Advances in Bioscience and Biotechnology
Vol.3 No.4(2012), Article ID:21844,8 pages DOI:10.4236/abb.2012.34053
Malate dehydrogenase in plants: Its genetics, structure, localization and use as a marker
![]()
The Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia
Email: yurs@bionet.nsc.ru
Received 5 May 2012; revised 20 June 2012; accepted 4 July 2012
Keywords: loci; alleles; isozymes; markers; compartments
ABSTRACT
This article is an overview of literature data on the structure, properties, functions and genetic control of the enzyme malate dehydrogenase (MDH) in plants. In most of the plant entities studied, this enzyme is highly polymorphic, which means that malate dehydrogenase has multiple molecular forms. It has been found that MDH polymorphism in each species is genetically determined by several loci with multiple alleles. A readily identifiable phenotypic manifestation and a high activity of malate dehydrogenase in diverse organs and tissues make this enzyme a reliable and convenient genetic marker, which can effectively be used in special, ecological and population genetics.
1. INTRODUCTION
The discovery of isozymes [1,2] has provided researchers with simple and reliable marker traits that can be used for studying the widest class of biological phenomena, and so all-new possibilities for marking genes and genetic systems have been opened up. An advantage of isozyme markers is that they themselves perform functions in metabolism, and so these functions can now be identified using not only genes or genomic loci, but also using isozyme markers. Isozymes can be used both as markers for phylogenetic analysis and identification of introgressions and as markers for detection of species differentiation, cultivar identification and analysis of populations. The most prominent advantages of isozymes as genetic markers are their codominant inheritance, readily identifiable phenotypic manifestations and that isozymes themselves are easily identifiable. One of the most extensively studied isozyme systems are dehydrogenases, including malate dehydrogenase.
NAD-dependent malate dehydrogenase (MDH, L-malate: NAD oxidoreductase; EC 1.1.1.37) is an enzyme very commonly occurring in animals, plants and microorganisms. MDH catalyzes the oxidation (dehydration) of L-malate to oxaloacetate in the presence of NAD+ as a cofactor:
СООН-СН2-СНОН-СООН + NAD+ =
L-malate
СООН-СН2-СО-СООН + NADH + Н+
oxaloacetate [3].
MDH is a well-studied enzyme. In the cell, malate dehydrogenase exists in various molecular forms. Two MDH forms have been found in most of the plant objects studied: mitochondrial MDH (m-MDH) and cytoplasmic MDH (s-MDH) [4-8]. Mitochondrial MDH functions as part of the Krebs cycle, while soluble (cytoplasmic) MDH can be involved in acid metabolism in plant tissues, in autotrophic carbon dioxide fixation in higher plants and in more other metabolic pathways [9-11]. In plant tissues, MDH has additionally been found in glyoxysomes [6,7, 12,13], peroxisomes [14,15] and microsomes [16-18]. The microbody form of MDH is involved in photorespiration and the glyoxylate cycle [19].
2. THE STRUCTURE AND PROPERTIES
Studies of the structure of the malate dehydrogenase molecule in maize, celery, rice, avocado and other plants have demonstrated that each MDH molecule consists of two subunits, that is, based on its quaternary structure, MDH is a dimer [6,20-23].
Malate dehydrogenase is best studied in maize [5,11, 24-31]. In that plant, MDH is located in chloroplasts [11], microbodies, mitochondria and the cytoplasm [10]. Much bigger differences in primary structure are found between cytoplasmic and mitochondrial form in the same plant than between mitochondrial forms of MDH in different genera [10,31,32]. This conclusion was reached by comparing the reactions of different MDH isozymes to antisera each raised against the product of each homozygous Mdh locus. The antiserum raised against purified mitochondrial MDH was cross-reactive with microbody MDH; however, it was not reactive with soluble MDH [10, 31,32].
Studies of the biochemical properties of mitochondrial and cytoplasmic MDH in maize have demonstrated that these two forms have different thermal stability, different optimum рН values, different kinetic properties and other differences [7,8]. Isoelectric focusing has identified differences between cytoplasmic and mitochondrial MDH isoforms in grape. The cytoplasmic forms have рН values ranging from 4.0 to 5.5, while the mitochondrial forms are less acidic (рН 5.5 - 7.5) and have a molecular weight of 80000 Da [33]. Differences have also been found between the respective molecular properties of glyoxysomal and mitochondrial MDH in watermelon cotyledons [13,34]. In that plant, the molecular weight of glyoxysomal MDH is 67,000 Da, while that of mitochondrial MDH is 74,000 Da. At high MDH concentrations, it has been observed that the glyoxysomal enzyme aggregates, while the mitochondrial enzyme does not. These enzymes differ in isoelectric points, thermal stability, optimum рН values and other characteristics [13,34].
