT and MT fragments as competitors (comp.) at the relative concentrations indicated. OsARF24-DNA complexes are indicated by the arrowhead.

Figure 6. Phenotype of transgenic rice overexpressing the OsARF24 cDNA. (a) Comparison of gross morphology between wild-type (left) and Act::OsARF24 transgenic (right) rice. Bar, 20 cm. (b) Leaf morphology of wild-type (left) and Act::OsARF24 transgenic (right) rice. Bar, 1 cm. (c) Phyllotaxis of Act::OsARF24 transgenic rice. Numbers indicate leaf number from top to bottom.
an ARF that functions as a transcriptional repressor [13, 14]. In our experiments, transgenic rice overexpressing the OsARF24 cDNA showed auxin-deficient phenotypes including dwarf stature, narrow leaf, and aberrant phyllotaxis. In addition, the expression levels of auxin-response genes OsIAA1 and OsGH3-1 were decreased in these transformants. These results strongly support the hypothesis that OsARF24 acts as a repressor ARF. However, the Tos17 retrotransposon insertion mutant of OsARF24, osarf24-1, also showed both auxin-deficient
 (a) (b) (c)
(a) (b) (c)
Figure 7. Relative mRNA levels of OsARF24, OsIAA1, and OsGH3-1 in the leaves of transgenic rice overexpressing the OsARF24 cDNA. Expression levels were normalized against the values obtained for Histone H3. The value obtained from wild-type rice leaves was arbitrarily set at 1.0. Each column represents mean ± s.d. of 3 biological repeats.
phenotypes and decreased levels and auxin responses of OsIAA1 and OsGH3-1 expression. Although another rice ARF, OsARF23, shows high amino acid sequence similarity with OsARF24, the expression of OsARF23 and OsARF24 is regulated by different mechanisms in various organs of wild-type rice plants and in response to IAA treatment. Based on these results, we consider that OsARF23 and OsARF24 do not function redundantly in rice.
Most ARF proteins contain a C-terminal dimerization domain related to domains III and IV in Aux/IAA proteins [4,7,10]. The C-terminal dimerization domains in both ARF and Aux/IAA proteins are protein-protein interaction domains that allow homoand heterodimerization of ARF proteins and hetero-dimerization among ARF and Aux/IAA proteins [4,9,10]. Although ARF repressors can dimerize via their C-terminal dimerization domains, ARF repressor-Aux/IAA and ARF represssorARF activator interactions are much weaker than ARF activator-Aux/IAA and ARF activator-ARF activator interacttions, and it remains unclear whether Aux/IAAs interact with ARF repressors, or whether ARF repressors interact with ARF activators, to regulate target gene expression in plants [3,10,12,14].
In the osarf24-1 mutant, a Tos17 insertion in OsARF24 altered the amino acid sequence in the C-terminal dimerization domain of the OsARF24 protein, and the C-terminal half of domain IV was truncated. This mutation may reduce the formation of repressor ARF dimers; however, this does not explain why the osarf24-1 mutant showed reduced sensitivity to auxin because both monomers and dimers of ARF repressors can target and repress the expression of auxin-response genes [3]. Another hypothesis is that the C-terminal dimerization domain, especially the C-terminal half of domain IV, affects the stability or the repressor activity of OsARF24. The Arabidopsis ARF1 repressor is targeted for proteasomal degradation via a different set of machinery than that used for Aux/IAA degradation [25]. In addition, we cannot exclude the possibility that the weak interactions found between ARF repressors and Aux/IAA proteins in vitro are enough to function in vivo, but that the truncated OsARF24 repressor protein in the osarf24-1 mutant is unable to interact with rice Aux/IAA proteins and thus constitutively represses auxin-response gene expression. We envisage that detailed analyses of the osarf24-1 mutant and OsARF24 protein will help to reveal the function of the C-terminal dimerization domain in ARF repressors.
4. Conclusion
The osarf24-1 mutant contains a truncation of domain IV in the C-terminal dimerization domain of OsARF24 protein. This mutant showed auxin-deficient phenotypes and reduced sensitivity to auxin. However, wild-type OsARF24 protein contains an SPL-rich repression domain and acts as a repressor ARF. These results imply that the C-terminal dimerization domain, especially the C-terminal half of domain IV, is essential for the proper regulation of OsARF24 function as a transcriptional repressor in rice.
