American Journal of Plant Sciences
Vol.4 No.5(2013), Article ID:32139,8 pages DOI:10.4236/ajps.2013.45128
Plant Long ncRNAs: A New Frontier for Gene Regulatory Control
![]()
Department of Biochemistry, Molecular Biology, Entomology and Plant Pathology, Mississippi State University, Mississippi State, USA.
E-mail: *zp7@BCH.msstate.edu
Copyright © 2013 Jian Zhang et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received March 8th, 2013; revised April 15th, 2013; accepted May 10th, 2013
Keywords: Long Non-Coding RNA; Plants, Polycomb Repressing Complex 2; Gene Regulation
ABSTRACT
Long non-coding RNA (lncRNA) refers to an over 200 nt functional RNA molecule that will not be translated into protein. Previously thought to be dark matters of the genome, lncRNAs have been gradually recognized as crucial gene regulators. Although tremendous progress has been made in animals and human, the study of lncRNAs in plant is still in its infancy. Here, we reviewed the biogenesis and regulation mechanisms of lncRNAs and summarized the achievements that have been made in plant lncRNA identification and functional characterization. Genome-wide identification has uncovered large amount of lncRNAs in Arabidopsis, Rice, Maize and Wheat, and more information from other plant species will be expected with the aid of deep sequencing technologies. Similar to other species, LncRNAmediated gene regulation also widely exists in plants, even though only a few functionally characterized examples are available. Up to now, at least four divergent lncRNA-mediated regulation mechanisms have been unraveled, including target mimicry, transcription interference, PRC2 associated histone methylation and DNA methylation. lncRNAs may be involved in the regulation of flowering, male sterility, nutrition metabolism, biotic and abiotic stress response in plants.
1. Introduction
Classic central dogma indicates a flow of genetic information from DNA to RNA to protein. RNA molecules are the only messengers that pass information from DNA to protein, which ultimately decides the cellular function and phenotype. With the discovery of non-coding RNAs in the past decades, the classic central dogma has been greatly extended to encompass the developing roles of RNAs. A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into protein. Transcriptomic analyses by whole genome tiling arrays or transcriptome sequencing have revealed that 70% - 90% of the mammalian genome is transcriptionally active, but only 1% - 2% code for proteins, suggesting that a large proportion of mammalian RNAs are ncRNAs [1-3]. In the model plant Arabidopsis (Arabidopsis thaliana), less than 50% of its genome is capable of coding proteins [4]. In addition to the structural ncRNA such as transfer RNAs, ribosomal RNAs, small nuclear RNAs and small nucleolar RNAs, some of the ncRNAs are believed to play regulatory roles in eukaryotes. Based on the lengththe regulatory ncRNAs can be further divided into small ncRNAs (sncRNA, shorter than 200 nt) and long ncRNA (lncRNA, longer than 200 nt) [2,5,6]. Small regulatory RNAs, such as micro RNAs and small interfering RNAs, have been extensively studied and are well-known for their important roles in post-transcriptional and trancriptional regulations. However, the regulatory function of lncRNA, which takes 80% of the ncRNAs, largely remains unknown.
2. Biogenesis and Regulation Mechanism of lncRNA
The biogenesis of lncRNAs is very similar to that of protein-coding mRNAs and sncRNAs, most of the lncRNAs are transcribed by RNA polymerase II in eukaryotes. However, many novel lncRNAs have been found to be transcribed by RNA polymerase III, which was previously thought to only transcribe infrastructure RNAs like tRNA and 5S RNA [7]. Large-scale complementary DNA (cDNA) sequencing projects such as FANTOM (Functional Annotation of Mammalian cDNA) has revealed that lncRNAs have many common features of mRNAs, including 5’capping, splicing, and polyadenylation [1]. Most of the lncRNAs are localized in the nucleus with a few exceptions that localized in cytosolic fractions [8,9]. lncRNAs can originate from intronic, exonic, intergenic, intragenic, promoter regions, 3’-and 5’ UTR, and enhancer sequences and can be transcribed in either the sense or antisense direction [10]. Natural Antisense Transcripts (NATs) is referred to the antisense transcripts of protein-coding transcripts. NATs are broadly grouped into two categories based on whether they act in cis or in trans [11]. The so-called cis-NATs are transcribed from the same loci as sense transcripts and therefore have perfect match with the sense transcripts. On the contrary, trans-NATs are transcribed from different genomic loci and usually display only partial complementarity with the sense transcript. NATs are widespread in eukaryotes with evidences showing that between 5% - 30% of transcriptional units in diverse eukaryotes have been found to have cis-NATs [12]. Conley et al. (2008) have found each protein-coding locus is associated with an average of 6 cis-NATs in Human [13]. Some literature also shows that NATs represent a significant portion of the transcriptome in plants [14-16]. Recent studies have implied important roles of NATs in the regulation of gene transcription. For example, the human inherited form of α- thalassemia is caused by the silencing of the hemoglobin α-2 gene through cis-NAT action [17].
