Advances in Microbiology
Vol.2 No.4(2012), Article ID:25851,8 pages DOI:10.4236/aim.2012.24063
Mass Spectrometric Identification of Propionibacterium Isolates Requires Database Enrichment
1Unité de Recherche sur les Maladies Infectieuses Tropicales et Emergentes (URMITE), Aix-Marseille Université, Marseille, France
2Fédération de Microbiologie Clinique, Hôpital de la Timone, Marseille, France
Email: *pierre-edouard.fournier@univmed.fr
Received July 30, 2012; revised August 22, 2012; accepted September 7, 2012
Keywords: Propionibacterium; Propionibacterium acnes; MALDI-TOF; Mass Spectrometry
ABSTRACT
Propionibacterium species are mostly environmental bacteria, some being commensal of mammals including humans, and sometimes pathogenic. These bacteria are poorly identified using routine laboratory methods. Recently, Matrix Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF-MS) has emerged as a rapid and efficient method to identify bacterial species. We evaluated the use of MALDI-TOF-MS for identification of all validated Propiobibacterium species. Only four of the 15 tested reference strains (26.7%) were correctly identified at the species level, and P. acnes, the most common human pathogenic species was not identified. When applying MALDI-TOF-MS to 48 P. acnes strains, only 18.7% were correctly identified, suggesting an intraspecific variability of proteic profiles among Propionibacterium strains. However, by enriching the Bruker database with spectra from five of these strains and re-testing the other 43 strains against this new database, 93.0% were correctly identified. Our study demonstrates that MALDI-TOF-MS may be used for the identification of Propionibacterium isolates but requires a database enrichment in spectra from additional isolates.
1. Introduction
Propionibacterium sp. are non spore-forming, Grampositive, anaerobic or microaerophilic bacteria, first described in 1909. To date, fourteen species and 2 subspecies are validated
(http://www.bacterio.cict.fr/p/propionibacterium.html). On the basis of their natural habitat, i.e., environment or human flora, species are divided into two main groups. Environmental species include Propionibacterium freundenreichii, P. acidipropionici, P. jensenii and P. thoeni that are associated to dairy products, and P. cyclohexanicum, P. microaerophilum and P. australiense that were isolated from spoiled orange juice, olive mill wastewater and bovine lesions, respectively. The remaining seven species are part of the human commensal flora. As such, Propionibacterium sp. were described as bacteria of no or low pathogenicity for many years. Currently, they are often considered as contaminant bacteria of blood and body fluid cultures but are able to cause serious human infections. The most common pathogenic species for humans is P. acnes, which belongs to the normal flora of the skin, oral cavity, large intestine, conjunctiva, and external ear [1]. It is primaly recognized for its role in acne vulgaris [2]. However, P. acnes was also involved in other human infections, mainly as an opportunistic pathogen in immunocompromised patients and in post-operative infections, notably in presence of prosthetic material [3]. In particular, P. acnes was found to be the causative agent of spondylodiscitis, central nervous system infections, endocarditis, endophtalmitis, bone and joint infections [4] and SAPHO syndrome (synovitis, acne, pustulosis, hyperostosis, and osteitis) [5]. P. avidum was only described in splenic abscess, perianal abcess, sacroileitis, osteomyelitis, and more recently in breast abscess [6]. P. granulosum was demonstrated to cause bacteremia, endocarditis and endophtalmitis [7,8]. To date, P. propionicum was isolated from lacrymal apparatus infections [9], and P. acidifaciens from the human oral cavity [10]. In addition, two former Propionibacterium species, P. innocuum and P. lymphophilum, isolated from the human skin [11] or associated with certain types of lymphadenopathies [12], were reclassified into the Propioniferax and Propionimicrobium genera, respectively [13,14].
Propionibacterium sp. grow anaerobically, on 5% sheep blood agar and produce β-haemolytic colonies after a minimum of 48 hours of incubation. Phenotypic identification of these bacteria is time-consuming and unreliable at the species level [15]. In contrast, molecular methods provide a reliable but time-consuming and expensive species identification [16] that may not be adapted to routine bacterial identification in most laboratories.