As has been demonstrated by exposing MDH in maize to various inhibitors, mitochondrial MDH isozymes are synthesized on cytoplasmic ribosomes [24]. It has been reported that the mitochondrial and glyoxysomal forms of MDH are synthesized as higher molecular weight of predecessors, which, as they are transported to organelles, lose parts of their polypeptide chains: a 8000-Da fragment is cleaved off each glyoxysomal predecessor and a 3300- Da fragment is cleaved off each mitochondrial predecessor, to lead to the observed functional subunits. Cytoplasmic MDH has not been observed to be synthesized as a predecessor [35].
Malate dehydrogenase has also been studied in other plant entities: amaranth [36], avocado [22,37], barley [38- 40], beans [41,42], cedar [43], celery [21,23], cereus [44], citrus [45,46], Corydalis soli da [47], Cynosurus cristatus [48], eucalyptus [49], oats [50], peach [51], pecan [52], pepper [53], petunia [54], pine [55-58], prickly pear [59], rice [20], soybean [60], wheat [38,61,62].
Not only do the multiple molecular forms of MDH in plants have differences as indicated, but they also have things in common. In most of the entities studied, the molecular weight of MDH ranges from 60,000 Da to 70,000 Da. In starch gel electrophoresis, the malate dehydrogenase forms that are contained in organelles move from anode to cathode in an ordered manner: the closest to the cathode end is microbody MDH, the second closest is mitochondrial MDH and the farthest away is cytoplasmic MDH [63]. Another point in common has been revealed by immunological studies: in most eukaryotes, mitochondrial MDH has similar structural properties, which suggests a high degree of evolutionary preservation [31]. Based on the above considerations and considering the fact that different and highly specific forms of this enzyme are present in almost each main subcellular component, it may be concluded that MDH is a unique model enzyme, which can be used in studying crosstalk between organelles [11].
3. THE GENETIC CONTROL OF MALATE DEHYDROGENASE
The polymorphism and specificity of multiple molecular forms of NAD-MDH are typical of both different tissues of a single plant and different compartments of a single cell. Additionally, the polymorphism of molecular forms has been revealed for each compartment. Seven mitochondrial and six cytoplasmic MDH isoforms have been identified in maize (all resolved electrophoretically) [27, 28,30,32,64] three soluble and three mitochondrial MDH isoforms, in soybean [60]; four soluble and five mitochondrial MDH isoforms, in Arabidopsis [65]; two soluble and two mitochondrial MDH isoforms, in pine [57]; four mitochondrial and three microsomal MDH isoforms, in sugar beet [16-18] and so on. As is known, the molecular heterogeneity of enzyme forms either is determined genetically or is the result of epigenetic changes.
The first evidence of the genetic origin of MDH polymorphism has been found in maize by Longo and Scandalios [6]. Reciprocal hybrids were obtained by crosses between two inbred maize lines, 59 and Oh 51A, which carry different mitochondrial forms of MDH. The isozyme spectra of these hybrids were identical and each contained both parental and hybrid forms of MDH. Segregation of F2 plants into two parental and one hybrid phenotype was Mendelian.
To date, malate dehydrogenase isozymes have been better studied in maize than in any other plant. There are two models for describing the genetic control of MDH in maize. According to Goodman’s and the associates’ model, mitochondrial MDH is encoded by three structural genes (Mdh1, Mdh2 and Mdh3), while cytoplasmic MDH is encoded by two genes (Mdh4 and Mdh5). When mitochondrial forms of MDH interact, they form intergenic and interallelic heterodimers. The isoforms that are located in different subcellular organelles do not form hybrid dimers [31,32]. The structural genes for MDH are in a variety of linkage groups: Mdh1 is localized to chromosome 8; Mdh2, to the distal end of the long arm of chromosome 6; and Mdh3, to the distal end of the long arm of chromosome 3 [31,64]. McMillin and Scandalios suggest a different interpretation [27,28]. These models use different nomenclatures for and different substantiations of separate MDH variants (Table 1). One of the reasons the isozyme spectra are interpreted differently is the use of different buffer systems in electrophoresis. The use of Tris-citrate buffer (рН 7.0) (which is McMillin’s and Scandalios’ choice) results in a relatively weak separation of isozymes, which, in Goodman’s and Stuber’s opinion [30], may lead to misinterpretation.
Table 1 provides a listing of plant objects, for which the genetic control of malate dehydrogenase has been studied. The inheritance of MDH in maize, eucalypt, sugar beet was determined by a classical method of genetic analysis including hybridization of contrasting forms and analysis of segregation in F2 and backcross plants. In wheat, the inheritance of the isozymes was determined by comparing their spectra in normal and aneuploid forms. For some plants (pine, cedar, peach, to mention a few), analyses were performed for the correspondence between the frequencies of phenotypic classes in panmictic populations and the theoretical frequencies calculated by the HardyWeinberg equation and isozyme spectra in somatic tissue and pollen were compared. It was found that, in addition to the structural loci that control MDH synthesis, maize carries the Mmm gene, pine carries the Mdhm gene, and, cedar carries the Mdh1m gene, each modifying both electrophoretic mobility and enzymatic activity [30,43, 55]. In addition to maize, the chromosomal localization of Mdh loci has been determined in barley, wheat and petunia [30,32,38-39,54,62,66].