Various mechanisms have been proposed for lncRNA in the regulation of gene transcription [18]. The regulation mechanism is to a large extent determined by the genomic location of lncRNA transcription. Cis-NATs or exotic lncRNAs derived from protein-coding locis usually take a “transcriptional interference” effect in regulation. Because the promoters of the target gene and lncRNA are close to each other, the transcription initiated from both promoters may be co-regulated. This coregulation may occur via the competition for RNA ploymerase II and other initiation factors or via the premature termination of elongation complex from both sense and antisense promoters due to the interaction with each other [19]. However, there is also a report showing that lncRNA can enhance the accessibility of target genes to RNA polymerase, rather than competing for the RNA polymerase [20]. Some lncRNAs can bind to the promoter DNA of target gene to form a RNA-dsDNA triplex, thus physically block the preinitiation complex from the target gene promoter [21]. Other lncRNAs may also regulate target genes in the transcriptional level by controlling the transcription factor subcellular localization or inhibiting the RNA polymerase activities [22-24].
Other than the regulation at the transcriptional level, lncRNAs also post-transcriptionally modulate various aspects of mRNA processing, including pre-mRNA alternative splicing, transport, translation, and degradation. For instance, translation of Zeb2 mRNA needs the retention of its long 5’UTR intron due to an internal ribosome entry site in it. A natural antisense transcript (NAT) that overlaps the 5' splice site in the intron can prevent the intron from being spliced out, thus keep the Zeb2 translated [25]. Besides alternative splicing, lncRNAs may also regulate target mRNA stability via a trans-acting manner. Similar to microRNAs, this requires complementary base paring with target mRNA so that a double strand RNA duplex can be formed, and then the RNA duplex can be processed into endo-siRNAs to degrade the target mRNA [26].
Recently, emerging evidence has shown that lncRNAs may play essential roles in the epigenetic control of gene expression, including Tsix/RepA/Xist, HOTAIR and COLDAIR [27-30]. X-inactive specific transcript (Xist) RNA is one of the earliest examples that lncRNA regulate gene transcription by modifying the chromatin status [30]. In female mammals, one of the two copies of the X chromosome is inactivated to maintain the same dosage of gene products as males. Xist, an lncRNA of 17kb in mouse or 19kb in human, is a major effector in X chromosome silencing. Xist is expressed in the inactive X-chromosome during early embryonic stem cell differentiation and coated on the chromosome region. Subsequently, the coated Xist recruits Polycomb repressor complex 1 (PRC1), which mediates histone H2A lysine 119 ubiquitinylation (H2AK119ub1), and PRC2, which mediates histone H3 lysine 27 trimethylation (H3K27 me3) to enrich the suppressing chromatin modifications. It is believed that RNA coating together with the histone modification and DNA methylation build the epigenetic memory for the mitotically heritable X-chromosome inactivation [18].
3. lncRNAs in Plants
Compared with human and animals, the transcriptomic identification of lncRNA in plants is still in its infancy. Fortunately, some dramatic progress has been made in the last few years with the aid of the novel highthroughput sequencing techniques. By sequencing the full length cDNA from various tissues and hybridizing RNA populations to whole genome arrays (WGA), Yamada et al. (2003) analyzed the transcripitional activity of Arabidopsis genome [4]. About 7600 (30%) of the annotated genes in their study were found to have significant antisense RNA expression. Interestingly, the sense and antisense RNAs for many genes displayed a tissuespecific expression pattern, indicating that these transcripts are biologically functional. Another study showed that 76 Arabidopsis ncRNAs were identified through in silica genome-wide analysis of full-length cDNA databases, including 5 siRNA precursors, 14 natural antisense transcipts of protein-coding genes [31]. According to the stress expression profile, 22 lncRNAs were found to be related to abiotic stress response. Ectopic expression of two lncRNAs affected Arabidopsis differentiation and growth responses to abiotic stresses. Very recently, a deep sequencing of mRNA for assessing sense and antisense transcripts that were derived from different abiotic stresses and normal conditions have uncovered 3819 putative rice cis-NATs, out of which 2292 are potential small RNA precursors and 503 may be related with abiotic stress response. The deep sequence data from isolated epidermal cells of rice seedlings, a special type of homogenous cells, showed that 54.0% of cis-NATs were expressed simultaneously [32]. lncRNAs appears to be widespread in Maize as well. Boerner and McGinnis (2012) attempted to identify, classify and localize potential lncRNAs in maize genome by using a computational pipeline [33]. Totally 1802 lncRNAs were identified from 18,668 full-length cDNA sequences with over 200 bp in length, 60% of which are annotated to be small RNA precursors. Cis-NATs take a proportion of 20% of the lncRNAs derived from the genic region. In wheat, Xin et al. (2011) also identified 71 powdery mildew infection responsive lncRNAs and 77 heat shock responsive lncRNAs respectively [34].