In recent years, matrix-assisted laser desorption ionisation time-of-flight mass spectrometry (MALDI-TOF MS) has been increasingly be used for the identification of bacterial isolates on the basis of their peptidic spectra [17], and more recently for the direct identification of bacteria in clinical specimens [18]. MALDI-TOF MS was applied with success to identify Arthrobacter species [19], Bacteroides sp. [20], Bartonella [21], Burkholderia cepacia [22], Campylobacter species [23], Clostridium species [24], Coxiella burnetii [25], Erwinia species [26], Escherichia coli [27], Francisella tularensis [28], Helicobacter pylori [29], Legionella species [30], Listeria species [31], Neisseria species [32], Salmonella species [33], staphylococci [34], Vibrio species [35], viridans streptococci [36], Yersinia enterocolitica [27], nonfermenting bacteria [37,38], and oral anaerobic bacteria [39]. Moreover, recent studies emphasized that MALDI-TOF MS identification was an efficient and cost-effective method for rapid and routine identification of bacterial isolates [17,40-42]. To date, the Bruker database contains spectra from 3769 bacteria including 35 spectra for Propionibacterium spp.
In the present study, in order to determine whether MALDI-TOF MS could be used as a routine tool for identifying clinical isolates of Propionibacterium sp., we first determined the peptidic spectrum of a representative strain of all validated species that have been cultured. Then, we attempted mass spectrometric identification of a collection of P. acnes clinical isolates, the most frequently pathogenic Propionibacterium species.
2. Materials and Methods
2.1. Propionibacterium Strains
Type strains from each of the 12 validated Propionibacterium species as well as P. innocuum and P. lymphophilum, were included in our study (Table 1). For P. freundenreichii, we included a type strain from each of the two validated subspecies, i.e., subsp. freundenreichii and shermanii. All strains were grown on Columbia blood agar containing 5% whole sheep blood (BioMerieux, Marcy l’étoile, France) at 37˚C in anaerobic atmosphere.
In addition, in order to estimate the validity of MALDITOF MS for P. acnes identification, the main pathogenic Propionibacterium species, 48 additional P. acnes isolates from clinical samples were tested by MS. P. acnes isolates were cultivated on Columbia blood agar at 37˚C in anaerobic atmosphere as described above from blood (12), cardiac valve biopsies (2), surgically-removed pace-makers (3), a lung biopsy (1), lymph node biopsies (3), bone biopsies (3), cerebrospinal fluid (1), skin biopsy samples (23). The identification of these strains was

Table 1. Propionibacterium, Propioniferax and Propionimicrobium reference strains used in this study.
confirmed by PCR amplification and sequencing of the 16S RNA, as previously described [43]. MS identification was performed as a blind test after anonymization of the isolates.
2.2. MALDI-TOF-MS
For each strain, four isolated colonies were harvested in 20 μL of sterile water. One μL of each mixture was deposited on a target plate (Bruker Daltonics, Bremen, Germany) in a separate well, and allowed to dry at room temperature. Two μL of matrix solution made of alphacyano-4-hydroxycinnamic acid (Sigma) saturated with 2.5% trifluoroacetic acid and 50% acetonitrile, was then added and allowed to co-cristallize with the sample. Samples were processed using the Autoflex II MALDITOF-MS spectrometer (Bruker Daltonics) with the Flex Control software (Bruker Daltonics). Positive ions were extracted with an accelerating voltage of 20 kV in linear mode. Each spectrum was the sum of the ions obtained from 200 laser shots performed in five different regions of the same well. The spectra have been analyzed in an m/z range of 2000 to 20,000. The analysis was performed with the Flex Analysis and BioTyper softwares and calibrated with protein calibration standard I (Bruker Daltonics). The