Food and Nutrition Sciences
Vol. 2 No. 4 (2011) , Article ID: 5444 , 10 pages DOI:10.4236/fns.2011.24044
Detection, Identification and Characterization of Staphylococci in Street Vend Foods —Characterization of Staphylococcus Isolates
![]()
Human Resource Development, Central Food Technological Research Institute, Mysore, India.
Email: psnegi@cftri.res.in
Received January 13th, 2011; revised March 28th, 2011; accepted April 7th, 2011.
Keywords: Staphylococcus, 16S rRNA Gene Sequence, RFLP, Phylogenetic Tree
ABSTRACT
In the present investigation the diversity of the Staphylococcus species in different street vend food samples was studied. A total of 35 staphylococcal food isolates comprising of various species from 14 different street vend food samples were identified and characterized phenotypically. Staphylococcus aureus was found to be the most prevalent species in these foods. A PCR-RFLP analysis based on 16S rRNA gene was used for identification of Staphylococcus species. Isolates showing distinct RFLP pattern for AluI restriction digestion were selected for nucleotide sequence analysis. Phylogenetic tree constructed using the multiple alignments of 16S rRNA gene sequences of isolates showed a hotspot region of 169 bp and the relationship among species was evaluated by bootstrap values generated in phylogenetic analysis. 16S rRNA gene sequences allowed bacterial identification that was reproducible and more accurate than that obtained by phenotypic testing. 16S rRNA gene sequence analysis would be helpful in timely and correct identification of pathogens.
1. Introduction
Members of Staphylococci are wide spread in nature and have been isolated sporadically from a wide range of environmental sources such as air, water, soil and plant surfaces, meat, poultry and dairy products [1]. They are capable of causing mild to life threatening diseases, which also includes food borne illnesses. Several species in this genus are having capability to produce a wide range of heat stable enterotoxins, and the main members of this genus, S. aureus is considered the third most important cause of foodborne diseases in the world among the reported foodborne pathogens [2-4].
Several Staphylococcus species other than S. aureus are reported to produce enterotoxins [5]. The coagulase positive S. hyicus and S. intermedius are the predominant non-S. aureus species, which have been shown to produce staphylococcal enterotoxins (SEs) and are involved in staphylococcal food poisoning outbreaks [6-9]. Among the coagulase negative species, S. cohnii, S. epidermidis, S. xylosus and S. haemolyticus have been isolated from ewe’s milk and were found to produce one or the other SEs [10]. Other non Staphylococcus aureus species such as S. saprophyticus, S. warneri, S. chromogenes and S. lentus isolated from healthy goat milk and dry-cured hams were found to have the enterotoxigenic potential [11-14].
Screening for Staphylococcus species among various ready to serve food products available in the street vend shops is important for epidemiological reasons. As most of these products are sold in open conditions, this screening will also indicate the level of hygiene followed by these vendors. Therefore, the aim of the present investigation was to study the distribution of various Staphy-lococcus species in street vend food.
2. Materials and Methods
2.1. Isolation of Staphylococcus Species from Different Food Sources
Street vend food samples were collected from Mysore city limits in a sterilized container, trans-ported under cold condition to the laboratory and screened for the presence of Staphylococcus species within an hour. The isolation of Staphylococcus species from different food samples was performed according to the conventional procedure by serial dilution in sterile 0.85% saline and pour plating on Baird Parker Agar supplemented with Egg Yolk emulsion (5%) and saturated potassium tellurite (0.3%) solution. The plates were incubated under aerobic conditions at 37˚C for 24 to 48 h. From each plate, gray to black colonies with lecithinase positive as well as negative activity were chosen for further identification of species by biochemical and gene based methods. Staphylococcus aureus (FRI 722), S. saprohyticus (ATCC 15305), S. epidermidis (ATCC 12228), S. xylosus (ATCC 35033) and S. hemolyticus (ATCC 29978) were used as reference strains.
2.2. Conventional Biochemical Test
Biochemical identification and characterization of isolates were done after confirmation of Gram reaction and catalase test, and only positive isolates were selected for further study. Biochemical tests included production of DNase, TNase, phosphatase, nitrate reductase and acetoin; haemolytic activity (sheep blood); tube coagulase test; antibiotic sensitivity test to novobiocin; utilization of D-mannitol and salt tolerance test at 10% and 15% NaCl concentration [15]. On the basis of biochemical test results, isolates were identified to species level with the aid of Bergey’s Manual of Systematic Bacteriology [1].
2.3. DNA Extraction
Genomic DNA was isolated as described previously (A. K. Saxena, Manual of DNA Sequencing and Microbial Identification, National Training Programme, September 1 - 7, 2008, National Bureau of Agriculturally Important Microorganisms, Mau Nath Bhanjan, India) with slight modification. In brief, the bacterial cultures were grown in pre sterilized brain heart infusion broth (pH-7) overnight at 37˚C and harvested by centrifuging it at 10,000 rpm for 5 min at room temperature to collect the pellet. Pellet was washed with 1 ml TE (10 mM Tris-HCl, pH 8.0 and 1 mM EDTA pH 8.0) and resuspended in 0.5 ml SET buffer (75 mM NaCl, 25 mM EDTA pH 8.0, 20 mM Tris-HCl pH 8.0) containing 100 μg lysozyme and incubated at 37˚C for 1 h. To this 1/10 volume of 10% SDS and 100 μg of proteinase K was added and incubated at 55˚C for 1 h. After incubation, 1/3 volume of 5 M NaCl and equal volume of phenol: chloroform: isoamylalcohol (25:24:1) were added and incubated at 37˚C for further 30 min. After centrifuging it at 5000 rpm for 15 min at 4˚C, aqueous layer was collected in a fresh eppendorf tube and precipitated with ethanol. The pellet obtained by centrifugation at 10,000 rpm for 5 min at 4˚C was washed with 70% ethanol by centrifugation at 10,000 rpm for 5 min at 4˚C, air dried and dissolved in TE buffer. The concentration and quality of genomic DNA was calculated by the spectrophotometer reading at 260 nm. Integrity of the DNA was checked by running in 0.8% agarose gel.
