Advances in Microbiology
Vol.3 No.6A(2013), Article ID:38616,9 pages DOI:10.4236/aim.2013.36A011
Alternative Nitrogenases in Anabaena variabilis: The Role of Molybdate and Vanadate in Nitrogenase Gene Expression and Activity
Department of Biology, University of Missouri-St. Louis, St. Louis, USA
Email: *thiel@umsl.edu
Copyright © 2013 Teresa Thiel, Brenda S. Pratte. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received August 30, 2013; revised September 2, 2013; accepted September 15, 2013
Keywords: Cyanobacteria; V-Nitrogenase; Molybdate; Vanadate; Tungstate
ABSTRACT
Anabaena variabilis ATCC 29413 has two distinct nitrogenases that function in heterocysts, a conventional Mo-nitrogenase and an alternative V-nitrogenase. Synthesis of these two enzymes was repressed in cells growing with a source of fixed nitrogen, such as ammonium; however, the V-nitrogenase was also repressed by Mo. Expression of the V-nitrogenase which was not affected by V and expression of the Mo-nitrogenase was not affected by the presence or absence of either Mo or V. In the absence of both Mo and V in an environment lacking fixed nitrogen, cells became starved for both metals; however, low levels of nitrogen fixation and slow growth persisted. A mutant lacking the V-nitrogenase was still able to grow very slowly in Moand V-free medium; however, loss of the Mo-nitrogenase in a nifDK1 mutant abolished the residual growth, suggesting that only the Mo-nitrogenase functioned under these conditions to support slow growth. The addition of vanadate, molybdate, or tungstate, which is transported by the molybdate transporter, to cells starved for these metals resulted in an increase in nitrogenase activity within two hours after the addition of the metal and this increase required new protein synthesis. While tungstate functioned about as well as vanadate in supporting acetylene reduction, the cells were not able to grow any better with tungstate than with no added metal. A mutant lacking the V-nitrogenase showed no increase in nitrogenase activity upon addition of tungstate, suggesting that the V-nitrogenase was able to incorporate tungstate. Tungstate was able to substitute for molybdate in repressing transcription of a Mo-transport gene, but it did not repress transcription of the vnfH gene, which was repressed by Mo. The availability of Mo and V plays an important role in controlling whether the Moor the V-nitrogenase is used for nitrogen fixation.
1. Introduction
In filamentous heterocyst-forming cyanobacteria, aerobic nitrogen fixation occurs exclusively in specialized microoxic cells called heterocysts that differentiate at semiregularly spaced intervals in the filament in response to nitrogen deprivation (reviewed [1-3]). Under these conditions, nitrogenase is synthesized only in heterocysts, which protect the oxygen-labile enzyme from oxygen by preventing oxygen-evolving photosystem II activity, increasing respiration and synthesizing a thick glycolipid layer that limits oxygen diffusion into the cell [4,5]. The differentiation of heterocysts is a complex process that begins with the perception of nitrogen starvation by the vegetative cells in a filament that initiates a cascade of factors that ultimately result in the development of a mature heterocyst with nitrogenase activity [3,6].
Anabaena variabilis ATCC 29413 has two distinct nitrogenases that function in heterocysts, a conventional Mo-nitrogenase and an alternative V-nitrogenase. The latter enzyme functions in heterocysts only under conditions of both nitrogen and molybdate limitation, when the Mo-nitrogenase is not functional [7]. The V-nitrogenase, first characterized in Azotobacter vinelandii [8-10], is similar in structure to the Mo-nitrogenase but it has a FeV-cofactor in place of the FeMo-cofactor [10]. In addition to these two enzymes, there is another Mo-nitrogenase in A. variabilis, encoded by the nif2 gene cluster, which is expressed in vegetative cells, but only under strictly anoxic conditions [11,12].
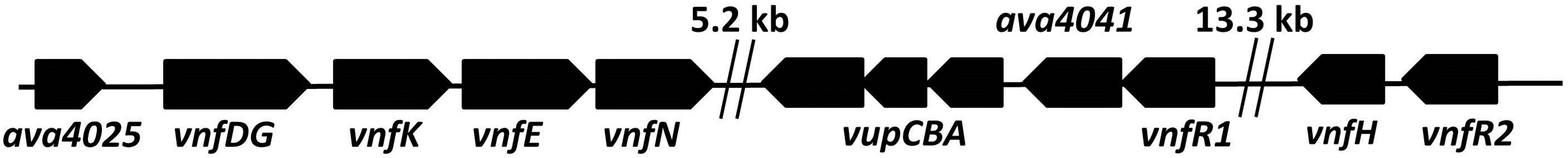
In A. variabilis the V-nitrogenase genes, vnfDG (a single gene), vnfK, vnfE, vnfN, the vupABC genes encoding a high-affinity vanadate transport system as well as vnfH (dinitrogenase reductase), are part of a large cluster of Mo-repressed genes located within a region of about 40 kb in the genome [7,13-15] (Figure 1(a)). The vnfDGKEN genes, which form a single operon with the upstream gene, ava4025 (whose function is not known), are under the dual control of two recently described Modependent repressors, vnfR1 and vnfR2 [16]. The vnfR1 gene is located upstream of the vupABC genes while vnfR2 is immediately upstream of vnfH (Figure 1(a)).
While the Mo-nitrogenase functions completely independently in the V-nitrogenase genes, the converse is not true. Synthesis of the V-nitrogenase depends on the activity of at least some of the nif1 genes including nifB1, nifS1, nifU1 [17] and possibly one or more of the poorly characterized genes downstream of nifN1. We have also shown that either nifH1 or vnfH can function interchangeably as dinitrogenase reductase for the Moor the V-nitrogenase [14].