data obtained with the four replicates were added to minimize random effect. The presence and absence of peaks were considered as fingerprints for a particular isolate. A maximum of 100 peaks with a signal-to-noise (S/N) ratio of 3 were selected in the range of 3000 - 15,000 Da. Afterwards the main spectra were generated as a reference using all spectra given for a single microorganism. In general, 75 peaks were picked automatically, which occurred in at least 25% of the spectra and with a mass deviation of 200 ppm. A reference spectrum was determined for each species. The profiles were analyzed and compared using the MaldiBioTyper 3.0 software (Bruker Daltonik GmbH). The identification process starts with preprocessing of unknown spectra and generation of peak lists. Unknown spectra were identified by the MALDI Biotyper software by comparison with reference spectra. Peaks of unknown spectra were compared and aligned with peaks of reference spectra. In the first step, unknown spectra are calibrated to the reference spectra to reach the highest grade of homology. After calibration each matching peak of the unknown spectrum receives a dedicated point value. The peak of the unknown spectrum will receive the same score value like the corresponding peak of the reference spectra. From the cumulative score value of the unknown spectrum and the maximum score of reference spectra a final score value for the unknown spectrum will be calculated. For the blind test, the 48 tested P. acnes strains were compared to the URMS (Unité des Rickettsies Mass Spectrometry) database that contained the complete Bruker database added with reference spectra from the 14 tested validated species (Table 1). Subsequently, in order to evaluate whether an enrichment of the database in P. acnes spectra would improve MALDI-TOF identification of isolates from this species, we added to the URMS database reference spectra from five of the 48 strains (with best scores against P. acnes of 1.5, 1.6, 1.8, 2.0 and 2.1, respectively), and re-tested the remaining 43 isolates against the new database (Table 2). Identification at the species level was obtained when a tested strain exhibited highest log scores ≥ 1.9 against the correct species for at least 2 replicates, and genus identification was confirmed when a tested strain exhibited highest log scores ≥ 1.7 but <1.9 against any Propionibacterium species. The complete set of reference spectra obtained in the present study is available in the URMS database online (http://www.ifr48.com).
3. Results
3.1. Type Strains
MALDI-TOF-MS spectra were obtained from all 15 strains from the 12 validated Propionibacterium species
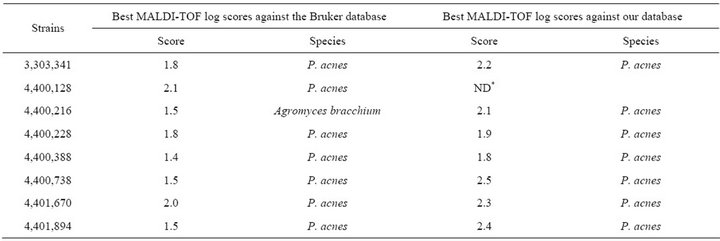
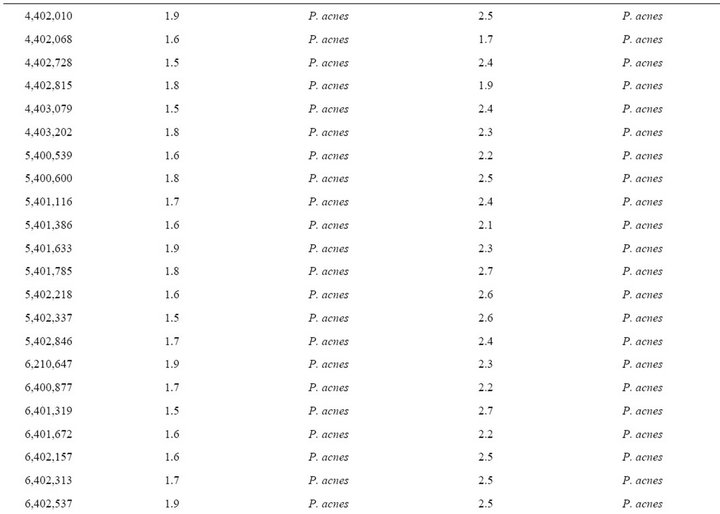

Table 2. Forty-eight Propionibacterium acnes strains from clinical specimens used in this study.
and P. innocuum and P. lymphophilum. All tested species and subspecies exhibited distinct proteic profiles (Figure 1). When compared to 3670 bacteria within the Bruker database, only four/15 representative Propionibacterium strains (26.7%) were correctly identified at the species level despite the presence in the database of spectra from 12 species (scores ≥ 1.9 for P. acidipropionici, P. avidum, P. granulosum and P. lymphophilum) (Table 1). Another five strains (33.3%) were correctly identified at the genus level only (scores ≥ 1.7 and <1.9 for P. australiense, P. cyclohexanicum, P. freudenreichii subsp. shermanii, P. jensenii and P. thoenii) although their highest scores were against the correct species. The remaining six strains (40%) were not identified (scores < 1.7), includeing P. acidificiens, P. innocuum and P. propionicum for which no spectra were available in the Bruker database, and P. acnes, P. freudenreichii subsp. freudenreichii, and P. microaerophilum which exhibited highest scores against their respective species. All obtained spectra were added to the URMS database.