Despite the identification of lncRNAs in plant genomes, our understanding of the functions of lncRNAs remains rather poor. So far, most of our knowledge in lncRNA regulation is from FLC , a master repressor of flowering in Arabidopsis. Vernalization is the acquisition of certain plant's ability to flower or germinate in the spring by exposure to the prolonged cold of winter. Prolonged cold treatment induces the expression of VIN3 (Vernalization insensitive 3), which can recruit the chromatin remodeling complex PRC2 to deposit the suppressing H3K27me3 modification on the FLC locus, thus stably maintain the FLC gene in a “shut down” status. By using a single nucleotide resolution array which covers both strands of FLC and 50kb adjacent region, Swiezewski et al. (2009) indentified a serial of cold induced antisense transcripts covering the entire FLC locus and named them as COOLAIR (cold induced antisense intragenic RNA) [35]. Like many other lncRNAs in mammals, COOLAIR are capped, have poly A tails and are alternatively spliced. Supported by the evidence that the reaction to cold of COOLAIR was even earlier than VIN3, the earliest factor in the polycomb silencing mechanism, COOLAIR is believed to negatively regulate FLC sense transcription in a polycombindependent manner. This conclusion is also reinforced by the fact that COOLAIR remains transcribed in the absence of VIN3. However, COOLAIR repression effect is reversible after the plants moved back from cold to warm conditions, suggesting that COOLAIR only transiently suppresses the FLC , and polycomb machinery is indispensable for the construction of epigenetic memory of FLC inactivation. Because the COOLAIR and nascent FLC expression levels are negatively correlated, and a reduction in occupancy of RNA polII at FLC was observed with the presence of COO LAIR, it seemed that COOLAIR suppresses FLC by a transcription interference manner. Thus, a regulation model of FLC in vernalization was speculated with the following possible path: COOLAIR induction during cold, transient suppression of FLC sense transcription, up-regulation of VIN3, recruitment of polycomb repression complex 2 by VIN3, epigenetic memory construction of FLC inactivation (Figure 1(A)). Although, it has been demonstrated that COOLAIR suppresses FLC through transcription interference, in particular promoter interference, the possible involvement of any post-transcriptional machinery still needs to be explored in future studies.
COLDAIR is another class of lncRNA derived from FLC locus [27]. Divergent from COOLAIR, COLDAIR is a sense transcript starting from the first intron of FLC, and it is approximately 1100 bases long with 5’ cap but no obvious polyA tail was found in the 3’ end, which are typical features of lncRNAs that are transcribed by RNA polymerase V and IV. Nevertheless, COLDAIR is transcribed by RNA polymerase II like many other lncRNAs in mammals. The COLDAIR expression is induced by cold exposure, reaches a peak after 20 days of treatment, and returns back to the prevernalized level after 30 days of cold. This kind of expression pattern is very similar to COOLAIR, except the peak expression comes 10 days later than COOLAIR [35]. The COLDAIR knock-down plants showed late flowering after vernalization, which can be explained by the phenomena that there is no enrichment of H3K27me3 on the FLC chromatin. More interestingly, RNA binding assay showed that COLDAIR specifically interact with CLF (Curly Leaf), a key component of PRC2 complex for H3K27me3 modification. Hence, it is very likely that COLDAIR negatively modulates FLC via a polycomb-dependent model. COLDAIR may play a role in the recruitment of PRC2 to FLC chromatin to trigger the epigenetic memory establishment of FLC silencing by vernalization (Figure 1(B)).