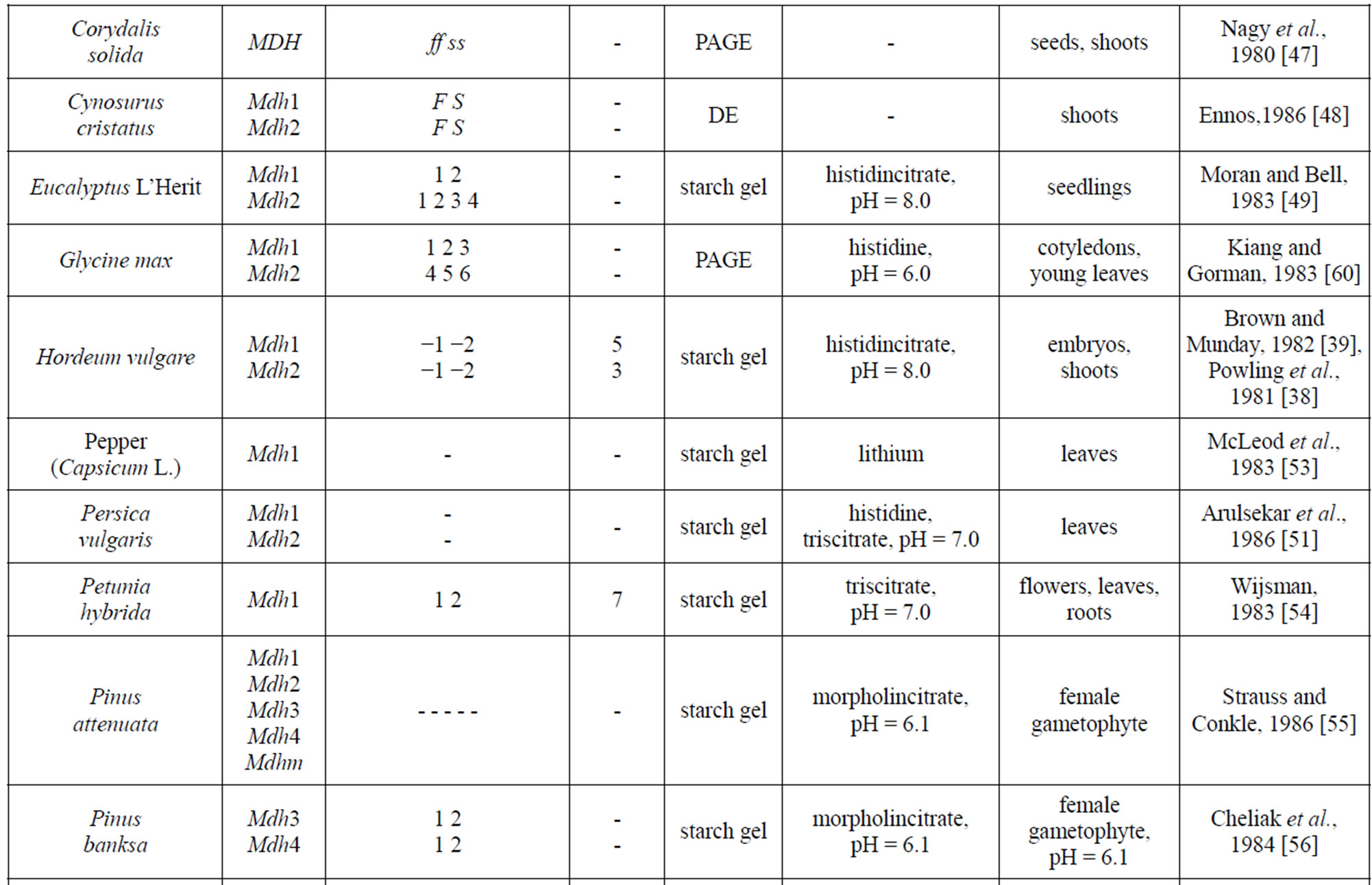
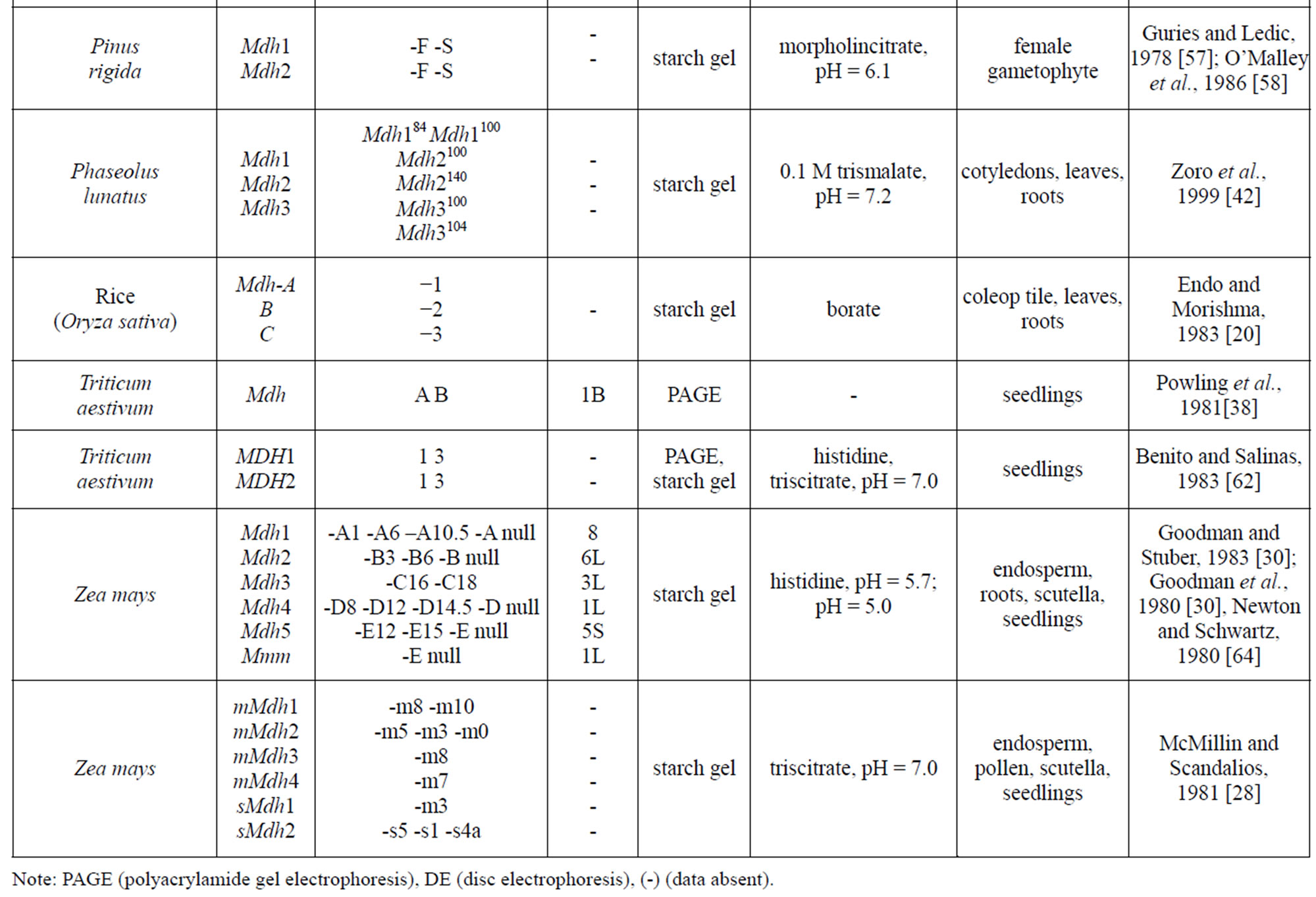
Table 1. Genetic control of NAD-dependent malate dehydrogenase in plants.
4. THE EXPRESSION OF LOCI CONTROLLING MDH IN PLANTS
Changes in isozyme spectra that occur as tissue undergoes growth and differentiation have been described in many plants and for different enzyme systems. The same is true of malate dehydrogenase spectra. In maize, cytoplasmic and mitochondrial MDH forms are active in roots, seedlings, endosperm, corymbs, hypocotyls, coleoptiles, leaves and pollen [6,31,33,64], while microbody MDH is active in endosperm and scutellum [6,10,24,25,31]. An increase in the number of MDH isozymes during caryopsis maturation has been demonstrated [11]. A de novo synthesis of MDH isozymes has been detected in developing maize seedlings [67]. The disappearance and emergence of separate MDH forms have been observed in developing wheat plants [66] and Arabidopsis thaliana [65].
The organ specificity of NAD-MDH in sugar beet has been studied in detail. Isozyme spectra have been observed in dry seeds, seedlings, leaves, stems, roots, bracts, buds, ovaries and pollen [68]. The specificity is characterized both by the number of isozymes and their relative activity in the organs studied (seeds, seedlings, leaves, bracts, buds, ovaries and ovules) and by a total lack of isozymes on the electropherograms (stems, roots). However, it should be noted that the lack of some MDH isozymes on electropherograms does not necessarily imply that the corresponding locus is inactive in those organs: it is possible that the isozymes are present at below the minimum detectable amount. The genetic determination of MDH isozymes in sugar beet allows tissue specificity to be interpreted as the result of the differential activity of the genes that control malate dehydrogenase at different stages of ontogenesis. It is possible that the genetic information coming from two different alleles is not implemented simultaneously. For example, in the leaves of sugar beet line P-83-3 plants, which are all heterozygous for the Mor1 locus with the Mor1-N and Mor1-P alleles, three MDH isozymes (nn, np and pp) should be revealed, which is as expected of the spectrum of a dimeric enzyme in heterozygotes. However, in young leaves, the Моr1-N allele is not activated, while later on, in normally developed leaves, all the three isozymes will appear. Asynchrony in the activation of parental alleles in heterozygotes during ontogenesis has also been reported [69].