3.2. Clinical Isolates
MALDI-TOF MS spectra were obtained for the 48 blindtested clinical strains. When compared to the Bruker database, only 9/48 (18.7%) P. acnes clinical strains were correctly identified at the species with scores > 1.9, level, and another 15/48 (31.2%) were correctly identified at the genus level with scores > 1.7 but <1.9 (Table 2). Another P. acnes strain was misidentified as Agromyces bracchium with a score of 1.5. All 23 remaining P. acnes strains (47.9%) exhibited highest scores < 1.7 against P. acnes for at least 2/4 spectra.
When tested against the URMS database enriched with spectra from 5 P. acnes clinical strains, 40/43 P. acnes clinical isolates (93.0%) were correctly identified at the species level with scores > 1.9 (Table 2). The remaining 3 strains exhibited scores > 1.7 against P. acnes as best hit, but <1.9.
4. Discussion
Long considered as culture contaminants, Propionibacterium species have recently been recognized as human pathogens in a variety of diseases. In particular, the incidence of opportunistic and postoperative bone infections caused by P. acnes has increased [44]. However, the routine identification of these bacteria is time-consuming and unreliable using phenotypic methods, or expensive using molecular methods, which stimulated the development of efficient identification tools that would enable rapid and adequate management of patients. Over recent years, MALDI-TOF mass spectrometry has emerged as a rapid (less than an hour) and reliable tool for the identification of bacterial strains [17,40,42,45] and has successfully been applied to a variety of genera [19-39]. In an analysis of out of 1660 bacterial isolates by MALDITOF, Seng et al. tested 60 Propionibacterium sp. against the Bruker database (Bruker) and obtained an identification of only 45% of them at the genus level [17]. Such a low identification rate may be due to various factors, including: 1) the absence of reference spectra for a given
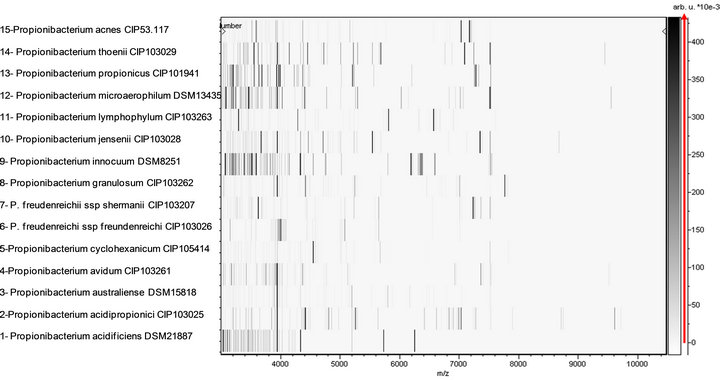
Figure 1. Virtual gel view of MS reference spectra from validated Propionibacterium species.
species; and 2) an insufficient number of reference spectra for a given species [46].
At the time the study was performed, the Bruker database contained reference spectra from 3769 bacterial isolates, including 35 spectra from validated Propionibacterium, Propioniferax and Propionimicrobium species. By comparison with the 12 validated Propionibacterium species, the P. acidificiens and P. propionicum spectra were missing from the Bruker database, as was that from P. innocuum, which explains why the isolates from these three species that we tested were not identified at the species level (Table 1), in contrast with other isolates. However, only four strains (26%) were correctly identified at the species level with a significant score, suggesting an intra-species proteomic variability. In addition, despite the use of a reference strain, no reliable identification was obtained for P. acnes, neither at the species nor genus level. Such a low efficiency of MALDI-TOF to identify Propionibacterium strains was confirmed by our blind test of 48 P. acnes strains, where only 18.7% were correctly identified at the species level, 50% not even being identified at the genus level (Table 2). Subsequently, in order to estimate whether such a poor sensitivity, particularly for P. acnes, the most frequent human pathogen among Propionibacterium species, could be improved by enriching the database in spectra, we added the spectra from 5 strains to our database. The result of this enrichment was that 40 of the 43 remaining clinical isolates were identified at the species level with scores > 1.9, thus increasing the sensitivity of MALDI-TOF for the identification of this species to 93.0%. The five strains used to enrich the database were selected so that their individual scores in the blind test ranged from 1.5 to 2.1. MALDI-TOF MS was known to be rapid and cost effective for bacterial identification. However, our study demonstrates that this method, to be efficiently used for the identification of Propionibacterium isolates, requires a database enrichment in isolates for the species of interest. The spectra obtained for all tested strains are freely available in our URMS database (http://www.ifr48.com).