Plant lncRNA was also reposted to regulate gene expression via a target mimicry mechanism [36]. The non-protein coding gene IPS1 (Induced By Phosphate Starvation1) from Arabidopsis thaliana is highly induced by phosphate starvation. Accumulation of IPS1 can reduce the phosphate content in shoot of plants by upregulating the expression of PHO2, a negative regulator
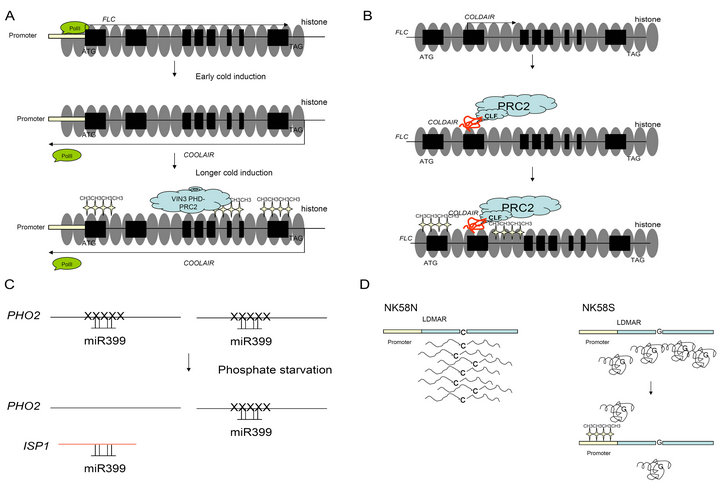
Figure 1. Schematic representation of four types of lncRNA regulation mechanism in plants. (A) COOLAIR regulates FLC in a transcription interference model. Top: FLC is transcribed by RNA Polymerase II before prolonged cold exposure; Middle: early cold treatment induced the expression of COOLAIR, which interferes with the RNA polymerase II binding to the FLC promoter, thus transiently suppresses the FLC expression; Bottom: after longer cold exposure, VIN3 recruits PRC2 complex to deposit H3K27me3 modification on FLC loci; (B) COLDAIR regulates FLC in a PRC2 associated histone modification model. Top: COLDAIR is induced by cold treatment. Middle: COLDAIR recruit PRC2 complex to the FLC loci; Bottom: PRC2 complex deposit H3K27me3 on the FLC loci; (c) ISP1 regulates PHO2 in a target mimicry model. Top: under normal growth condition, miR399 specifically binds to PHO2 and degrades PHO2 mRNA; Bottom: ISP1 competitively binds with miR399 to arrest its degradation function on PHO2; (D) LDMAR regulates the transcription of itself by a DNA methylation model. In NK58N, LDMAR is normally expressed. In NK58S, the C-to-G mutation altered the secondary structure of LDMAR and leads to the promoter DNA methylation, which reduced the LDMAR expression responsible for PSMS in NK58S.
for phosphate uptake. Interestingly, miR399, which specifically degrades PHO2, was found partially complementary with conserved 23-nt motif of IPS1. The imperfect complementation allows IPS1 to form a RNA duplex with miR399, instead of being degraded by miR399, thus arrest miR399 for its normal function in phosphate translocation. Meanwhile, modified miR399-cleavable IPS1 did not own any inhibitory activity on miR-399. Therefore, it seems that the uncleavable IPS1 competes with PHO2 for miR399 to keep a balance of functional miR 399 in phosphate absorption (Figure 1(C)).
In rice, an lncRNA LDMAR was cloned for controlling PSMS (Photo-sensitive male sterility). Originated from an elite japonica rice variety Nongken 58N (NK 58N), Nongken 58S (NK58S) was a spontaneous mutant exhibiting PSMS, i.e. its pollen becomes completely sterile when grown under long-day conditions, whereas the pollens are viable under short-day growth conditions. By using a position cloning strategy, Ding et al. (2011) successfully cloned the LDMAR gene, in which a C-to-G mutation caused the PSMS in NK 58S [37]. LDMAR is 1236 bases in length and non-protein-coding, essentially an lncRNA. Pollen fertility under long-day conditions for both NK58N and NK58S requires a high dosage of LDMAR. However, the transcript abundance of LDMAR in NK 58S is lower than NK58N under long-days, possibly due to the fact that the DNA in the promoter region of NK 58S LDMAR is more methylated than that
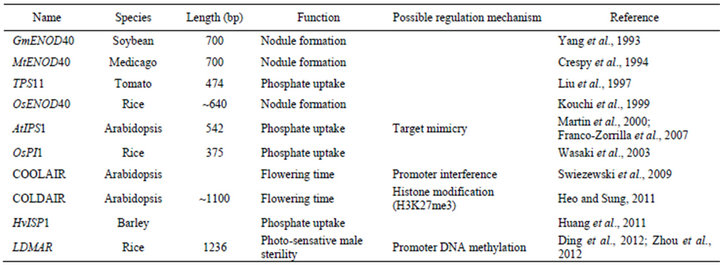
Table 1. A summary of the reported lncRNA genes in plants.
of NK 58N LDMAR. Based on the findings, the authors proposed that the C-to-G mutation altered the secondary structure of LDMAR in NK58S, and the structural alteration brought DNA methylation in the promoter region, which suppressed the LDMAR expression, and the insufficient LDMAR eventually led to the sterility of NK 58S under long-days (Figure 1D). Later, Psi-LDMAR, a siRNA derived from the sense strand of LDMAR promoter region was also found to be responsible for the regulation of DNA methylation in this regoin [38]. Closely following Ding et al’s work (2011), another group narrowed the essential sequence for PMSA in LDMAR locus to a 136-nt small RNA [39]. Nevertheless, the data presented so far has not excluded the possibility that either the 1236 nt lncRNA or 136 nt small RNA or both are the functional form of LDMAR [40].