In all nitrogen-fixing bacteria, the nif gene clusters include many genes, several of which are involved in production and insertion of the FeMo-cofactor (reviewed in [18]). In nitrogen-fixing cyanobacteria, the major nif gene cluster contains nifB, fdxH nifS, nifU, nifH, nifD, nifK, nifE, nifN, nifX, nifW and several other genes whose functions are not yet understood (Figure 1(b)). In vegetative cells, the nifD gene is interrupted by an 11-kb excision element whose site-specific removal by XisA (encoded within the excision element) from the chromosome in heterocysts late in the differentiation process is essential for nitrogenase expression [19-21]. In the sequenced genomes of Anabaena sp. PCC 7120 and A. variabilis ATCC 29413, nifV (homocitrate synthase), nifP (serine O-acetyltransferase), nifZ, and nifT are located 17 kb downstream from the 3’ end of the major nif cluster. A mutation in nifV has no effect on nitrogen fixation in Anabaena sp. PCC 7120 [22], but fdxH is required for optimum nitrogenase activity [23]. There are no recognizable homologs of nifM, nifY nifF or nifQ in the genomes of the sequenced cyanobacteria.
Mo is an element with five oxidation states; however, only the soluble Mo(VI) form can be transported and metabolized by cells. Molybdate is taken up in many bacteria by a high-affinity ABC-type transport system, first characterized in E. coli, which is encoded by the modABC genes. ModA is the periplasmic molybdatebinding protein, ModB is the membrane component of the permease, while ModC, on the cytoplasmic side of the membrane, provides the energizer function [24,25]. In A. variabilis, modA is distant from the fused modBC gene (Figure 1(c)) but both modA and modBC are induced by low concentrations of molybdate in the medium [26,27]. Mutants in either of these genes resulted in strains that were unable to fix nitrogen using the Monitrogenase, but were not impaired in their ability to use the V-nitrogenase [26]. Vanadate transport in A. variabilis is mediated by an ABC-type vanadate transport system, vupABC, with a Km for vanadate of about 3 × 10−9 M [15]. The vupABC genes, like the genes for the Vnitrogenase, are repressed by molybdate, and also like the latter genes the vanadate transport genes are expressed only in heterocysts [16].
Regulation of the major cluster of nif genes is poorly understood in cyanobacteria, which lack homologs of the NifA transcriptional activator found in nitrogen-fixing proteobacteria [28]. While it has been reported that the nifBSUHDKEN locus of Anabaena comprises at least three transcripts; nifB-fdxN-nifS-nifU [29], nifHDK [30,31], and nifEN [32], we have shown that the nifB1S1- U1H1D1K1 genes in A. variabilis are under the control of the nifB1 promoter and we have more recent unpublished evidence that the entire contiguous nif1 cluster in A. variabilis depends on the promoter upstream of nifB1
 (a)
(a)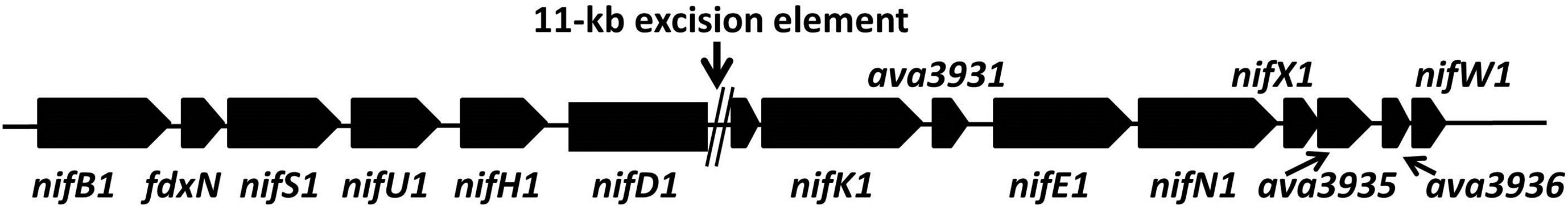 (b)
(b)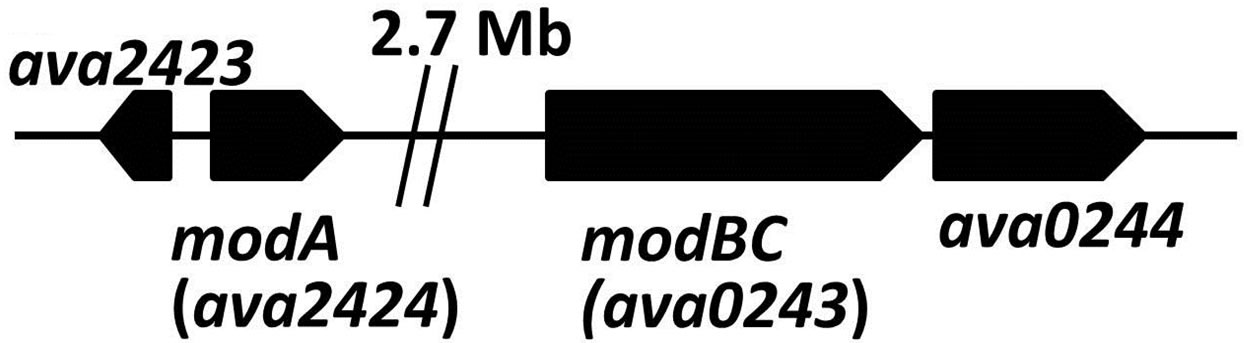 (c)
(c)
Figure 1. Maps showing (a) the vnf-vupABC regions, (b) the nif1 operon and (c) the two mod regions of the chromosome of A. variabilis, based on the genome sequence. Gaps are indicated by double vertical lines.