REFERENCES
- I. Brook and E. H. Frazier, “Infections Caused by Propionibacterium Species,” Reviews of Infectious Diseases, Vol. 13, No. 5, 1991, pp. 819-822. doi:10.1093/clinids/13.5.819
- C. Dessinioti and A. D. Katsambas, “The Role of Propionibacterium acnes in Acne Pathogenesis: Facts and Controversies,” Clinics in Dermatology, Vol. 28, No. 1, 2010, pp. 2-7. doi:10.1016/j.clindermatol.2009.03.011
- P. Y. Levy, F. Fenollar, A. Stein, F. Borrione, E. Cohen, B. Lebail and D. Raoult, “Propionibacterium acnes Postoperative Shoulder Arthritis: An Emerging Clinical Entity,” Clinical Infectious Diseases, Vol. 46, No. 12, 2008, pp. 1884-1886. doi:10.1086/588477
- A. L. Perry and P. A. Lambert, “Propionibacterium acnes,” Letters in Applied Microbiology, Vol. 42, No. 3, 2006, pp. 185-188. doi:10.1111/j.1472-765X.2006.01866.x
- T. Schaeverbeke, L. Lequen, B. B. De, L. Labbe, C. M. Bebear, Y. Morrier, B. Bannwarth, C. Bebear and J. Dehais, “Propionibacterium acnes Isolated from Synovial Tissue and Fluid in a Patient with Oligoarthritis Associated with Acne and Pustulosis,” Arthritis & Rheumatism, Vol. 41, No. 10, 1998, pp. 1889-1893. doi:10.1002/1529-0131(199810)41:10<1889::AID-ART23>3.0.CO;2-F
- S. Panagea, J. E. Corkill, M. J. Hershman and C. M. Parry, “Breast Abscess Caused by Propionibacterium avidum Following Breast Reduction Surgery: Case Report and Review of the Literature,” Journal of Infection, Vol. 51, No. 5, 2005, pp. e253-e255. doi:10.1016/j.jinf.2005.04.005
- C. Branger, B. Bruneau and P. Goullet, “Septicemia Caused by Propionibacterium granulosum in a Compromised Patient,” Journal of Clinical Microbiology, Vol. 25, No. 12, 1987, pp. 2405-2406.
- R. Chaudhry, B. Dhawan, A. Pandey, S. K. Choudhary and A. S. Kumar, “Propionibacterium granulosum: A Rare Cause of Endocarditis,” Journal of Infection, Vol. 41, No. 3, 2000, p. 284. doi:10.1053/jinf.2000.0728
- J. S. Brazier and V. Hall, “Propionibacterium propionicum and Infections of the Lacrimal Apparatus,” Clinical Infectious Diseases, Vol. 17, No. 5, 1993, pp. 892-893. doi:10.1093/clinids/17.5.892
- J. Downes and W. G. Wade, “Propionibacterium acidifaciens sp. nov., Isolated from the Human Mouth,” International Journal of Systematic and Evolutionary Microbiology, Vol. 59, No. 11, 2009, pp. 2778-2781. doi:10.1099/ijs.0.010470-0
- D. G. Pitcher and M. D. Collins, “Phylogenetic Analysis of Some LL-Diaminopimelic Acid-Containing Coryneform Bacteria from Human Skin: Description of Propionibacterium innocuum sp. nov,” FEMS Microbiology Letters, Vol. 68, No. 3, 1991, pp. 295-300.
- J. C. Torrey, “Bacteria Associated with Certain Types of Abnormal Lymph Glands,” The Journal of Medical Research, Vol. 34, 1916, pp. 65-80.