2.4. PCR Amplification of 16S rRNA and Purification
16S rRNA Universal primer forward 5’ AGA GTT TGA TCC TGG CTC AG 3’ and reverse 5’ AAG GAG GTG ATC CAG CCG CA 3’ were synthesized from Sigma Aldrich (Bangalore, India). A reaction mixture containing approximately 1 ng of template DNA, 2.5 μl of 10X buffer, 20 pM concentration of each PCR primer, 10 mM dNTP mix and 1 U of Taq polymerase (Bangalore Genei, Bangalore, India) in a total of 25 μl was prepared by adding sterile milli Q water for all the reactions. The PCR was carried out in a Thermal Cycler (Mastercycler, Eppendorf, Hamburg, Germany). The amplified product of 16S rRNA gene was purified with the PCR purification kit following the standard protocol as supplied by the manufacturer (HiMedia, Mumbai, India). The purification involved adding binding buffer to the PCR mix and centrifuging through filter tubes. The unincorporated nucleotides were removed by adding wash buffer and centrifugation at 10,000 rpm for 2 min. The PCR products were eluted using elution buffer by centrifugation at 10,000 rpm for 1 min.
2.5. Restriction Endonuclease Digestion
The 16S rRNA amplified products were digested with BamHI, HindIII, EcoRI, AluI and HaeIII restriction enzymes (Bangalore Genei, Bangalore, India). PCR product was digested separately with 10 U of each enzyme for 3 h at 37˚C in a circulating water bath (Julabo, Germany). Reaction was stopped by incubating the samples at 65˚C for 5 min [16]. Digested samples were electrophoresed on 2.5% agarose gel and photographed by gel documentation system (Vibler Lourmat, Marne-la-Vallee, France).
2.6. Cluster Analysis
Different fragments on the gel were numbered sequentially, followed by scoring the samples based on presence and absence of fragments (present 1, absent 0) and compared according to the genetic distance method. The strains were clustered by the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) with the aid of NTSYS software (Applied Biostatistics Inc V.1.07).
2.7. Sequence Analysis of 16S rRNA Gene
The purified product was sequenced in an automated DNA sequencing facility (Bangalore Genei, Bangalore, India). The results were processed into sequence data with sequence analysis Chromas software (version 2.33; Technelysium Pvt. Ltd; http://www.technelysium.com.au/chromas.html) and the partial sequences were combined into a single consensus sequence by finding multiple matching sub segments in both forward and reverse sequences with the aid of William Pearson’s lalign program (http://www.ch.embnet.org/software/lalign_form.html).
A single sequence analysis data obtained was blasted in NCBI database for the identification of isolates. These sequences were compared with the reported Staphylococcus species 16S rRNA gene sequences available in the GenBank database (http://www.ncbi.nlm.nih.gov/Genbank/index.html). Based on the highest percentage homology to the reported sequences, the food isolates were identified. Multiple sequence alignments were performed using clustalW (Kyoto University, Bioinformatics Center; http://align.genome.jp/). Phylogenetic tree and Bootstrapping were constructed based on the 16S rRNA gene sequences using CLC Main workbench software (version 5.6.1, CLC bio, www.clcbio.com).
2.8. Nucleotide Sequence Accession Numbers
The 16S rRNA sequences determined for the food isolates were submitted to the eMBL database (http://www.ebi.ac.uk/embl/Submission/).
3. Results
Based on differences in morphological characteristics on BPA such as big to pin point colonies showing grayish black to black colour with lecithinase positive as well as negative activity, a total of 35 native isolates were selected from 14 street vend food samples in Mysore city area. The results of biochemical characterization of these isolates showed that S. aureus was the most frequently isolated species among different food samples accounting for 20.0% the population studied. The other Staphylococcus isolates were distributed as S. cohnii (17.14%); S. sciuri and S. intermedius (11.42% each); S. hyicus (8.57%); S. chromogenes and S. intermedius (8.43% each); S. epidermidis; S. xylosus and S. felis (5.70% each); and S. haemolyticus; S. delphini; S. pseudointermedius; S. saprophyticus and S. chromogenes (2.85% each) (Table 1).