In addition to the lncRNAs reviewed above, a few other lncRNAs from plant were also reported, including the Enod40 related to soil bacteria-plant interaction in Medicago and Soybean [41,42]; IPS1 related to phosphate metabolism in tomato, Medicago and rice [43-45] (Table 1).
4. Epigenetic Regulation of Genes by Polycomb Associated LncRNAs
Polycomb-group proteins (PcG) are a family of proteins that can regulate the target genes expression by remodeling the structures of chromatin associated with the target genes. PcG proteins function by forming three principle types of hetero multimeric complexes, including polycomb repressive complex 1 (PRC1), polycomb repressive complex 2 (PRC2) and Pho RC. The PRC2 complex is mostly conserved among eukaryotes and has been extensively studied [46]. It has been demonstrated that PRC2 catalyzes trimethylation on the lysine 27 of histone 3 (H3K27me3) to suppress the chromatin associated gene expression, while PRC1 represses gene expression by catalyzing the monoubiquitination of histone H2A at lysine 119 (H2AK119ub).
It should be noted that PRC2 is recruited by lncRNAs in several reported cases, implying a widespread of the lncRNA-PRC2 gene silencing mechanism in different species [47]. So far, the question concerning how is PRC 2 recruited to the right chromatin region has not been fully addressed. Tsai et al. (2010) suggsted that lncRNAs may serve as scaffolds by providing binding surfaces for polycomb complex assembling, thereby specifying the pattern of histone modifications on target genes in a temporaland spatial-manner [29].
Genome-wide identification of lncRNAs that associated with polycomb complex, especially PRC2, will be extremely helpful for the understanding of lncRNAmediated epigenetic regulation and the molecular function of lncRNAs. Up to now, some research has been done in this field. Khali et al. (2009) tried to identify human PRC2-associated lncRNAs by using a method of RIP (RNA co-immunoprepcipitation) followed by hybridization to tilling array [48]. As a result, around 20% of lncRNAs were bound by PRC2, and additional lncRNAs were bound by other chromatin modifying complexes. Employing a RNA immunoprecipitation sequencing (RIPSeq) method, over 9000 PRC2-interacting RNAs, half of which are non-coding RNAs, were identified in mouse [49]. It is very likely that Ezh2 bridged the RNA with PRC2 for interactions. By using a novel method Chromatin Isolation by RNA Purification (ChIRP), Chu et al. (2011) revealed the focal and sequence-specific distribution patterns of three lncRNA occupancy sites in Drosophila and human genomes [50]. Drosophila roX2 RNA favors the male X-linked gene region; Human telomerase RNA TERC distributed more on the telomeres, while HO TAIR lncRNA preferentially occupies a GA-rich DNA motif. These featured distribution patterns suggested that lncRNAs regulate chromatin status in a sequence-specific manner. Unfortunately, to the best of our knowledge, no work on the PRC2 associated lncRNA identification in plants has been reported so far.
5. Conclusions and Perspectives
Although the study in plant lncRNAs started much later than human and animals, tremendous progresses have been made in the past few years. With the aid of new, affordable deep sequencing technologies and bioinformatics, we can anticipate a knowledge explosion of plant lncRNAs identifications in the following years. Currently, we still rely on the bioinformatic prediction of open reading frame to determine if a RNA is coding or noncoding. It becomes extremely complicated to clearly distinguish coding RNA from non-coding RNA, since coding RNA can be short with a small ORF, while noncoding RNA derived from genic region may contain long ORF with homology to known proteins [51]. Therefore, the proteomic identification of plant proteins will be necessary to at least exclude the coding RNAs in the transcriptome. Meanwhile, how to remove the transcript noise from the transcriptome remains to be a great challenge in lncRNA identification.
With only a few lncRNAs functionally characterized so far, the function of lncRNAs in plants is largely a mystery. The emerging role of lncRNA in human, animals as well as plant development have indicated that lncRNA-mediated gene regulation widely exists and is conserved among eukaryotes. Thus, the knowledge gained from other organisms may be applied to plants. For examples, since some of the cellor tissue-specific expression patterns are epigenetically controlled by lnc RNAs through a PRC2 complex recruiting mechanism, it is rationale to speculate that some of the plant development processes are modulated via the same mechanism. On the other hand, the new techniques such as RIP-Seq and ChIRP, which have been successfully used to identify lncRNA interactive DNA, RNA and proteins in other species, can definitely be used in plants and facilitate the functional characterization of plant lncRNAs.
6. Acknowledgements
The research is supported by a USDA grant (2005- 3560415357 and 2011-67013-30125) to Zhaohua Peng.