- E. Stackebrandt, P. Schumann, K. P. Schaal and N. Weiss, “Propionimicrobium gen. Nov. A New Genus to Accomodate Propionibacterium lymphophilum (Torrey 1916) Johnson and Cummins 1972, 1057AL as Propionimicrobium lymphophilum comb. nov.,” International Journal of Systematic and Evolutionary Microbiology, Vol. 52, 2002, pp. 1925-1927. doi:10.1099/ijs.0.02188-0
- A. Yokota, T. Tamura, M. Takeuchi, N. Weiss and E. Stackebrandt, “Transfer of Propionibacterium innocuum Pitcher and Collins 1991 to Propioniferax gen. nov. as Propioniferax innocua comb. nov,” International Journal of Systematic Bacteriology, Vol. 44, No. 3, 1994, pp. 579- 582. doi:10.1099/00207713-44-3-579
- L. V. Holdeman, E. P. Cato and W. E. C. Moore, “Anaerobe Laboratory Manual,” Virginia Polytechnic Institute and State University, Blacksburg, 1977.
- M. Nakamura, I. Kametani, S. Higaki and T. Yamagishi, “Identification of Propionibacterium acnes by Polymerase Chain Reaction for Amplification of 16S Ribosomal RNA and Lipase Genes,” Anaerobe, Vol. 9, 2003, pp. 5- 10. doi:10.1016/S1075-9964(03)00061-1
- P. Seng, M. Drancourt, F. Gouriet, B. La Scola, P. E. Fournier, J. M. Rolain and D. Raoult, “Ongoing Revolution in Bacteriology: Routine Identification of Bacteria by Matrix-Assisted Laser Desorption Ionization Time-ofFlight Mass Spectrometry,” Clinical Infectious Diseases, Vol. 49, No. 4, 2009, pp. 543-551. doi:10.1086/600885
- S. B. La and D. Raoult, “Direct Identification of Bacteria in Positive Blood Culture Bottles by Matrix-Assisted Laser Desorption Ionisation Time-of-Flight Mass Spectrometry,” Plos One, Vol. 4, No. 11, 2009, p. e8041. doi:10.1371/journal.pone.0008041
- M. Vargha, Z. Takats, A. Konopka and C. H. Nakatsu, “Optimization of MALDI-TOF MS for Strain Level Differentiation of Arthrobacter Isolates,” Journal of Microbiological Methods, Vol. 66, No. 3, 2006, pp. 399-409. doi:10.1016/j.mimet.2006.01.006
- E. Nagy, T. Maier, E. Urban, G. Terhes and M. Kostrzewa, “Species Identification of Clinical Isolates of Bacteroides by Matrix-Assisted Laser-Desorption/Ionization Time-of-Flight Mass Spectrometry,” Clinical Microbiology and Infection, Vol. 15, No. 8, 2009, pp. 796-802. doi:10.1111/j.1469-0691.2009.02788.x
- P. E. Fournier, C. Couderc, S. Buffet, C. Flaudrops and D. Raoult, “Rapid and Cost-Effective Identification of Bartonella Species Using Mass Spectrometry,” Journal of Medical Microbiology, Vol. 58, No. Pt 9, 2009, pp. 1154- 1159. doi:10.1099/jmm.0.009647-0
- E. Vanlaere, K. Sergeant, P. Dawyndt, W. Kallow, M. Erhard, H. Sutton, D. Dare, B. Devreese, B. Samyn and P. Vandamme, “Matrix-Assisted Laser Desorption Ionisation-Time-of-Flight Mass Spectrometry of Intact Cells Allows Rapid Identification of Burkholderia cepacia Complex,” Journal of Medical Microbiology, Vol. 75, No. 2, 2008, pp. 279-286. doi:10.1016/j.mimet.2008.06.016
- R. Kolinska, M. Drevinek, V. Jakubu and H. Zemlickova, “Species Identification of Campylobacter jejuni ssp. jejuni and C. coli by Matrix-Assisted Laser Desorption/ Ionization Time-of-Flight Mass Spectrometry and PCR,” Folia Microbiologica, Vol. 53, No. 5, 2008, pp. 403-409. doi:10.1007/s12223-008-0061-7
- A. Grosse-Herrenthey, T. Maier, F. Gessler, R. Schaumann, H. Bohnel, M. Kostrzewa and M. Kruger, “Challenging the Problem of Clostridial Identification with Matrix-Assisted Laser Desorption and Ionization-Timeof-Flight Mass Spectrometry (MALDI-TOF MS),” Anaerobe, Vol. 14, No. 4, 2008, pp. 242-249. doi:10.1016/j.anaerobe.2008.06.002
- L. Skultety, L. Hernychova, E. Bereghazyova, K. Slaba and R. Toman, “Detection of Specific Spectral Markers of Coxiella burnetii Isolates by MALDI-TOF Mass Spectrometry,” Acta Virologica, Vol. 51, No. 1, 2007, pp. 55-58.