To confirm the phenotypic identification of the food isolates, definitive species identification was done on the basis of PCR-RFLP and sequence data analysis for the 16S rRNA gene. All Staphylococcal food isolates and reference strains generated a single PCR product of about 1500 bp length for 16S rRNA gene. The PCR products were then digested with BamHI, HindIII, EcoRI, HaeIII and AluI for RFLP analysis. It was observed that majority of the isolates and reference standards namely S. aureus (FRI 722), S. epidermidis (ATCC 12228), S. saprophyticus (ATCC 15305), S. haemolyticus (ATCC 29978) and S. xylosus (ATCC 35033) had identical restriction profiles with BamHI, HaeIII, HindIII and EcoRI, but these strains yielded different digestion patterns when subjected to AluI restriction digestion. On the basis of scoring, presence and absence of fragments (present 1, absent 0), phylogenetic tree was constructed with the help of NTYSIS software and isolates found at different branching nodes were selected for further sequence analysis. When comparing with the reference standards to the food isolates using biochemical tests it was observed that even the isolates of same species were forming in different nodes (Figure 1) this indicates that either there is a difference among the Staphylococcus species or biochemical test is not so accurate for identification at species level. Therefore, 14 isolates that were found at different nodes were selected for sequence analysis and the data were compared with NCBI database (Table 2). The highest similarity with a best match to the reported sequence data were S. cohnii (99%); S. sciuri (99%); S. saprophyticus (97%); S. saprophyticus subsp saprophyticus (98%) and S. gallinarum (96%) indicating high degree of similarity between the food isolates and the reported species. These sequences were deposited in EMBL database under accession No FN646065 - FN646078.