REFERENCES
- P. Carninci, et al., “The Transcriptional Landscape of the Mammalian Genome,” Science, Vol. 309, No. 5740, 2005, pp. 1559-1563. doi:10.1126/science.1112014
- P. Kapranov, et al., “RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription,” Science, Vol. 316, No. 5830, 2007, pp. 1484-1488. doi:10.1126/science.1138341
- T. R. Mercer, M. E. Dinger and J. S. Mattick, “Long Non-Coding RNAs: Insights into Functions,” Nature Reviews Genetics, Vol. 10, 2009, pp. 155-159. doi:10.1038/nrg2521
- K. Yamada, et al., “Empirical Analysis of Transcriptional Activity in the Arabidopsis Genome,” Science, Vol. 302, No. 5646, 2003, pp. 842-846. doi:10.1126/science.1088305
- Y. Jia, W. Liu, A. Li, L. Yang and X. Zhan, “Intrinsic Noise in Post-Transcriptional Gene Regulation by Small Non-Coding RNA,” Biophysical Chemistry, Vol. 143, No. 1-2, 2009, pp. 60-69. doi:10.1016/j.bpc.2009.04.001
- U. A. Orom, et al., “Long Noncoding RNAs with Enhancer-Like Function in Human Cells,” Cell, Vol. 143, 2010, pp. 46-58. doi:10.1016/j.cell.2010.09.001
- G. Dieci, G. Fiorino, M. Castelnuovo, M. Teichmann and A. Pagano, “The Expanding RNA Polymerase III Transcriptome,” Trends in Genetics, Vol. 23, No. 12, 2007, pp. 614- 622. doi:10.1016/j.tig.2007.09.001
- R. Louro, T. El-Jundi, H. I. Nakaya, E. M. Reis and S. Verjovski-Almeida, “Conserved Tissue Expression Signatures of Intronic Noncoding RNAs Transcribed from Human and Mouse Loci,” Genomics, Vol. 92, 2008, pp. 18-25. doi:10.1016/j.ygeno.2008.03.013
- T. R. Mercer, M. E. Dinger, S. M. Sunkin, M. F. Mehler and J. S. Mattick, “Specific Expression of Long Noncoding RNAs in the Mouse Brain,” Proceedings of the National Academy of Sciences of USA, Vol. 105, No. 2, 2008, pp. 716-721. doi:10.1073/pnas.0706729105
- L. Nie, et al., “Long Non-Coding RNAs: Versatile Master Regulators of Gene Expression and Crucial Players in Cancer,” American Journal of Translational Research, Vol. 4, No. 2, 2012, pp. 127-150.
- J. Chen, et al., “Over 20% of Human Transcripts Might Form Sense-Antisense Pairs,” Nucleic Acids Research, Vol. 32, No. 16, 2004, pp. 4812-4820. doi:10.1093/nar/gkh818
- M. Lapidot and Y. Pilpel, “Genome-Wide Natural Antisense Transcription: Coupling Its Regulation to Its Different Regulatory Mechanisms,” EMBO Report, Vol. 7, 2006, pp. 1216-1222. doi:10.1038/sj.embor.7400857
- A. B. Conley and I. K. Jordan, “Epigenetic Regulation of Human Cis-Natural Antisense Transcripts,” Nucleic Acids Research, Vol. 40, No. 4, 2008, pp. 1438-1445. doi:10.1093/nar/gkr1010
- S. R. Henz, J. S. Cumbie, K. D. Kasschau, J. U. Lohmann, J. C. Carrington, D. Weigel and M. Schmid, “Distinct Expression Patterns of Natural Antisense Transcripts in Arabidopsis,” Plant Physiology, Vol. 144, No. 3, 2007, pp. 1247-1255. doi:10.1104/pp.107.100396
- L. Li, et al., “Global Identification and Characterization of Transcriptionally Active Regions in the Rice Genome,” PLoS One, Vol. 2, No. 3, 2007, e294. doi:10.1371/journal.pone.0000294
- J. Ma, D. J. Morrow, J. Fernandes and V. Walbot, “Comparative Profiling of the Sense and Antisense Transcriptome of Maize Lines,” Genome Biology, Vol. 7, 2006, p. R22. doi:10.1186/gb-2006-7-3-r22
- Y. Zhang, X. S. Liu, Q. R. Liu and L. Wei, “GenomeWide in Silico Identification and Analysis of Cis Natural Antisense Transcripts (cis-NATs) in Ten Species,” Nucleic Acids Research, Vol. 34, No. 12, 2006, pp. 3465- 3475. doi:10.1093/nar/gkl473
- C. P. Ponting, P. L. Oliver and W. Reik, “Evolution and Functions of Long Noncoding RNAs,” Cell, Vol. 136, No. 4, 2009, pp. 629-641. doi:10.1016/j.cell.2009.02.006