[33]. The 5’ transcript for nifH that was identified at position -123 relative to the start codon in Anabaena sp. strain PCC7120 [31,34] and in A. variabilis it is actually a transcriptional processing site [33] while the 5’ transcript for nifB in Anabaena sp. strain PCC 7120 at -179 relative to the start codon [29] is a true transcriptional start site in A. variabilis [33].
2. Materials and Methods
2.1. Growth
The strain A. variabilis FD, a derivative of A. variabilis 29413, that can grow at 40˚C and supports the growth of bacteriophages better than the parent strain [35] and FD derivatives were grown photoautotrophically in an eightfold dilution of the medium of Allen and Arnon [36] (AA/8) or AA/8 supplemented with 5.0 mM NH4Cl and 10 mM N-tris (hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) pH 7.2, at 30˚C with illumination at 120 μE∙m−2∙s−1. Mo-free medium was prepared from stocks scrubbed free of contaminating Mo using activated charcoal [37]. The microelement stock was prepared without Mo, but was not treated with activated charcoal. This medium was then used to grow A. variabilis for about one week, allowing the cells to biologically “scrub” the medium of all remaining Mo and V. To remove traces of Mo, glassware was treated with 1% Count-Off (New England Nuclear) and 10 mM ethylene diamine tetraacetic acid for 24 h and then thoroughly rinsed with deionized water purified through a Millipore water purification system. Na3VO4 or Na2WO4 stocks were made in the scrubbed medium, to prevent Mo contamination. The purity of the Na2WO4 was >99.995%, ensuring that no contaminating Mo or V was inadvertently added with the tungstate. For growth assays, OD720 was measured during exponential growth over two-three cell divisions, which required that growing cells be diluted to an OD720 of about 0.04 in appropriate medium (2.0 ml of diluted culture in each well of a 12 well plate) at the start of the experiment. Growth was observed to be exponential up to an OD720 of about 0.2 and then became linear, but growing eventually to OD720 of about 2.5, as the cells became light limited by self-shading. Growth was measured every 3 - 6 hours and the exponential growth data were used to calculate the growth rate constant using the formula µ = ln (N2) − ln (N1)/(t2 − t1) where N = OD720 and t = time in days. Growth experiments were done using quadruplicate cultures; the average and standard deviation are shown.
2.2. Construction of lacZ Fusion Strains
Strains BP193 (vnfDG:lacZ translational fusion) and BP221 (nifH1:lacZ translational fusion) were made by single-crossover integration of plasmids pBP193 and pBP221 into the chromosome of A. variabilis, following transfer by conjugation as described previously [38]. These two plasmids were constructed by fusing a 296-bp region upstream of vnfDG (primers vnfDR, 5’-ATACCCGGGTAGTTTTAGTGGCATTCCTTTATT-3’ and vnfD296-L, 5’-GAAGATCTCACTTAATTTATCGGCTTTTCA-3’), or a 399-bp region upstream of nifH1 (primers NifH1p-L399 5’-GAAGATCTTCAAACTACTACTGCAAGG-3’ and NifH1p-R, 5’-ATACCCGGGTAATGTTTTCGTCAGTCAT-3’) in-frame with lacZ in a pBR322-based plasmid with characteristics that have been previously described (pBP288, [39]). The only difference between pBP193 and pBP221 compared to pBP288 is that pBP288 produces transcriptional fusions to lacZ while the plasmids described here are translational fusions using lacZ from plasmid pPL2A [40]. Like pBP288, the relevant characteristics of pBP193 and pBP221 include: 1) a promoterless lacZ gene for assaying promoter activity in vivo, 2) a 6.5-kb ntcA region of A. variabilis that allows homologous recombination, 3) a 1.1-kb npt gene from pRL648 interrupting ntcA, ensuring only one functional copy of ntcA after recombination, 4) a 1.0-kb Ώ streptomycin/spectinomycin-resistance cassette, and 5) BglII and SmaI sites to allow for cloning of promoter fragments in-frame with the lacZ gene.
2.3. Acetylene Reduction Assays
Nitrogenase activity was measured in vivo by acetylene reduction assays. 5-ml aliquots of cultures at an OD720 of about 0.15 were added to 16-ml Hungate tubes. The tubes were sealed with gas-tight stoppers, injected with 1.0 ml acetylene gas and shaken at 30˚C in the light for 60 min. 250 μl samples of gas were injected into a Shimazu gas chromatograph equipped with a 6 ft Poropak N column. The column temperature was either 65˚C (for detection of both ethylene and ethane, characteristic of the V-nitrogenase) or 80˚C (for detection of only ethylene, characteristic of the Mo-nitrogenase).
2.4. Determination of 15N2 Fixation
The incorporation of 15N2 into cells was determined in triplicate using 15 ml of cells at an OD720 of about 0.2 that were incubated in an atmosphere of 85% argon, 15% 15N2 for 2 h at 30˚C in the light. Cells were collected by centrifugation for 15N analysis as described by [41].
2.5. Determination of Intracellular Mo or V Concentration
Cells were grown at 30˚C in the light in AA/8 medium lacking Na2MoO4 and Na3VO4 or supplemented with 1.0 μM of either metal, washed 3 times in metal-free medium, resuspended at a concentration of about 5 × 109 cells∙ml−1 and lysed by multiple cycles of freeze thawing. A highspeed supernatant was analyzed by plasma emission spectroscopy to determine the amount of Mo or V in the cells.