Multiple alignments of 16S rRNA gene data sequences of Staphylococcus food isolates were obtained using ClustalW programme. Sequence similarity of 16S rRNA gene among 14 different Staphylococcus species ranged from 30% - 97% with a mean similarity of 63.5% (Table 3). These values are consistently lower and indicate high discrimination among the different Staphylococcus species and the most variability among all the food isolates was observed at 595 to 763 bp region of 169 bp (Table 4). The pair wise comparisons among different Staphylococcal species showed a wide variability (3 to 70%). The highest percentage similarity was observed between the species S. cohnii and S. saprophyticus isolates (96.5%), S. cohnii and S. sciuri isolates (92%) and S. saprophyticus and S. sciuri isolates (91.5%), and the least similarity was observed between S. saprophyticus subsp saprophyticus and S. simulans (30%).
The relationships among species of the genus Staphylococcus were confirmed by phylogenetic analysis based on the 16S rRNA gene sequencing, and the topology of the tree was evaluated by bootstrap values. The phylogenetic tree (Figure 2) showed that Staphylococcus species were divided in distinct subgroups. The group consisting of S. cohnii, S. saprophyticus subsp saprophyticus and S. xylosus was characterized as being novobiocin resistant, and it formed a monophyletic clade in the phylogenetic tree with 89% of the bootstrap value. S. cohnii has a relatively deep subline with in a cluster group of S. saprophyticus subsp saprophyticus. According to our phylogenetic tree another closely related group consists of S. aureus, S. simulans, S. felis and S. sciuri,
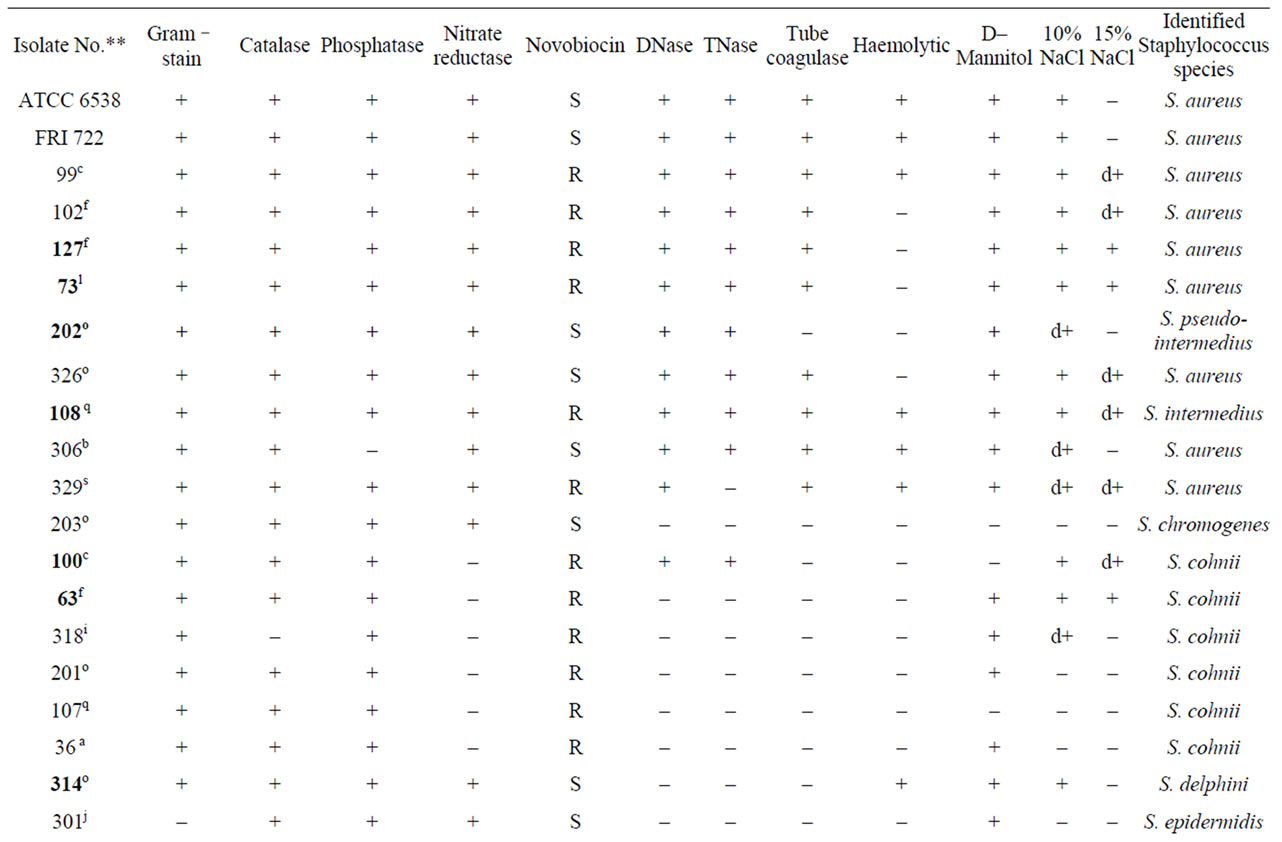
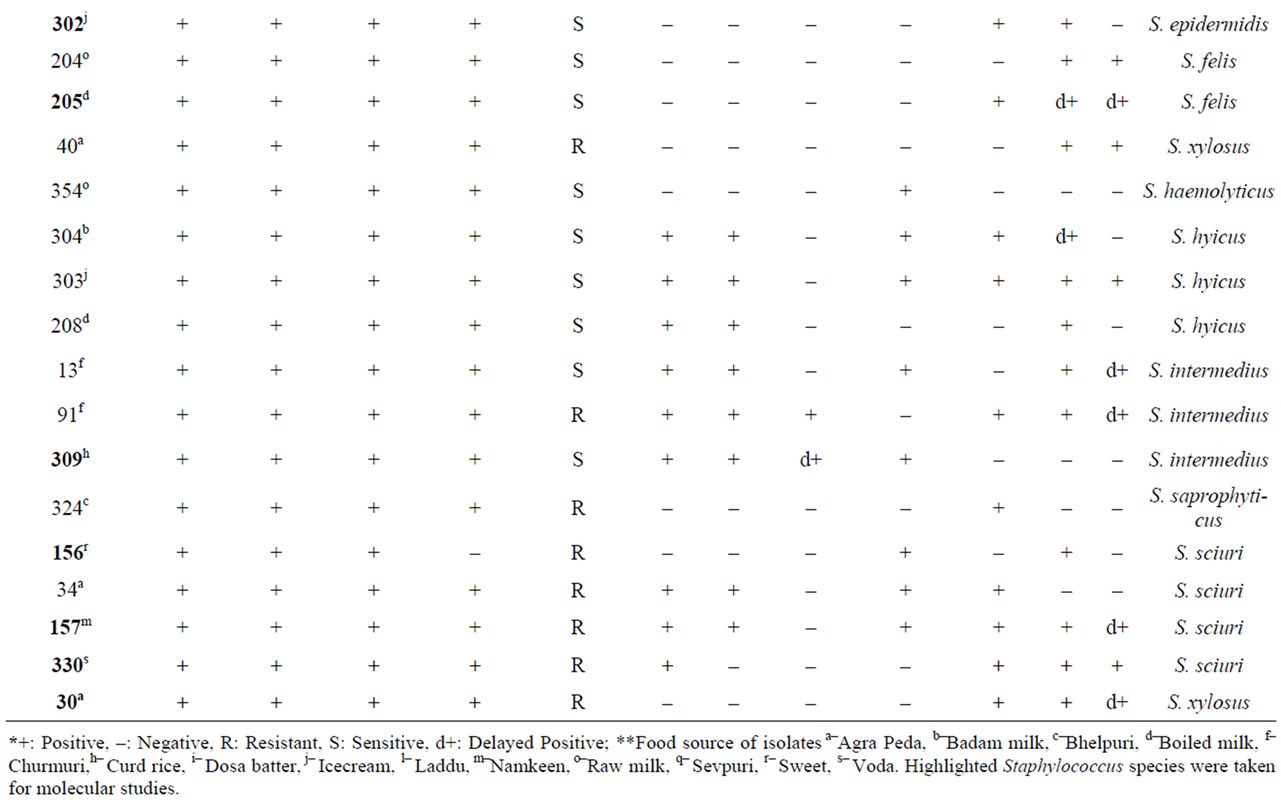
Table 1. Biochemical characterization of Staphylococcus food isolates*.