- A. Mazo, J. W. Hodgson, S. Petruk, Y. Sedkov and H. W. Brock, “Transcriptional Interference: An Unexpected Layer of Complexity in Gene Regulation,” Journal of Cell Science, Vol. 120, 2007, pp. 2755-2761. doi:10.1242/jcs.007633
- K. Hirota, T. Miyoshi, K. Kugou, C. S. Hoffman, T. Shibata and K. Ohta, “Stepwise Chromatin Remodelling by a Cascade of Transcription Initiation of Non-Coding RNAs,” Nature, Vol. 456, No. 7281, 2008, pp. 130-134. doi:10.1038/nature07348
- I. Martianov, A. Ramadass, A. Serra Barros, N. Chow and A. Akoulitchev, “Repression of the Human Dihydrofolate Reductase Gene by a Non-Coding Interfering Transcript,” Nature, Vol. 445, 2007, pp. 666-670. doi:10.1038/nature05519
- P. D. Mariner, R. D. Walters, C. A. Espinoza, L. F. Drullinger, S. D. Wagner, J. F. Kugel and J. A. Goodrich, “Human Alu RNA Is a Modular Transacting Repressor of mRNA Transcription during Heat Shock,” Molecular Cell, Vol. 29, No. 4, 2008, pp. 499-509. doi:10.1016/j.molcel.2007.12.013
- V. T. Nguyen, T. Kiss, A. A. Michels and O. Bensaude, “7SK Small Nuclear RNA Binds to and Inhibits the Activity of CDK9/Cyclin T Complexes,” Nature, Vol. 414, No. 6861, 2001, pp. 322-325. doi:10.1038/35104581
- A. T. Willingham, A. P. Orth, S. Batalov, E. C. Peters, B. G. Wen, P. Aza-Blanc, J. B. Hogenesch and P. G. Schultz, “A Strategy for Probing the Function of Noncoding RNAs Finds a Repressor of NFAT,” Science, Vol. 309, No. 5740, 2005, pp. 1570-1573. doi:10.1126/science.1115901
- M. Beltran, I. Puig, C. Pena, J. M. Garcia, A. B. Alvarez, R. Pena, F. Bonilla and A. G.de Herreros, “A Natural Antisense Transcript Regulates Zeb2/Sip1 Gene Expression during Snail1-Induced Epithelial-Mesenchymal Transition,” Genes and Development, Vol. 22, 2008, pp. 756- 769. doi:10.1101/gad.455708
- D. E. Golden, V. R. Gerbasi and E. J. Sontheimer, “An Inside Job for siRNAs,” Molecular Cell, Vol. 31, No. 3, 2008, pp. 309-312. doi:10.1016/j.molcel.2008.07.008
- J. B. Heo and S. Sung, “Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA,” Science, Vol. 331, No. 6013, 2011, pp. 76-79. doi:10.1126/science.1197349
- J. L. Rinn, et al., „Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs,” Cell, Vol. 129, No. 7, 2007, pp. 1311- 1323. doi:10.1016/j.cell.2007.05.022
- M. C. Tsai, et al., “Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes,” Science, Vol. 329, No. 5992, 2010, pp. 689-693. doi:10.1126/science.1192002
- J. Zhao, B. K. Sun, J. A. Erwin, J. J. Song and J. T. Lee, “Polycomb Proteins Targeted by a Short Repeat RNA to the Mouse X Chromosome,” Science, Vol. 322, No. 5902, 2008, pp. 750-756. doi:10.1126/science.1163045
- B. Ben Amor, et al., “Novel Long Non-Protein Coding RNAs Involved in Arabidopsis Differentiation and Stress Responses,” Genome Research, Vol. 19, 2009, pp. 57-69. doi:10.1101/gr.080275.108
- T. Lu, et al., “Strand-Specific RNA-seq Reveals Widespread Occurrence of Novel Cis-Natural Antisense Transcripts in Rice,” BMC Genomics, Vol. 13, 2012, p. 721. doi:10.1186/1471-2164-13-721
- S. Boerner and K. M. McGinnis, “Computational Identification and Functional Predictions of Long Noncoding RNA in Zea Mays,” PLoS One, Vol. 7, No. 8, 2012, e43047. doi:10.1371/journal.pone.0043047
- M. Xin, et al., “Identification and Characterization of Wheat Long Non-Protein Coding RNAs Responsive to Powdery Mildew Infection and Heat Stress by Using Microarray Analysis and SBS Sequencing,” BMC Plant Biology, Vol. 11, 2011, p. 61. doi:10.1186/1471-2229-11-61
- S. Swiezewski, F. Liu, A. Magusin and C. Dean, “ColdInduced Silencing by Long Antisense Transcripts of an Arabidopsis Polycomb Target,” Nature, Vol. 462, No. 7274, 2009, pp. 799-802. doi:10.1038/nature08618
- J. M. Franco-Zorrilla, et al., “Target Mimicry Provides a New Mechanism for Regulation of Microrna Activity,” Nature Genetics, Vol. 39, No. 8, 2007, pp. 1033-1037. doi:10.1038/ng2079