2.6. Northern Blots
RNA was isolated using Tri-Reagent (Sigma) as previously described [33]. DNA was removed using a Turbo DNA-free kit (Ambion, Austin, TX). RNA (about 20 μg) was separated in a 1.5% formaldehyde-agarose gel and transferred to NytranSupercharge (Schleicher & Schuell). Blots were prehybridized in ULTRAhyb (Ambion) containing 100 μg∙ml−1 of sheared herring testes DNA for two hours at 42˚C and then hybridized overnight at 42˚C to radiolabeled nifH1, vnfH, modA, or rnpB (encoding RNase P) [42]. DNA probes were added to prehybridization buffer (ULTRAhyb containing sheared herring testes DNA). Blots were imaged using a Molecular Dynamics STORM 820 phosphoimager and ImageQuant software (Molecular Dynamics). PCR products amplifying a region of the nifH1, vnfH, modA, or rnpB gene were used to make DNA probes. 32P-labeled PCR probes were obtained from the following primer sets: nifH1, nifH1-L (5’-CGGCATGACCTATTGGTAGC-3’) and nifH1234- R (5’-GGTGARATGATGGCGATGTAYGC-3’); vnfH, nifH3-L (5’-AGCTTCCAGTGCTTGAGCTT-3’) and nifH1234-R; modA, modAint-L, (5’-TTTTGGCAGTCGGTTTACCATTGT-3’) and modAint-R (5’-GCCGCGTTAACATTTCTGCTACTT-3’); and rnpB, RNaseP-L (5’-AGAGTAGGCGTTGGCGGTTGC-3’) and RNasePR (5’-ATTGCTTTACACGAGGGCGATTAT-3’). Approximately 12.5 ng of PCR product was labeled with a32P-dCTP by random primer extension using the Ambion Klenow kit.
2.7. In Situ Localization of lacZ Expression
For in situ localization of lacZ expression cells were washed twice with water and fixed for 15 min at 25˚C with 0.04% glutaraldehyde. Cells were washed twice with water to remove glutaraldehyde and incubated in the dark at 37˚C for 30 minutes with 100 µM 5-dodecanoylaminofluorescein di-β-D-galactopyranoside in 25% dimethyl sulfoxide. Excess substrate was removed by washing twice with water, and cells were resuspended in Vectashield® (Vector Laboratories), an antibleaching agent. The cells were imaged on a Zeiss epifluorescence microscope using a fluorescein filter set (excitation, 450 - 490 nm; dichroic, 510 nm; barrier, 520 nm) with a 560- nm shortpass filter. Images were taken using a Retiga EXi (QImaging) cooled charge-coupled device camera with IP Labs 4.0 software (BD Biosciences).
3. Results
3.1. Nitrogenase Activity in Moand V-Deficient Cells
The Mo-nitrogenase and the V-nitrogenase both function in heterocysts, but not usually at the same time, since the V-nitrogenase is not made in the presence of Mo [14]. The Mo-nitrogenase genes are made in the absence of Mo, but the enzyme cannot function in the absence of the Mo-cofactor. In A. variabilis, the Mo-nitrogenase was better at fixing nitrogen than the V-nitrogenase, as measured both by cell growth and by 15N2 reduction (Table 1). The V-nitrogenase supported slower growth and lower levels of 15N2 reduction than the Mo-nitrogenase, but it was particularly inefficient at acetylene reduction compared to the Mo-nitrogenase (Table 1). The V-nitrogenase is capable of further reducing ethylene to ethane, which is not characteristic of the Mo-nitrogenase [43]. The relative inefficiency of the V-nitrogenase compared to the Mo-nitrogenase probably explains why it is not made unless Mo is not present in the cells.
We have observed slow but persistent growth of cells in a medium that has less than 10−10 M Mo (<10 ppt as measured by ICP-MS, data not shown). While growth was slow, it was also accompanied by both acetylene reduction and by 15N2 reduction (Table 1) indicating that one or both nitrogenases were still functional, despite the very low external metal concentration, which had a profound effect on metal concentrations inside the cell. The
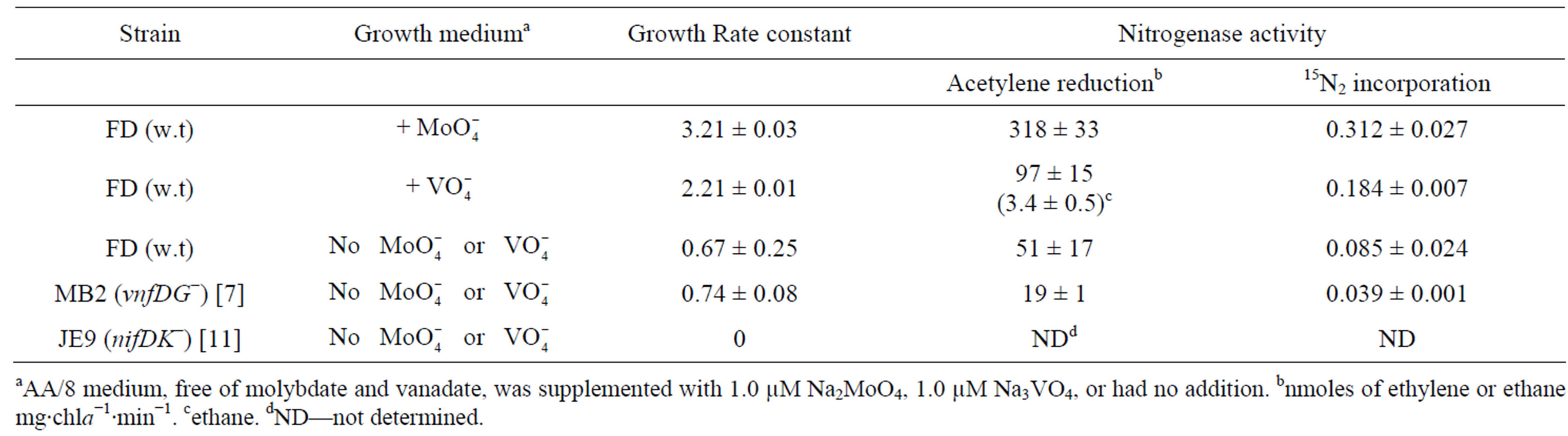
Table 1. Nitrogenase activity in cells grown with or without molybdate or vanadate.