Thus, malate dehydrogenase represents an example of how useful isozymes can be for studying regulatory systems in eukaryotes. Genetic analysis allows both structural and regulatory genes to be comparatively easily localized. Collectively, all this provides an opportunity to obtain reliable information on the regulation of gene activity during ontogenesis and to gain a better insight into cell differentiation pathways in developing organisms [69-71].
5. THE USE OF MALATE DEHYDROGENASE ISOZYMES IN PLANT POPULATION GENETIC STUDIES
Isozyme spectra can serve the basis for cultivar identification and registration purposes, for documenting the genetic resources of cultivated plants and their wild relatives with a view to their consideration, preservation and use in breeding. The results of such work depend upon how well the genetic control of the enzymes involved is studied.
Thus, due to fundamental efforts by Goodman and the colleagues [33], 25,000 maize plants in 125 races and synthetic cultivars from South and North America have been analyzed; inbred lines and their F1 and F2 hybrids, synthetic and various commercial and experimental hybrids have been studied; and a catalog of genotypes containing various combinations of allelic variants of malate dehydrogenase, has been developed. Considerable differentiation has been found in the frequency of different MDH genotypes among the objects studied. For example, the Mdh1-A6, Mdh2-B6 and Mdh3-C16 genotypes have been found in 21 lines; Mdh1-A6, Mdh2-B3, Mdh3-C16 have been found in 12 lines; while there are many MDH genotypes each of which is confined to a single line or hybrid. It is possible that this differentiation is because some lines have common origin, while the others are of unique origin. However, it is not excluded that the commonest genotypes have advantages over the others, and so the former have had selective preferences. This malate dehydrogenase example provides further support to the view that the use of loci with multiple alleles is extremely beneficial for breeding, ecological genetics and population genetics.
NAD-dependent malate dehydrogenase isozymes have also been used for identification of inbred sugar beet lines in the collection maintained in the Laboratory of Plant Population Genetics of the Institute of Cytology and Genetics (Novosibirsk, Russia). A total of 394 lines have been analyzed, and to each line the corresponding genotype has been assigned based on the isozyme spectrum of malate dehydrogenase. The frequencies of nine genotypic classes, normally observed when the character being studied is controlled by two loci with two alleles at each locus, have been determined [16,72]. The respective frequencies of the genotype classes for these two alleles were strongly different. The ratio of genotypic classes was 285NN:93NP:16PP at the Mor1 locus and 238SS:136FS: 20FF at the Mor2 locus. The structure of wild and cultivated amaranth populations has been studied in terms of the frequencies of the alleles at the loci controlling isozymes, including those at the Mdh1 locus [73]. Alongside other isozymes, malate dehydrogenase was used for identification of 57 apple tree cultivars [74]. One of the polymorphic MDH loci has been used in breeding programs with 290 peach cultivars surveyed [51].The genetic structure of a collection of 570 cedar trees from three California regions has been described in terms of the frequencies of multiple alleles at two loci encoding malate dehydrogenase: Mdh1 with seven alleles and Mdh2 with three alleles [43].
All the above findings indicate that a high level of malate dehydrogenase activity in diverse organs and tissues, and the presence of loci with multiple alleles, codominant inheritance and a clear phenotypic manifestation of isozyme spectra. Therefore, this enzyme is a reliable and useful genetic marker in population and genetic studies.
6. CONCLUSION
The large number of species with MDH studied and used in experiments aimed at describing and characterizing various processes provides evidence that this enzyme represents a highly efficient genetic and physiological marker. In molecular biology, it is important that enzymes with their isozyme systems and protein cofactors represent extremely important biological and genetic markers with many promising uses for theory and practice.
REFERENCES
- Hunter, R.L. and Markert, C.L. (1957) Histochemical demonstration of enzymes separated by zone electrophoresis in starch gel. Science, 125, 1294-1295. doi:10.1126/science.125.3261.1294-a
- Markert, C.L. and Moller, F. (1959) Multiple forms of enzymes: Tissue, ontogenetic and species specific patterns. Proceedings of the National Academy of Sciences, 45, 753-763. doi:10.1073/pnas.45.5.753
- Dixson, M. and Webb, E.C. (1982) Enzymes. Mir, Moscow.