- J. Ding, et al., “A Long Noncoding RNA Regulates Photoperiod-Sensitive Male Sterility, an Essential Component of Hybrid Rice,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 109, No. 7, 2011, pp. 2654-2659. doi:10.1073/pnas.1121374109
- J. Ding, J. Shen, H Mao, W. Xie, X. Li and Q. Zhang, “RNA-Directed DNA Methylation Is Involved in Regulating Photoperiod-Sensitive Male Sterility in Rice,” Molecular Plant, Vol. 5, No. 6, 2012, pp. 1210-1216. doi:10.1093/mp/sss095
- H. Zhou, et al., “Photoperiodand Thermo-Sensitive Genic Male Sterility in Rice Are Caused by a Point Mutation in a Novel Noncoding RNA That Produces a Small RNA,” Cell Research, Vol. 22, No. 4, 2012, pp. 649-660. doi:10.1038/cr.2012.28
- D. Zhu and X. W. Deng, “A Non-Coding RNA Locus Mediates Environment-Conditioned Male Sterility in Rice,” Cell Research, Vol. 22, No. 5, 2012, pp. 791-792. doi:10.1038/cr.2012.43
- M. D. Crespi, E. Jurkevitch, M. Poiret, Y. d’AubentonCarafa, G. Petrovics, E. Kondorosi and A. Kondorosi, “Enod40, a Gene Expressed During Nodule Organogenesis, Codes for a Non-Translatable RNA Involved in Plant Growth,” The Embo Journal, Vol. 13, No. 21, 1994, pp. 5099-5112.
- W. C. Yang, et al., “Characterization of GmENOD40, A Gene Showing Novel Patterns of Cell-Specific Expression during Soybean Nodule Development,” The Plant Journal, Vol. 3, No. 4, 1993, pp. 573-585. doi:10.1046/j.1365-313X.1993.03040573.x
- S. H. Burleigh and M. J. Harrison, “A Novel Gene Whose Expression in Medicago Truncatula Roots Is Suppressed in Response to Colonization by Vesicular-Arbuscular Mycorrhizal (VAM) Fungi and to Phosphate Nutrition,” Plant Molecular Biology, Vol. 34, No. 2, 1997, pp. 199-208. doi:10.1023/A:1005841119665
- C. Liu, U. S. Muchhal and K. G. Raghothama, “Differential Expression of TPS11, a Phosphate StarvationInduced Gene in Tomato,” Plant Molecular Biology, Vol. 33, No. 5, 1997, pp. 867-874. doi:10.1023/A:1005729309569
- J. Wasaki, R. Yonetani, T. Shinano, M. Kai and M. Osaki, “Expression of The OsPI1 Gene, Cloned from Rice Roots Using cDNA Microarray, Rapidly Responds to Phosphorus Status,” New Phytologist, Vol. 158, No. 2, 2003, pp. 239-248. doi:10.1046/j.1469-8137.2003.00748.x
- A. He, et al., “Polycomb Repressive Complex 2 Regulates Normal Development of The Mouse Heart,” Circulation Research, Vol. 110, No. 3, 2009, pp. 406-415. doi:10.1161/CIRCRESAHA.111.252205
- B. R Nallamilli, J. Zhang, H. Mujahid, B. M. Malone, S. M. Bridges and Z. Peng, “Polycomb Group Gene OsFIE2 Regulates Rice (Oryza sativa) Seed Development and Grain Filling via a Mechanism Distinct from Arabidopsis,” PLoS Genetics, Vol. 9, No. 3, 2013, Article ID: e1003322. doi:10.1371/journal.pgen.1003322
- A. M. Khalil, et al., “Many Human Large Intergenic Noncoding RNAs Associate with Chromatin-Modifying Complexes and Affect Gene Expression,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 106, No. 28, 2009, pp. 11667-11672. doi:10.1073/pnas.0904715106
- J. Zhao, et al., “Genome-Wide Identification of PolycombAssociated RNAs by RIP-seq,” Molecular Cell, Vol. 40, No. 6, 2010, pp. 939-953. doi:10.1016/j.molcel.2010.12.011
- C. Chu, K. Qu, F. L. Zhong, S. E. Artandi and H. Y. Chang, “Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions,” Molecular Cell, Vol. 44, No. 4, 2011, pp. 667-678. doi:10.1016/j.molcel.2011.08.027
- M. E. Dinger, K. C. Pang, T. R. Mercer and J. S.Mattick, “Differentiating Protein-Coding and Noncoding RNA: Challenges and Ambiguities,” PLoS Computational Biology, Vol. 4, No. 11, 2008, Article ID: e1000176. doi:10.1371/journal.pcbi.1000176
NOTES
*Corresponding author.