aAA/8 medium, free of molybdate and vanadate, was supplemented with 1.0 µM Na2MoO4, 1.0 µM Na3VO4, or had no addition. bnmoles of ethylene or ethane mg∙chla−1∙min−1. cethane. dND—not determined.
concentration of Mo in Moand V-starved cells (7 - 11 pg per 106 cells) was only 2% - 3% of that of Mo-replete cells (about 400 pg per 106 cells) while the concentration of V in Moand V-starved cells (1 - 7 pg per 106 cells) was about 5% compared to V-replete cells (about 150 pg per 106 cells). While these levels are low, they are apparently enough to provide some active enzyme. The cells are capable of storing metals, especially Mo, which means that the metal concentrations found in metal-replete cells are much higher than is required for enzyme activity. Despite the very low intracellular levels of these two metals, the cells were able to make at least one functional enzyme. We compared nitrogenase activity in Moand V-deprived cells in the wild-type strain, in a vnfDG mutant lacking the structural genes for the V-nitrogenase (MB2) and in a strain lacking the nifDK1 structural genes for the Mo-nitrogenase (JE9). Nitrogenase activity was present in the mutant lacking the V-nitrogenase (MB2); however, the absence of growth or enzyme activity in the nifDK1 mutant (JE9) suggested that the Mo-nitrogenase contributed most of the nitrogen fixation under Moand V-limiting growth conditions. This was supported by the fact that nitrogenase activity in the wild-type strain did not produce ethane from acetylene, which is characteristic of the V-nitrogenase. Thus it appears that under conditions of very low intracellular Mo and V, the Mo-nitrogenase functions at a level sufficient to provide slow growth that contributes to long-term survival during the environmental stress of metal deficiency.
3.2. Effect of Tungstate on Nitrogenase Activity
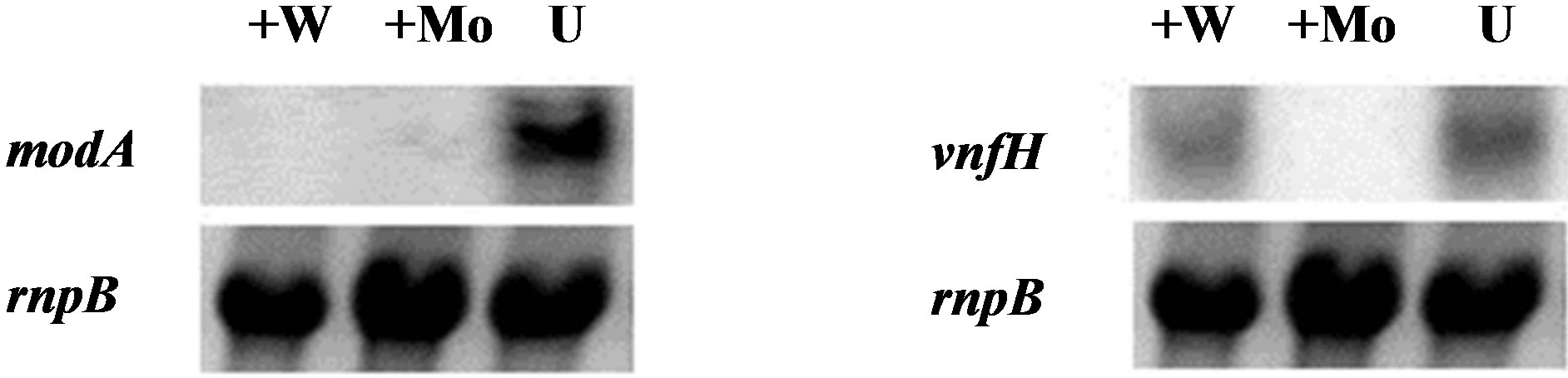
Since cells grown in a Moand V-free medium were able to make low levels of active Mo-nitrogenase and we have shown previously by immunoblots that Moand Vstarved cells produce Moand V-nitrogenase proteins [14] we wanted to determine the effect of adding back either of these two metals, or tungstate (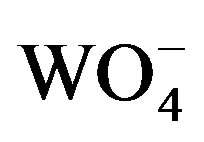 ), on nitrogenase activity. Molybdate and tungstate are very similar ions in terms of structure and size [44,45] and both