- Shannon, L.M. (1968) Plant isoenzymes. Annual Review of Plant Physiology, 19, 187-210. doi:10.1146/annurev.pp.19.060168.001155
- Scandalios. J.G. (1969) Genetic control of multiple forms of enzymes in plants: A review. Biochemical Genetics, 3, 37-79. doi:10.1007/BF00485973
- Longo, C.P. and Scandalios J.G. (1969) Nuclear gene control of mitochondrial malic dehydrogenase in maize. Proceedings of the National Academy of Sciences, 62, 104-111. doi:10.1073/pnas.62.1.104
- Yang, N.S. and Scandalios, J.G. (1974) Purification and biochemical properties of genetically defined malate dehydrogenase in maize. Archives of Biochemistry and Biophysics, 161, 335-353. doi:10.1016/0003-9861(74)90314-2
- Yang, N.S. (1975) Biochemical properties and development expression of genetically determined malate dehydrogenase isozymes in maize. In: Isozymes: III Developmental Biology, Academic Press, New York, 191-211.
- Lehninger, A.L. (1974) Biochemistry. Mir, Moscow.
- Ting, I.P., Fuhr I., Curry, R. and Zschoche, W.G. (1975) Malate dehydrogenase isozymes in plant: Preparation, properties and biological significance. In: Markert, C.L., Ed., Isozymes, Academic Press, New York, 369-384.
- Scandalios, J.G. (1974) Isoenzymes in development and differentiation. Annual Review of Plant Physiology, 25. 225-258. doi:10.1146/annurev.pp.25.060174.001301
- Breidenbach, R.W. and Beevers, H. (1967) Association of the glyoxylate cycle enzymes in a novel subcellular particle from castor bean endosperm. Biochemical and Biophysical Research Communications, 27, 462-469. doi:10.1016/S0006-291X(67)80007-X
- Walk, R.A. and Hock, B. (1977) Glyoxisomal and mitochondrial malate dehydrogenase of watermelon (Citrullus vulgaris) cotyledons. Planta, 136, 221-228. doi:10.1007/BF00385988
- Yamazaki, R.K. and Tolbert, N.E. (1969) Malate dehydrogenase in leaf peroxisomes. Biochimica et Biophysica Acta—Enzymology, 178, 11-20. doi:10.1016/0005-2744(69)90127-2
- Sadunishvili, T.A. and Nutsubidse, N.N. (1980) Intra cellular location of malate and glutamate dehydrogenases in lemon citrus-limon leaves. Soobsheniya Akademii Nauk Gruzinskoy SSR, 100, 693-696.
- Tarasova, R.S. (1988) Genetics and phenogenetics of malate dehydrogenase in sugar beet. Ph.D. Thesis, Institute of Cytology and Genetics, Novosibirsk.
- Yudina, R.S. and Levites, E.V. (2007) Malate dehydrogenase isozymes as markers of organelles physioligical state sugar beet (Beta vulgaris L.). Sugar Tech, 9, 67-71. doi:10.1007/BF02956916
- Yudina, R.S. and Levites, E.V. (2008) Subcellular localization of isozymes of NAD-dependent malate dehydrogenase in sugar beet Beta vulgaris L. Russian Journal of Genetics, 44, 1419-1423. doi:10.1134/S1022795408120053
- Gottlieb, L.D. (1982) Conservation and duplication of isozymes in plant. Science, 216, 373-380. doi:10.1126/science.216.4544.373
- Endo, T. and Morishma, H. (1983) Rise. In: Isozymes in Plant Genetics and Breeding. Elsevier, Amsterdam, 129- 146.
- Orton, T.J. (1983) Celery and celeriac (Apium graveolens L.). In: Isozymes in Plant Genetics and Breeding. Elsevier, Amsterdam, 351-356.
- Torres, A.M. (1983) Fruit trees. In: Isozymes in Plant Genetics and Breeding. Elsevier, Amsterdam, 401-421.
- Arus, P. and Orton, T.J. (1984) Inheritance patterns and linkage relationships of eight genes of celery (Apium graveolens). Journal of Heredity, 75, 11-14.
- Yang, N.S. and Scandalios, J.G. (1975) Cytoplasmic synthesis of soluble and mitochondrial malate dehydrogenase isozymes in maize. Archives of Biochemistry and Biophysics, 171, 575-585. doi:10.1016/0003-9861(75)90067-3
- Yang, N.S. and Scandalios, J.G. (1975b) De novo synthesis and developmental control of gene-controlled malate dehydrogenase isozymes in maize scutella. Biochimica et Biophysica Acta—Enzymology, 384, 293-306. doi:10.1016/0005-2744(75)90031-5
- Yang, N.S., Sorenson, J.G. and Scandalios, J.G. (1977) Genetic control of mitochondrial malate dehydrogenase: Evidence for duplicated chromosome segments. Proceedings of the National Academy of Sciences, 74, 310- 314. doi:10.1073/pnas.74.1.310
- McMillin, D.E. and Scandalios, J.G. (1980) Duplicated cytosolic malate dehydrogenase genes in Zea mays. Proceedings of the National Academy of Sciences, 77, 4866- 4870. doi:10.1073/pnas.77.8.4866
- McMillin, D.E. and Scandalios, J.G. (1981) Genetic analysis of two groups of duplicated genes coding for mitochondrial malate dehydrogenase in Zea mays: Possible origin of mMdh genes by chromosomal segment duplication. Molecular and General Genetics, 182, 211-221. doi:10.1007/BF00269660
- McMillin, D.E. and Scandalios, J.G. (1982) Genetic, immunological and gene dosage studies of mitochondrial and cytosolic MDH variante in maize. Journal of Heredity, 73, 177-182.