- S. Sauer, A. Freiwald, T. Maier, M. Kube, R. Reinhardt, M. Kostrzewa and K. Geider, “Classification and Identification of Bacteria by Mass Spectrometry and Computational Analysis,” Plos One, Vol. 3, No. 7, 2008, p. e2843. doi:10.1371/journal.pone.0002843
- D. Parisi, M. Magliulo, P. Nanni, M. Casale, M. Forina and A. Roda, “Analysis and Classification of Bacteria by Matrix-Assisted Laser Desorption/Ionization Time-ofFlight Mass Spectrometry and a Chemometric Approach,” Analytical and Bioanalytical Chemistry, Vol. 391, No. 6, 2008, pp. 2127-2134. doi:10.1007/s00216-008-2161-2
- E. Seibold, T. Maier, M. Kostrzewa, E. Zeman and W. Splettstoesser, “Identification of Francisella tularensis by Whole-Cell Matrix-Assisted Laser Desorption IonizationTime of Flight Mass Spectrometry: Fast, Reliable, Robust, and Cost-Effective Differentiation on Species and Subspecies Levels,” Journal of Clinical Microbiology, Vol. 48, No. 4, 2010, pp. 1061-1069. doi:10.1128/JCM.01953-09
- E. N. Ilina, A. D. Borovskaya, M. V. Serebryakova, V. V. Chelysheva, K. T. Momynaliev, T. Maier, M. Kostrzewa and V. M. Govorun, “Application of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry for the Study of Helicobacter pylori,” Rapid Communications in Mass Spectrometry, Vol. 24, No. 3, 2010, pp. 328-334. doi:10.1002/rcm.4394
- C. Moliner, C. Ginevra, S. Jarraud, C. Flaudrops, M. Bedotto, C. Couderc, J. Etienne and P. E. Fournier, “Rapid Identification of Legionella Species by Mass Spectrometry,” Journal of Medical Microbiology, Vol. 59, No. Pt 3, 2010, pp. 273-284. doi:10.1099/jmm.0.014100-0
- S. B. Barbuddhe, T. Maier, G. Schwarz, M. Kostrzewa, H. Hof, E. Domann, T. Chakraborty and T. Hain, “Rapid Identification and Typing of Listeria Species by MatrixAssisted Laser Desorption Ionization-Time of Flight Mass Spectrometry,” Applied and Environmental Microbiology, Vol. 74, No. 17, 2008, pp. 5402-5407. doi:10.1128/AEM.02689-07
- S. K. Gudlavalleti, A. K. Sundaram, J. Razumovski and V. Doroshenko, “Application of Atmospheric Pressure Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry for Rapid Identification of Neisseria Species,” Journal of Biomolecular Techniques, Vol. 19, No. 3, 2008, pp. 200-204.