are transported by the molybdate transport system [27]. Inside the cell, tungstate functioned like molybdate to repress transcription of the modA gene, which encodes the periplasmic-binding protein of the ModABC molybdate transporter (Figure 2(a)); however, it did not repress transcription of the vnfH gene, which was, however, repressed by molybdate (Figure 2(b)). Therefore, in cells starved for Mo and V, the addition of W should not inhibit the synthesis of the V-nitrogenase. Addition of 10−7 M molybdate, vanadate or tungstate to wild-type FD cells, previously starved for Mo and V, resulted in a substantial increase in nitrogenase activity, as measured by acetylene reduction activity, within 2 h of metal addition and activity increased rapidly over the next 6 h (Figure 3(a)). While Mo addition gave the most rapid and the highest
), on nitrogenase activity. Molybdate and tungstate are very similar ions in terms of structure and size [44,45] and both are transported by the molybdate transport system [27]. Inside the cell, tungstate functioned like molybdate to repress transcription of the modA gene, which encodes the periplasmic-binding protein of the ModABC molybdate transporter (Figure 2(a)); however, it did not repress transcription of the vnfH gene, which was, however, repressed by molybdate (Figure 2(b)). Therefore, in cells starved for Mo and V, the addition of W should not inhibit the synthesis of the V-nitrogenase. Addition of 10−7 M molybdate, vanadate or tungstate to wild-type FD cells, previously starved for Mo and V, resulted in a substantial increase in nitrogenase activity, as measured by acetylene reduction activity, within 2 h of metal addition and activity increased rapidly over the next 6 h (Figure 3(a)). While Mo addition gave the most rapid and the highest
 (a) (b)
(a) (b)
Figure 2. Northern blot analysis of the effect of molybdate and tungstate on expression of modA and vnfH. RNA was isolated 24 h after the addition of 10−6 M Na2WO4 (+W) or 10−6 M Na2MoO4 (+Mo), or no addition (U) to wild-type cells that had been grown for at least 10 generations in Mofree medium containing 10−6 M Na3VO4. Northern blots were hybridized with internal fragments from either (a) modA or (b) vnfH. The control for equal loading of RNA was rnpB.
 (a)
(a)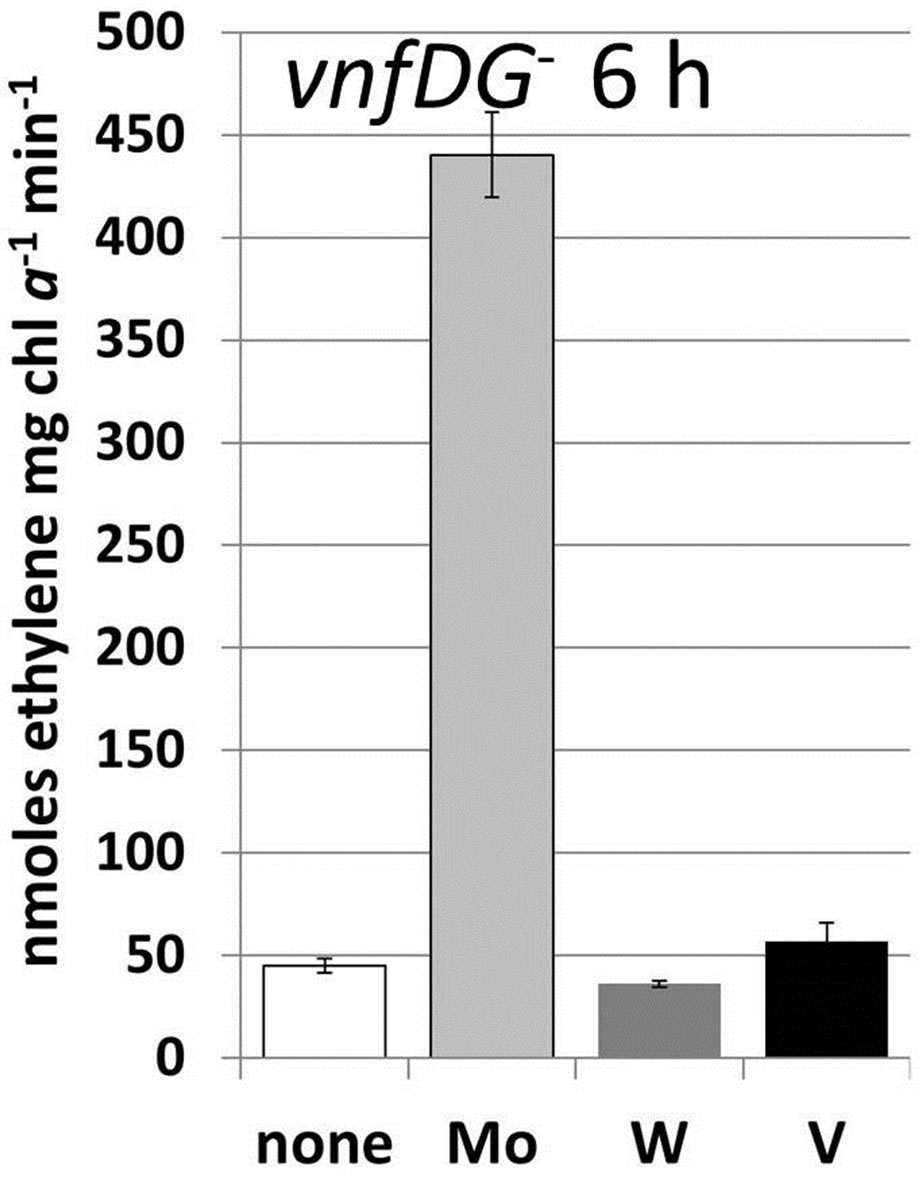 (b)
(b)