- Goodman, M.M. and Stuber, C.W. (1983) Maize. In: Isozymes in Plant Genetics and Breeding. Elsevier, Amsterdam, 1-33.
- Newton, K.J. (1983) Genetics of mitochondrial isozymes. In: Isozymes in Plant Genetics and Breeding. Elsevier, Amsterdam, 157-170.
- Goodman, M.M., Stuber, C.W., Lee, C.N. and Johnson, F.M. (1980) Genetics control of malate dehydrogenase isozymes in maize. Genetics, 94, 153-168.
- Taureilles-Saurel, C., Romieu, C.G., Robin, J.-P. and Flanzy, C. (1995) Grape (Vitis vinifera L.) malate dehydrogenase. I. intercellular compartmentation of the isoforms. American Journal of Enology Viticulture, 46, 22- 28.
- Walk, R.A., Michaeli, S. and Hock, B. (1977) Glyoxisomal and mitochond rial malate dehyddrogenase of watermelon (Citrullus vulgaris) cotyledons. Planta, 136, 211-220. doi:10.1007/BF00385987
- Gietl, C. and Hock, B. (1982) Organelle-bound malate dehydrogenase isoenzymes are synthe sized as higher molecular weight precursors. Plant Physiology, 70, 483- 487. doi:10.1104/pp.70.2.483
- Yudina, R.S., Zheleznova, N.B., Zakharova, O.V., Zheleznov, A.V. and Shumny, V.K. (2005) Isozyme analysis in genetic collection of amaranths (Amaranthus L.). Russian Journal of Genetics, 41, 1395-1400. doi:10.1007/s11177-006-0012-9
- Torres, A.M. (1984) Isozymes from avocado cotyledons. Journal of Heredity, 75, 300-302.
- Powling, A., Islam, A.K.M.R. and Sheferd, K.W. (1981) Isozymes in wheat-barley hybrid derivate lines. Biochemical Genetics, 19, 237-254. doi:10.1007/BF00504271
- Brown, A.H.D. and Munday, J. (1982) Population genetics structure and optimal sampling of land races of barley. Genetica, 40, 315-324.
- Linde-Laursen, I., Nelson, G. and Johansen, H.B. (1987) Distribution of isoenzyme markers at loci in a pedigree of European spring barley. Hereditas, 106, 241-251. doi:10.1111/j.1601-5223.1987.tb00258.x
- Habig, W. and Racusen, D. (1968) A malate dehydrogenase of high molecular weight, from bean leaves. Canadian Journal of Botany, 46, 719-720. doi:10.1139/b68-095
- Zoro, I.Bi., Maquet, A., Wathelet, B. and Baudoin, J.-P. (1999) Genetic control of isozymes in the primary gene pool Phaseolus lunatus L. Biotechnologie Agronomie Societe et Environnement, 3, 10-27.
- Harry, D.E. (1983) Identification of a locus modifjing the electrophoretic mobility of malate dehydrogenase isozymes in incense-cedar (Calocedrus decurrens), and its implications for population studies. Biochemical Genetics, 21, 417-434. doi:10.1007/BF00484435
- Machado, M.F.P.S., Prioli, A.J. and Mangolin, C.A. (1993) Malate dehydrogenase (MDH; EC1.1.1.37) isozymes in tissue and callus cultures of Cereus peruvianus (Cactaceae). Biochemical Genetics, 31, 167-172. doi:10.1007/BF02399923
- Torres, A.M., Soost, K.K. and Mau-Lastovisks, T. (1978) Leaf isozymes as genetic markers in citrus. American Journal of Botany, 68, 869-881. doi:10.2307/2442183
- Torres, A.M., Soost, K.K. and Diedenhoffen, U. (1982) Genetic and distinguishing nucellularfrom zygotic seedlings. Journal of Heredity, 73, 335-339
- Nagi, A.N., Siddiqui, M.O., Kocsis, Z.G. and Vida, G. (1980) In vitro dissociation-recombination of malate dehydroganase subunits in Corydalis solida. Theoretical and Applied Genetics, 58, 75-78.
- Ennos, R.A. (1986) Allozyme variation, linkage and duplication in the perrenial grass; Cynosurus cristatus. Journal of Heredity, 77, 61-62.
- Moran, G.F. and Bell, J.G. (1983) Eucalipthus. In: Isozymes in Plant Genetics and Breeding. Elsevier, Amsterdam, 423-442.