- R. Dieckmann, R. Helmuth, M. Erhard and B. Malorny, “Rapid Classification and Identification of Salmonellae at the Species and Subspecies Levels by Whole-Cell MatrixAssisted Laser Desorption Ionization-Time of Flight Mass Spectrometry,” Applied and Environmental Microbiology, Vol. 74, No. 24, 2008, pp. 7767-7778. doi:10.1128/AEM.01402-08
- E. Carbonnelle, J. L. Beretti, S. Cottyn, G. Quesne, P. Berche, X. Nassif and A. Ferroni, “Rapid Identification of Staphylococci Isolated in Clinical Microbiology Laboratories by Matrix-Assisted Laser Desorption IonizationTime of Flight Mass Spectrometry,” Journal of Clinical Microbiology, Vol. 45, No. 7, 2007, pp. 2156-2161. doi:10.1128/JCM.02405-06
- R. Dieckmann, E. Strauch and T. Alter, “Rapid Identification and Characterization of Vibrio Species Using Whole-Cell MALDI-TOF Mass Spectrometry,” Journal of Applied Microbiology, Vol. 109, No. 1, 2010, pp. 199- 211. doi:10.1111/j.1365-2672.2009.04647.x
- C. Friedrichs, A. C. Rodloff, G. S. Chhatwal, W. Schellenberger and K. Eschrich, “Rapid Identification of Viridans Streptococci by Mass Spectrometric Discrimination,” Journal of Clinical Microbiology, Vol. 45, No. 8, 2007, pp. 2392-2397. doi:10.1128/JCM.00556-07
- A. Mellmann, J. Cloud, T. Maier, U. Keckevoet, I. Ramminger, P. Iwen, J. Dunn, G. Hall, D. Wilson, P. Lasala, M. Kostrzewa and D. Harmsen, “Evaluation of MatrixAssisted Laser Desorption Ionization-Time-of-Flight Mass Spectrometry in Comparison to 16S rRNA Gene Sequencing for Species Identification of Nonfermenting Bacteria,” Journal of Clinical Microbiology, Vol. 46, No. 6, 2008, pp. 1946-1954. doi:10.1128/JCM.00157-08
- N. Degand, E. Carbonnelle, B. Dauphin, J. L. Beretti, B. M. Le, I. Sermet-Gaudelus, C. Segonds, P. Berche, X. Nassif and A. Ferroni, “Identification of Non Fermenting Gram Negative Bacilli Isolated in Cystic Fibrosis by Matrix Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry,” Journal of Clinical Microbiology, Vol. 46, 2008, pp. 3361-3367. doi:10.1128/JCM.00569-08
- C. S. Stingu, A. C. Rodloff, H. Jentsch, R. Schaumann and K. Eschrich, “Rapid Identification of Oral Anaerobic Bacteria Cultivated from Subgingival Biofilm by MALDITOF-MS,” Oral Microbiology and Immunology, Vol. 23, No. 5, 2008, pp. 372-376. doi:10.1111/j.1399-302X.2008.00438.x
- E. Carbonnelle, C. Mesquita, E. Bille, N. Day, B. Dauphin, J. L. Beretti, A. Ferroni, L. Gutmann and X. Nassif, “MALDI-TOF Mass Spectrometry Tools for Bacterial Identification in Clinical Microbiology Laboratory,” Clinical Biochemistry, Vol. 44, No. 1, 2010, pp. 104-109.
- A. Bizzini and G. Greub, “MALDI-TOF MS, a Revolution in Clinical Microbial Identification,” Clinical Microbiology and Infection, Vol. 16, No. 11, 2010, pp. 1614- 1619. doi:10.1111/j.1469-0691.2010.03311.x
- E. Bessede, M. Angla-Gre, Y. Delagarde, H. S. Sep, A. Menard and F. Megraud, “Matrix-Assisted Laser-Desorption/Ionization Biotyper: Experience in the Routine of a University Hospital,” Clinical Microbiology and Infection, Vol. 17, 2010, pp. 533-538. doi:10.1111/j.1469-0691.2010.03274.x.
- M. Drancourt, C. Bollet, A. Carlioz, R. Martelin, J. P. Gayral and D. Raoult, “16S Ribosomal DNA Sequence Analysis of a Large Collection of Environmental and Clinical Unidentifiable Bacterial Isolates,” Journal of Clinical Microbiology, Vol. 38, No. 10, 2000, pp. 3623-3630.
- N. Asseray, C. Papin, S. Touchais, P. Bemer, C. Lambert, D. Boutoille, B. Tequi, F. Gouin, F. Raffi, N. Passuti and G. Potel, “Improving Diagnostic Criteria for Propionibacterium acnes Osteomyelitis: A Retrospective Analysis,” Scandinavian Journal of Infectious Diseases, Vol. 42, No. 6-7, 2010, pp. 421-425. doi:10.3109/00365540903527330
- P. Seng, J. M. Rolain, P. E. Fournier, B. La Scola, M. Drancourt and D. Raoult, “MALDI-TOF-Mass Spectrometry Applications in Clinical Microbiology,” Future Microbiology, Vol. 5, 2010, pp. 1733-1754. doi:10.2217/fmb.10.127
- A. Croxatto, G. Prod’hom and G. Greub, “Applications of MALDI-TOF Mass Spectrometry in Clinical Diagnostic Microbiology,” FEMS Microbiology Reviews, Vol. 36, No. 2, 2011, pp. 380-407.
NOTES
*Corresponding author.