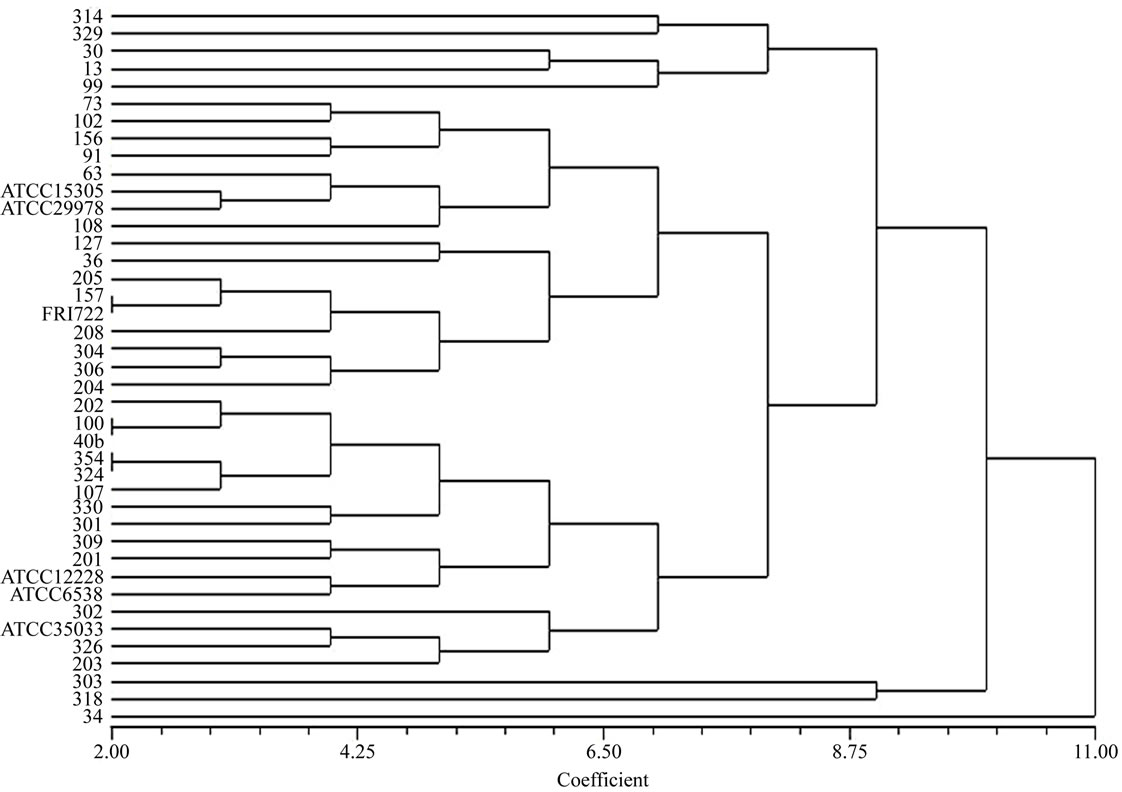
Figure 1. Unrooted Neighbour–joining phylogenetic tree of Staphy-lococcus food isolates constructed on the basis of digestion of 16S rRNA gene product with AluI restriction enzyme. The scale bar indicates the evolutionary distance value between the species.
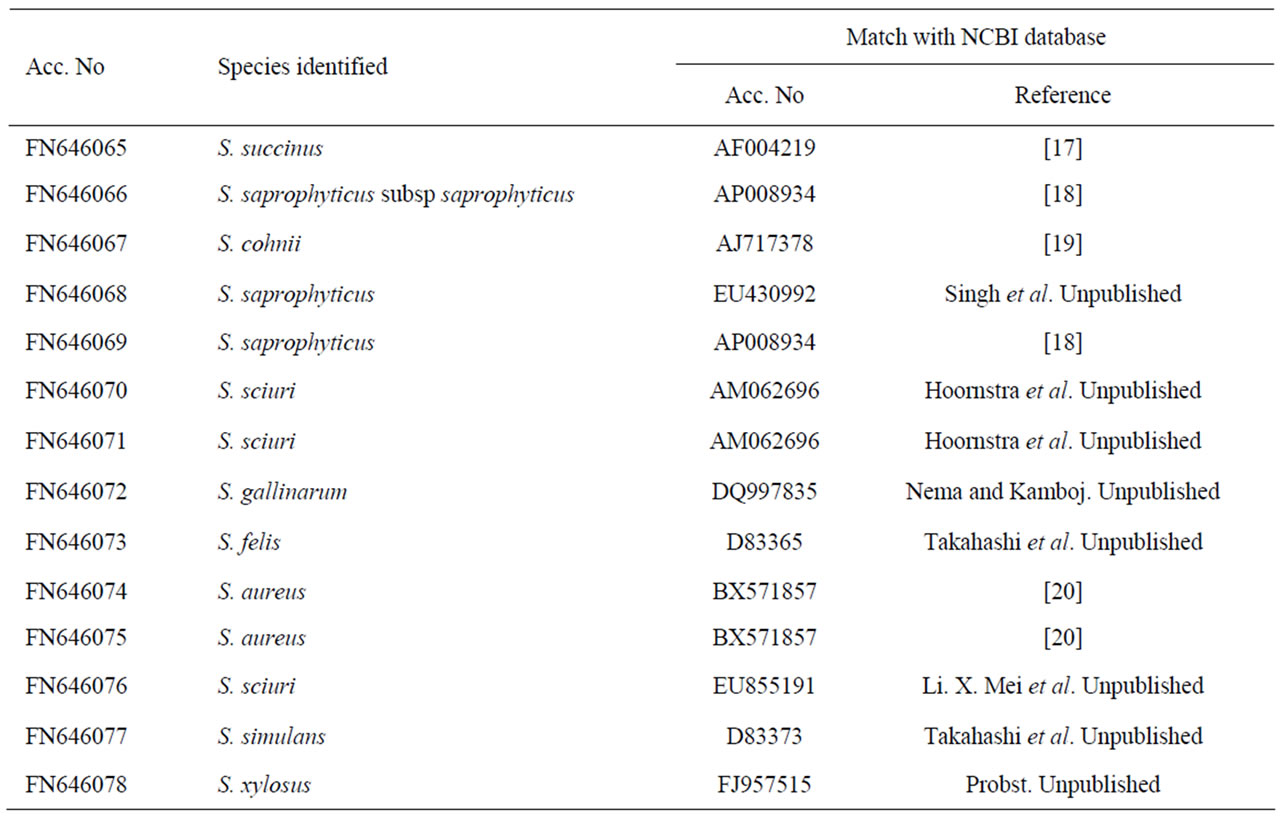
Table 2. Molecular identification of Staphylococcus food isolates.