5. Acknowledgements
We thank Asako Tokida-Segawa for technical assistance, and the GenBank project of the National Institute of Agrobiological Science in Japan for providing osarf24-1 (NE4013) mutant seeds and OsARF24 cDNA (AK- 067061). T. S. was supported by Grants-in-Aid for Young Scientists (Nos. 19688001 and 24780005) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- W. M. Gray, S. Kepinski, D. Rouse, O. Leyser and M. Estelle, “Auxin Regulates SCFTIR1-Dependent Degradation of AUX/IAA Proteins,” Nature, Vol. 414, No. 6861, 2001, pp. 271-276. doi:10.1038/35104500
- N. Zenser, A. Ellsmore, C. Leasure and J. Callis, “Auxin Modulates the Degradation Rate of Aux/IAA Proteins,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 98, No. 20, 2001, pp. 11795-11800. doi:10.1073/pnas.211312798
- T. J. Guilfoyle and G. Hagen, “Auxin Response Factors,” Current Opinion in Plant Biology, Vol. 10, No. 5, 2007, pp. 453-460. doi:10.1016/j.pbi.2007.08.014
- T. Ulmasov, G. Hagen and T. J. Guilfoyle, “ARF1, a Transcription Factor That Binds to Auxin Response Elements,” Science, Vol. 276, No. 5320, 1997, pp. 1865- 1868. doi:10.1126/science.276.5320.1865
- G. Parry and M. Estelle, “Auxin Receptors: A New Role for F-Box Proteins,” Current Opinion in Cell Biology, Vol. 18, No. 2, 2006, pp. 152-156. doi:10.1016/j.ceb.2006.02.001
- T. J. Guilfoyle, T. Ulmasov and G. Hagen, “The ARF Family of Transcription Factors and Their Role in Plant Hormone-Responsive Transcription,” Cellular and Molecular Life Sciences, Vol. 54, No. 7, 1998, pp. 619-627. doi:10.1007/s000180050190
- T. J. Guilfoyle and G. Hagen, “Auxin Response Factors,” Journal of Plant Growth Regulation, Vol. 20, No. 3, 2001, pp. 281-291. doi:10.1007/s003440010026
- T. Ulmasov, Z.-B. Liu, G. Hagen and T. J. Guilfoyle, “Composite Structure of Auxin Response Elements,” Plant Cell, Vol. 7, No. 10, 1995, pp. 1611-1623.
- T. Ulmasov, J. Murfett, G. Hagen and T. J. Guilfoyle, “Aux/IAA Proteins Repress Expression of Reporter Genes Containing Natural and Highly Active Synthetic Auxin Response Elements,” Plant Cell, Vol. 9, No. 11, 1997, pp. 1963-1971.
- T. Ulmasov, G. Hagen and T. J. Guilfoyle, “Dimerization and DNA Binding of Auxin Response Factors,” Plant Journal, Vol. 19, No. 3, 1999, pp. 309-319. doi:10.1046/j.1365-313X.1999.00538.x
- T. Ulmasov, G. Hagen and T. J. Guilfoyle, “Activation and Repression of Transcription by Auxin-Response Factors,” Proceedings of the National Academy of Sciences of the United States of America, Vo. 96, No. 10, 1999, pp. 5844-5849. doi:10.1073/pnas.96.10.5844
- S. B. Tiwari, G. Hagen and T. Guilfoyle, “The Roles of Auxin Response Factor Domains in Auxin-Responsive Transcription,” Plant Cell, Vo. 15, No. 2, 2003, pp. 533- 543. doi:10.1105/tpc.008417
- D. Wang, K. Pei, Y. Fu, Z. Sun, S. Li, H. Liu, K. Tang, B. Han and Y. Tao, “Genome-Wide Analysis of the Auxin Response Factors (ARF) Gene Family in Rice (Oryza sativa),” Gene, Vol. 394, No. 1-2, 2007, pp. 13-24. doi:10.1016/j.gene.2007.01.006
- C. Shen, S. Wang, Y. Bai, Y. Wu, S. Zhang, M. Chen, T. J. Guilfoyle, P. Wu and Y. Qi, “Functional Analysis of the structural Domain of ARF Proteins in Rice (Oryza sativa L.),” Journal of Experimental Botany, Vol. 61, No. 14, 2010, pp. 3971-3981. doi:10.1093/jxb/erq208
- S. Ohta, S. Mita, T. Hattori and K. Nakamura, “Construction and Expression in Tobacco of a β-Glucuronidase (GUS) Reporter Gene Containing an Intron within the Coding Sequence,” Plant and Cell Physiology, Vol. 31, No. 6, 1990, pp. 805-813.