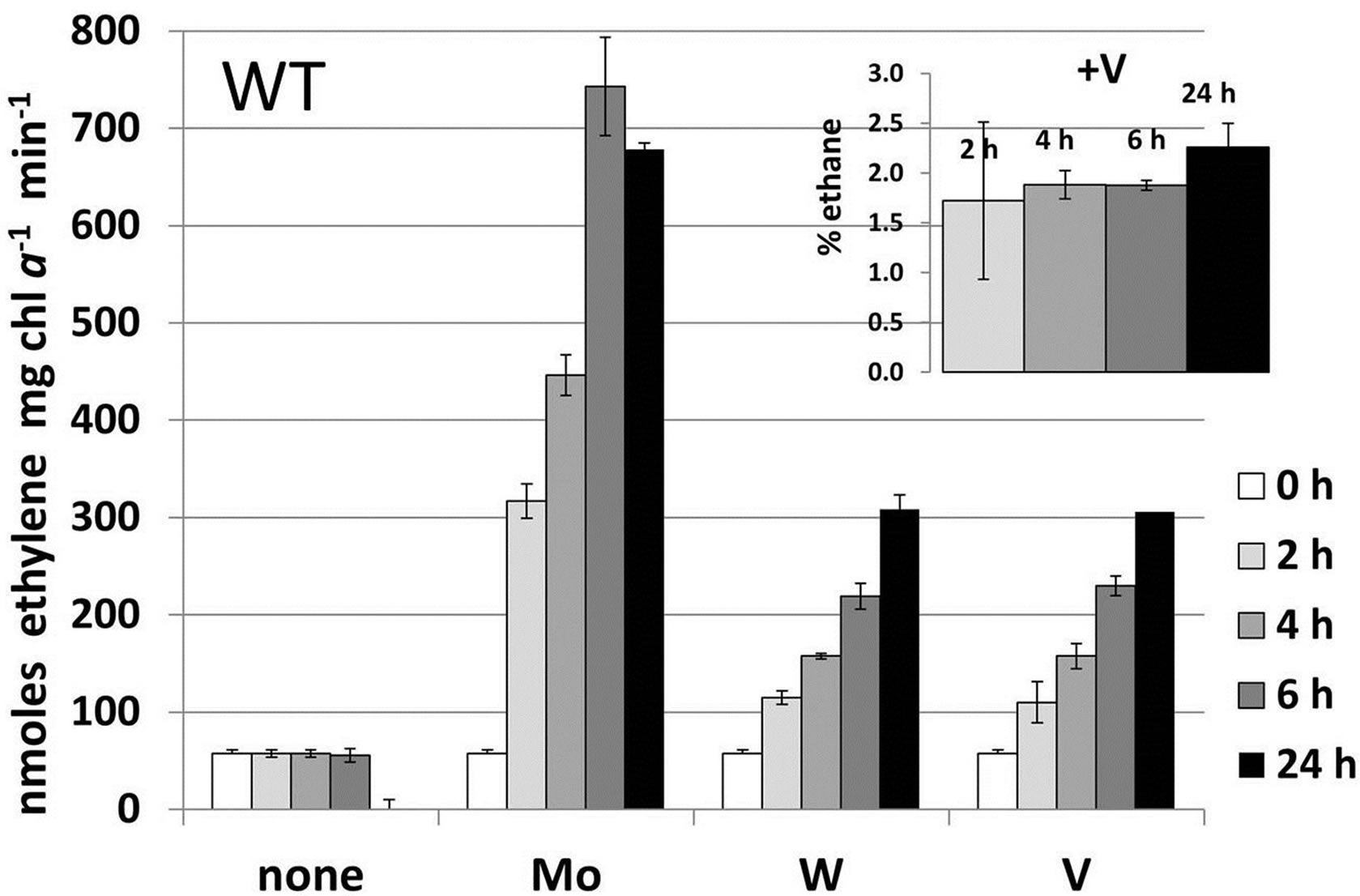
Figure 3. Metal-induced increase in nitrogenase activity in cells starved for molybdate and vanadate. A. variabilis strains FD (WT) and MB2 (vnfDG mutant) were grown in Moand V-free AA/8 medium for at least 10 generation to deplete internal stores of these metals and then Na2MoO4, Na3VO4 or Na2WO4 (all at 10−7 M) were added to these starved cells at 0 time. Acetylene reduction was measured for strain FD at 2 h, 4 h, 6 h and 24 h after metal addition and for strain MB2 at 6 h after metal addition. The data are the average of triplicate biological samples and the error bars represent the standard deviation. The inset shows the ethane (% of ethylene) produced by strain FD with added Na3VO4, which were the only samples that produced any ethane.
increase in nitrogenase activity, as expected from its superior activity in reducing acetylene compared to the Vnitrogenase (Table 1), W was about as effective as V in stimulating acetylene reduction activity. Only the addition of V resulted in the formation of ethane from acetylene (Figure 3(a), inset), which is characteristic of the V-nitrogenase [43]. Because of the structural similarities of molybdate and tungstate it seemed likely that tungstate was incorporated into the Mo-nitrogenase rather than the V-nitrogenase, so we determined the effect of addition of 10−7 M molybdate, vanadate or tungstate to strain MB2, a vnfDG mutant lacking the V-nitrogenase [7]. As expected, the addition of Mo strongly stimulated acetylene reduction by 6 h after addition while the addition of V had little effect (Figure 3(b)). Surprisingly, however, this mutant did not respond to the addition of tungstate (Figure 3(b)), suggesting that the V-nitrogenase is primarily responsible for the increase in acetylene reduction activity in the wild-type strain in response to tungstate. However, the V-nitrogenase that incorporated tungstate was incapable of reducing ethylene to ethane, suggesting that the enzyme did not function normally. This was supported by the fact that wild-type cells, starved for Mo and V, when supplemented with tungstate grew poorly over the 24 h period following addition of tungstate (growth rate constant = 0.49 ± 0.03) at a rate similar to the metal free culture (Table 1) suggesting that the W-nitrogenase functioned very poorly as a true nitrogenase.