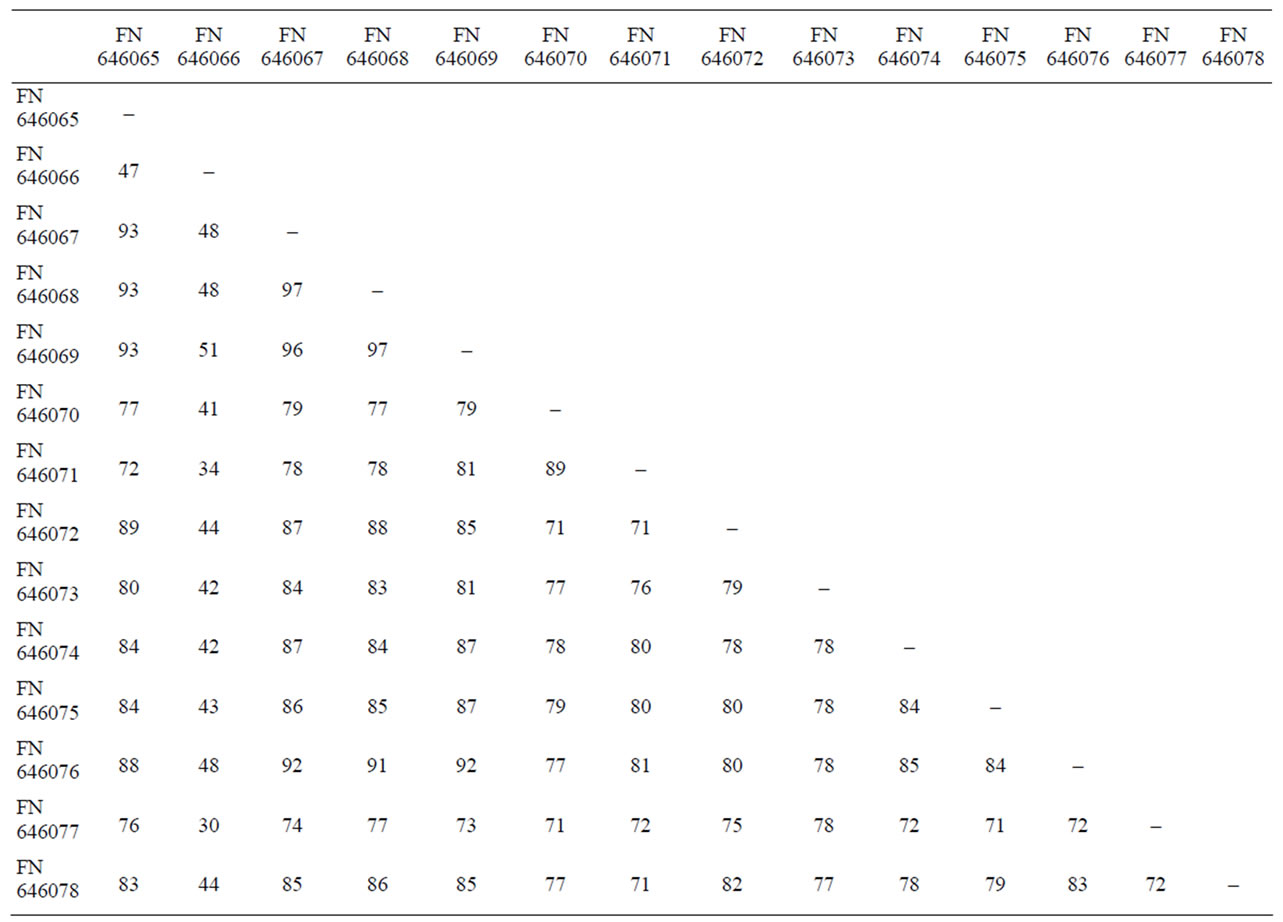
Table 3. Sequence similarity (%) of 16S rRNA gene of different Staphy-lococcus food isolates.
which showed a monophyletic clade of 100% bootstrap value. The phylogenetic tree showed different individual branches for isolates of S. saprophyticus and S. gallinarum.
4. Discussion
Using conventional biochemical test methods, S. aureus was identified as a major contaminant of street vends foods. Conventional biochemical methods used for the identification of Staphylococcus at species level are not so accurate as compared to molecular methods. Therefore, as an alternative multilocus methods such as RFLP analysis and 16S rRNA sequence analysis were used [21, 22]. With advances in molecular biology techniques, comparative DNA sequence analysis of genes of conserved macromolecules have become common in microbiology for taxonomic grouping of microorganisms, and the 16S rRNA gene is reported to be the most useful and extensively investigated taxonomic marker molecules [23]. 16S rRNA sequence analysis is more discriminative than other ribosomal regions for differentiating species and sub species of Staphylococcus, and it provides accurate identification of Staphylococcus at species level [24].