- Prise, S. and Kahler, A.L. (1983) Oats. In: Isozymes in Plant Genetics and Breeding. Elsevier, Amsterdam, 147- 167.
- Arulsekar, S., Parfitt, D.E., Beres, W. and Hansche, P.E. (1986) Genetics of malate dehydrogenase isozymes in the peach. Journal of Heredity, 77, 49-51.
- Marquard, R.D. and Skorpenske, R.G. (1989) Expression of heritable biochemical markers from various pecan tissues. Euphytica, 42, 65-70. doi:10.1007/BF00042616
- McLeod, M.J., Guttman, S.I. and Esbaugh, W.N. (1983) Peppers. In: Isozymes in Plant Genetics and Breeding. Elsevier, Amsterdam, 189-201.
- Wijsman, N.J.W. (1983) Petunia. In: Isozymes in Plant Genetics and Breeding. Elsevier, Amsterdam, 229-252.
- Srtauss, S.N. and Conkle, M.T. (1986) Segregation, linkage and diversity of allozymes in knobcone (Pinus attenuata). Theoretical and Applied Genetics, 72, 483-493. doi:10.1007/BF00289530
- Cheliak, W.M., Morgan, K., Dancik, B.P., Strobeck, C. and Yeeh, F.C.N. (1984) Segregationof allozymes in megagametophytes of viable seed from a natural population of jack pine, Pinus banksiana Lamb. Theoretical and Applied Genetics, 69, 145-151.
- Guries, R.P. and Ledic, F.T. (1978) Inheritance of some polymorphic isozymes in pitch pine (Pinus rigida Mill). Heredity, 40, 27-32. doi:10.1038/hdy.1978.3
- O’Malley, D.M., Guries, R.P. and Nordheim, E.V. (1986) Linkage analysis for 18 enzyme loci in Pinus rigida Mill. Theoretical and Applied Genetics, 72, 530-535. doi:10.1007/BF00289536
- Sternberg, L., Ting. I.P. and Hanscom, Z. (1979) Polymorphism of microbody malate dehydrogenase in Opuntia basilaris. Plant Physiology, 59, 329-330. doi:10.1104/pp.59.2.329
- Kiang, Y.T. and Gorman, M.B. (1983) Soybean. In: Isozymes in Plant Genetics and Breeding. Elsevier, Amsterdam, , 295-298.
- Vodenicharova, M.I., (1980) Genetics of isoenzymes in wheat species Uspekhi Sovremennoi Genetiki, 9, 171-182.
- Benito, C. and Salinas, J. (1983) The chromosomal location of malate dehyd rogenase isozymes in hexaploid wheat (Triticum aestivum L.). Theoretical and Applied Genetics, 64, 255-258. doi:10.1007/BF00303774
- O’Sullivan, S.A. and Wedding, R.T. (1972) Malate dehydrogenase isoenzymes from cotton leaves. Plant Physiology, 49, 117-123. doi:10.1104/pp.49.2.117
- Newton, K.J. and Schwartz, D. (1980) Genetic basis of the major malate dehydrogenase isozymes in maize. Genetics, 95, 425-442.
- Cammaerts, D and Jacobs, M. (1975) Study of the intracellular location and the genetic control of malate dehydrogenase isozymes in Arabidopsis thaliana. Plant Science Letters, 4, 249-256. doi:10.1016/0304-4211(75)90283-7
- Honold, G.R., Masko, V. and Stahman, M.A. (1967) NADand NADP-dependent malate dehydrogenase in wheat. Naturwissenschaften, 54, 169. doi:10.1007/BF00590845
- Scandalios, J.G. and Jang, N.S. (1973) Abstract. Genetics, 74, 242.
- Tarasova, R.S. (1987) Genetically determined forms of NAD-dependent MDH and their use in biochemical pass portization if inbred sugar beet lines. Genetica, 23, 1630- 1636.
- Korochkin, L.I. (1977) Gene interaction in development. Nauka, Moscow.
- Khavkin, E.E. (1969) Formation of metabolic system in growing plant cells. Nauka, Novosibirsk.
- Sozinov, A.A. (1985) Protein polymorphism in genetic and selection. Nauka, Moscow.
- Tarasova, R.S., Levites, E.V. and Maletsky, S.I. (1988) Isozyme as markers for identification of sugar beet inbred lines in the process of their development. Biochemical Identification of Varieties Materials III International Symposium ISTA, Leningrad, 240-243.
- Yudina, R.S., Ibragimova, S.S. and Zheleznova, N.B. (2008) A study of the structure of amaranth (Amaranthus L.) populations using isozyme spectra. Vestnik VOGIS, 12, 385-390.
- Biruk, A. and Kazlovskya, Z. (2008) Prospects for using of isozymes markers in identification of apple cultivars. Sodiinkyste ir darzininkyste. Mokslo Darbai, 27, 359- 364.