- Y. Hiei, S. Ohta, T. Komari and T. Kumashiro, “Efficient Transformation of Rice (Oryza sativa L.) Mediated by Agrobacterium and Sequence Analysis of Boundaries of the T-DNA,” Plant Journal, Vol. 6, No. 2, 1994, pp. 271- 282. doi:10.1046/j.1365-313X.1994.6020271.x
- K. Fujino, Y. Matsuda, K. Ozawa, T. Nishimura, T. Koshiba, M. W. Fraaije and H. Sekiguchi, “Narrow Leaf 7 Controls Leaf Shape Mediated by Auxin in Rice,” Molecular Genetics and Genomics, Vol. 279, No. 5, 2008, pp. 499-507. doi:10.1007/s00438-008-0328-3
- J. Qi, Q. Qian, Q. Bu, S. Li, Q. Chen, J. Sun, W. Liang, Y. Zhou, C. Chu, X. Li, F. Ren, K. Palme, B. Zhao, J. Chen, M. Chen and C. Li, “Mutation of the Rice Narrow Leaf 1 Gene, Which Encodes a Novel Protein, Affects Vein Patterning and Polar Auxin Transport,” Plant Physiology, Vol. 147, No. 4, 2008, pp. 1947-1959. doi:10.1104/pp.108.118778
- A. Nakamura, I. Umemura, K. Gomi, Y. Hasegawa, H. Kitano, T. Sazuka and M. Matsuoka, “Production and Characterization of Auxin-Insensitive Rice by Overexpression of a Mutagenized Rice IAA Protein,” Plant Journal, Vol. 46, No. 2, 2006, pp. 297-306. doi:10.1111/j.1365-313X.2006.02693.x
- M. Jain, N. Kaur, A. K. Tyagi and J. P. Khurana, “The Auxin-Responsive GH3 Gene Family in rice (Oryza sativa),” Functional & Integrative Genomics, Vol. 6, No. 1, 2006, pp. 36-46. doi:10.1007/s10142-005-0142-5
- Y. Song, L. Wang and L. Xiong, “Comprehensive Expression Profiling Analysis of OsIAA Gene Family in Developmental Processes and in Response to Phytohormone and Stress Treatments,” Planta, Vol. 229, No. 3, 2009, pp. 577-591. doi:10.1007/s00425-008-0853-7
- F. Waller, M. Furuya and P. Nick, “OsARF1, an Auxin Response Factor From Rice, Is Auxin-Regulated and Classifies as a Primary Auxin Responsive Gene,” Plant Molecular Biology, Vol. 50, No. 3, 2002, pp. 415-425. doi:10.1023/A:1019818110761
- H. Li, P. Johnson, A. Stepanova, J. M. Alonso and J. R. Ecker, “Convergence of Signaling Pathways in the Control of Differential Cell Growth in Arabidopsis,” Developmental Cell, Vol. 7, No. 2, 2004, pp. 193-204. doi:10.1016/j.devcel.2004.07.002
- G. Hagen and T. Guilfoyle, “Auxin-Responsive Gene Expression: Genes, Promoters and Regulatory Factors,” Plant Molecular Biology, Vol. 49, No. 3-4, 2002, pp. 373- 385. doi:10.1023/A:1015207114117
- J. Salmon, J. Ramos and J. Callis, “Degradation of the Auxin Response Factor ARF1,” Plant Journal, Vol. 54, No. 1, 2008, pp. 118-128. doi:10.1111/j.1365-313X.2007.03396.x
NOTES
*Corresponding author.