When compared with the 16S rRNA sequence analysis, biochemical tests were able to identify S. xylosus, S. sciuri and S. cohinii correctly, but misidentified other species. Two isolates of S. saprophyticus were misidentified biochemically as S. intermedius and S. aureus. The mismatching between biochemical and molecular identification may be due to the close relatedness among the species, as evident from the phylogenetic tree (Figure 2). This variability in results of biotyping and 16S rRNA sequence also has been reported in other study also [25]. Further, the identification to subspecies level is not possible by biochemical means and it is not possible to identify the S. saprophyticus subsp saprophyticus correctly by biochemical analysis. Molecular characterization is reported to be more accurate [23,26], and in present study also sequencing of the 16S rRNA fragment was discriminative enough to differentiate Staphylococcus food isolates at the sub species level. Kwok et al. [24] reported that neither 16S–23S rRNA intergenic space region nor genes such as hsp60 or sod A allow discrimi–



Table 4. 16S rRNA sequence analysis showing variable regions of different Staphylococcus food isolates.

Figure 2. Neighbour–joining phylogenetic tree of Staphylococcus food isolates constructed on the basis of 16S rRNA gene sequence.
nation at the sub species level, but results presented here show that 16S rRNA might be more discriminative target sequence to differentiate among subspecies. Pair wise comparison of the 16S rRNA gene sequences of 14 Staphylococcus food isolates ranged from 30% – 97% with mean similarity value of 63.5% (Table 3) indicating suitability of 16S rRNA gene for discrimination among the Staphylococcus food isolates, as the higher % similarity is less discriminatory [24]. While comparing relatedness among the food isolates, the phylogenetic tree (Figure 2) also showed distant groups indicating suitability of the method used for differentiation.
5. Conclusions
In present study, S. aureus was found to be the most prevalent species among the species of Staphylococcus identified in the street vend foods. Although pathogenicity of these species needs to be studied, their accurate identification is important, as many staphylococcal species are known for their enterotoxigenic potential. The 16S rRNA sequencing employed in this study for identification of Staphylococcus species may be useful for precise and timely identification and prevention of any untoward staphylococcal food poisoning incidences.
6. Acknowledgements
Authors are grateful to Dr. V. Prakash, Director, CFTRI and Dr. M. C. Varadaraj, Head, HRD, CFTRI for constant encouragement. YSR acknowledges the financial assistance in form of Senior Research Fellowship from Council of Scientific and Industrial Research, New Delhi for Ph. D. programme.
REFERENCES
- W. E. Kloos and K. H. Schleifer, “Genus IV Staphylococcus,” In: P. H. A Sneath, et al., Eds., Bergey’s Manual of Systematic Bacteriology, Vol. 2, Williams & Wilkins, Baltimore, 1986, pp. 1013-1035.
- H. Asperger and P. Zangerl, “Staphylococcus aureus,” In: H. Roginski, J. W. Fuquay and P. F. Fox, Eds., Encyclopedia of Dairy Sciences, Vol. 4, Academic Press and Elsevier Science, Amsterdam, 2003, pp. 2563-2569.
- J. A. Boerema, R. Clemens and G. Brightwell, “Evaluation of Molecular Methods to Determine Enterotoxigenic Status and Molecular Genotype of Bovine, Ovine, Human and Food Isolates of Staphylococcus aureus,” International Journal of Food Microbiology, Vol. 107, No. 2, 2006, pp. 192-201. doi:10.1016/j.ijfoodmicro.2005.07.008
- G. Normanno, A. Firinu, S. Virgilio, G. Mula, A. Dambrosio, A. Poggiu, et al, “Coagulase–Positive Staphylococci and Staphylococcus aureus in Food Products Marketed in Italy,” International Journal of Food Microbiology, Vol. 98, No. 1, 2005, pp. 73-79. doi:10.1016/j.ijfoodmicro.2004.05.008
- J. M. Jay, “Staphylococcal Gastroenteritis. Modern Food Microbiology,” 4th Edtion, Van Norstrand Reinhold, New York, 1992, pp. 455–478.
- A. A. Adesyn, S. R. Tatini and D. Hoover, “Production of Enterotoxin by Staphylococcus hyicus,” Veterenary Microbiology, Vol. 9, No. 5, 1984, pp. 487-495. doi:10.1016/0378–1135(84)90069–5
- K. Becker, B. Keller, C. Von Eiff, M. Brück, G. Lubritz, J. Etienne and G. Peters, “Enterotoxigenic Potential of Staphylococcus intermedius,” Applied and Environmental Microbiology, Vol. 67, No. 12, 2001, pp. 5551-5557. doi:10.1128/AEM.67.12.5551–5557.2001
- E. Y. Hirooka, E. E. Muller, J. C. Freitas, E. Vicente, Y. Yoshimot and M. S. Bergdoll, “Enterotoxigenicity of Staphylococcus intermedius of Canine Origin,” International Journal of Food Microbiology, Vol. 7, No. 3, 1988, pp. 185-191. doi:10.1016/0168–1605(88)90036–0
- F. M. Khambaty, R. W. Bennet and D. B. Shah, “Application of Pulse–Field Gel Electrophoresis to the Epidemiological Characterization of Staphylococcus intermedius Implicated in a Food–Related Outbreak,” Epidemiology and Infection, Vol. 113, No. 1, 1994, pp. 75-80. doi:10.1017/S0950268800051487
- L. Bautista, et al., “A Quantitative Study of Enterotoxin Production by Sheep Milk Staphylococci,” Applied and Environmental Microbiology, Vol. 54, No. 2, 1988, pp. 566-569.
- M. A. Al–Bustan, E. E. Udo and T. D. Chugh, “Enterotoxin Production by Coagulase–Negative Staphylococci in Restaurant Workers from Kuwait City May be a Potential Cause of Food Poisoning,” Journal of Medical Microbiology, Vol. 48, No. 9, 1999, pp. 819-823. doi:10.1099/00222615–48–9–819
- M. E. Marin, M. C. de la Rosa and I. Cornejo, “Enterotoxigenicity of Staphylococcus Strains Isolated from Spanish Dry–Cured Hams,” Applied and Environmental Microbiology, Vol. 58, No. 3, 1992, pp. 1067-1069.
- J. Valle, L. E. Gomez, S. Piriz, J. Goyache, J. A. Orden and S. Vadilo, “Enterotoxins Production by Staphylococcal Isolated from Healthy Goats,” Applied and Environmental Microbiology, Vol. 56, No. 5, 1990, pp. 1323- 1326.
- F. Vandenesch, S. J. Projan, B. Kreiswirth, J. Etienne and R. P. Novick, “Agr-Related Sequences in Staphylococcus lugdunensis,” FEMS Microbiology Letter, Vol. 111, No. 1, 1993, pp. 115-122. doi:10.1111/j.1574–6968.1993.tb06370.x
- T. L. Bannerman, “Staphylococcus, Micrococcus, and Other Catalase–Positive Cocci That Grow Aerobically,” In: P. R. Murray, et al., Eds., Manual of Clinical MicroBiology, ASM Press, Washington DC, 2003. pp. 384-404.
- D. W. Russell and J. Sambrook, “Molecular Cloning: A Laboratory Manual,” Cold Spring Harbor Laboratory, Cold Spring Harbor, 2001.
- L. H. Lambert, T. Cox, K. Mitchell, R. A. Rossello–Mora, C. D. Cueto, D. E. Dodge, P. Orkand and R. J. Cano, “Staphylococcus succinus sp. Nov, Isolated from Dominican Amber,” International Journal of Systematic Bacteriology, Vol. 48, No. 2, 1998, pp. 511-518.
- M. Kuroda, A. Yamashita, H. Hirakawa, M. Kumano, K. Morikawa, M. Higashide, A. Maruyama, Y. Inose, et al., “Whole Genome Sequence of Staphylococcus saprophyticus Reveals the Pathogenesis of Uncomplicated Urinary tract Infection,” Proceedings of the National Academy of Science, Vol. 102, No. 37, 2005, pp. 13272-13277. doi:10.1073/pnas.0502950102
- I. Tiago, A. P. Chung and A. Verissimo, “Bacterial Diversity in a Nonsaline Alkaline Environment: Heterotrophic aerobic Populations,” Applied and Environmental Microbiology, Vol. 70, No. 12, 2004, pp. 7378-7387. doi:10.1128/AEM.70.12.7378–7387.2004
- M. T. Holden, E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, et al. “Complete Genomes of Two Clinical Staphylococcus aureus Strains: Evidence for the Rapid Evolution of Virulence and Drug Resistance,” Proceedings of the National Academy of Science, Vol. 101, No. 26, 2004, pp. 9786- 9791. doi:10.1073/pnas.0402521101
- J. G. Neigel and A. C. Avise, “Phylogenetic Relationship of Mitochondrial DNA under Various Models of Speciation,” In: S. Karlin and E. Nevo, Eds., Evalution Process and Theory, Academic Press, New York, 1986, pp. 515- 534.
- N. Takahata and M. Nei, “Gene Genealogy and Variance of Interpopulational Nucleotides Difference,” Genetics, Vol. 110, No. 2, 1985, pp. 325-344.
- K. Becker, D. Harmsen, A. Mellmann, M. Christian, P. Schumann, G. Peters and C. von Eiff, “Development and Evaluation of a Quality–Controlled Ribosomal Sequence Database for 16S Ribosomal DNA–Based Identification of Staphylococcus Species,” Journal of clinical Microbiology, Vol. 42, No. 11, 2004, pp. 4988-4995. doi:10.1128/JCM.42.11.4988–4995.2004
- A. Y. C. Kwok, et al., “Species Identification and Phylogenetic Relationships Based on Partial HSP60 Gene Sequences within the Genus Staphylococcus,” International Journal of Systematic Bacteriology, Vol. 49, No. 3, 1999, pp. 1181-1192. doi:10.1099/00207713–49–3–1181
- M. Palka-Santini, S. Puzfeld, B. E. E. Cleven, M. Kronke and O. Krut, “Rapid Identification, Virulence Analysis and Resistance Profiling of Staphylococcus aureus by Gene Segment–Based DNA Microarrays: Application to Blood Culture Postprocessing,” Journal of Microbiological Methods, Vol. 44, No. 7, 2006, pp. 2389-2397.
- J. E. Clarridge III, “Impact of 16S rRNA Gene Sequence Analysis for Identification of Bacteria on Clinical Microbiology and Infectious Diseases,” Clinical Microbiology Review, Vol. 17, No. 4, 2004, pp. 840-862. doi:10.1128/CMR.17.4.840–862.2004