We determined whether the nitrogenase that was produced after the addition of any of these three metals to the wild-type strain required new protein synthesis in order to assemble the metal-containing enzyme or whether existing precursors could be assembled. The addition of chloramphenicol (50 μg∙ml−1) inhibited the increase in acetylene reduction at 5 h after the addition of molybdate, vanadate or tungstate (Figure 4) indicating that new protein synthesis was required for assembly of the newly acquired metal into nitrogenase.
3.3. Expression of Nitrogenases under Metal-Stress Conditions
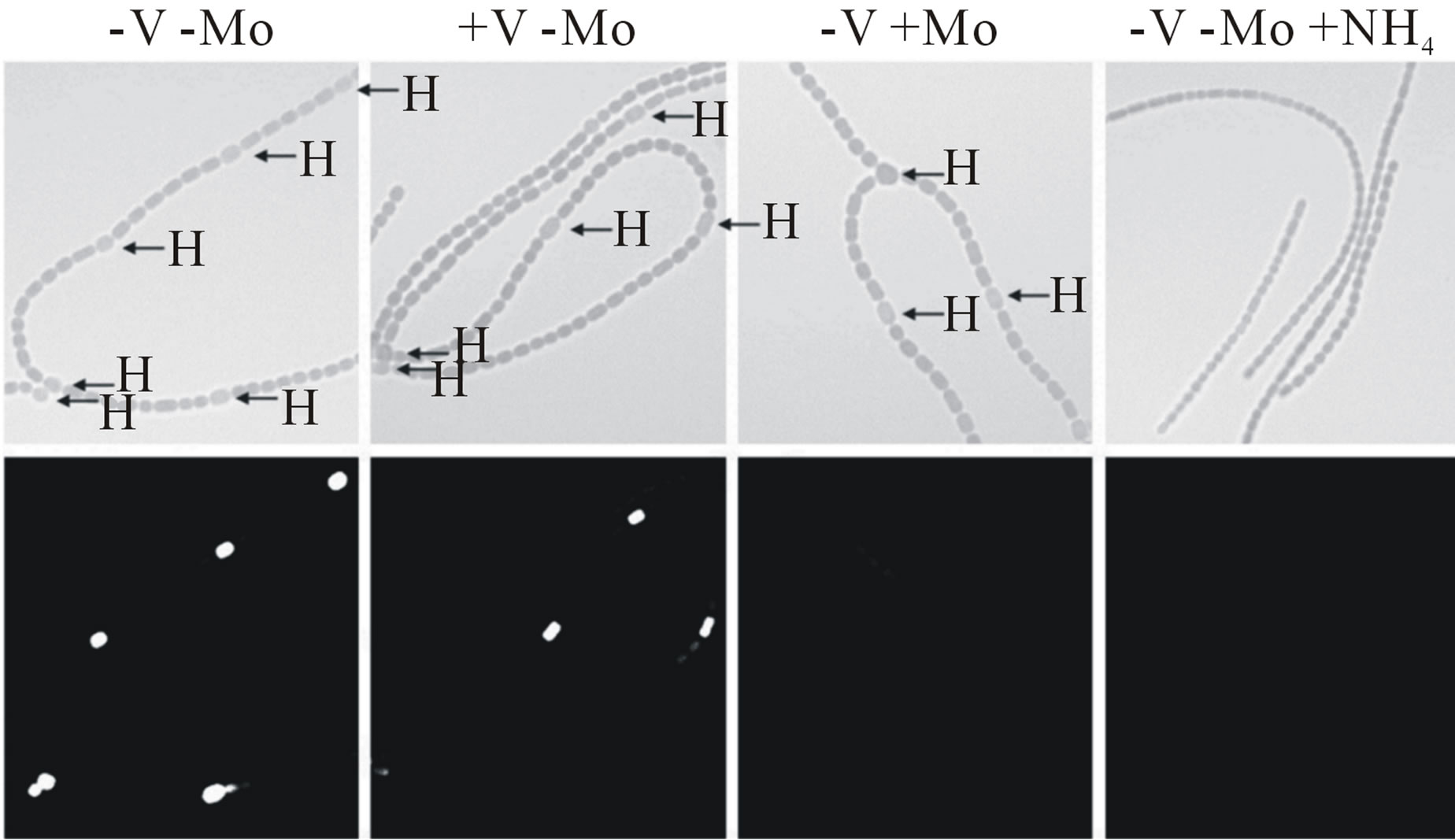
Molybdate represses the transcription of the vnfH gene (Figure 2(b)), but vanadate has no effect on transcription of the nifH1 gene [14]. We have shown previously that in the absence of both metals, cells produced both NifH1 and VnfH, as did cells grown with only vanadate, while cells grown only with molybdate made only the Mo-nitrogenase [14]. The effect of molybdate and vanadate on cell-specific nitrogenase expression was examined in strains BP193 and BP221 in which a promoterless lacZ gene was expressed from the region upstream of vnfDG (BP193) or nifH1 (BP221). Cell-specific expression of nifH1 or vnfDG was visualized in filaments using fluorescein-β-D-galactopyranoside, which fluoresces when
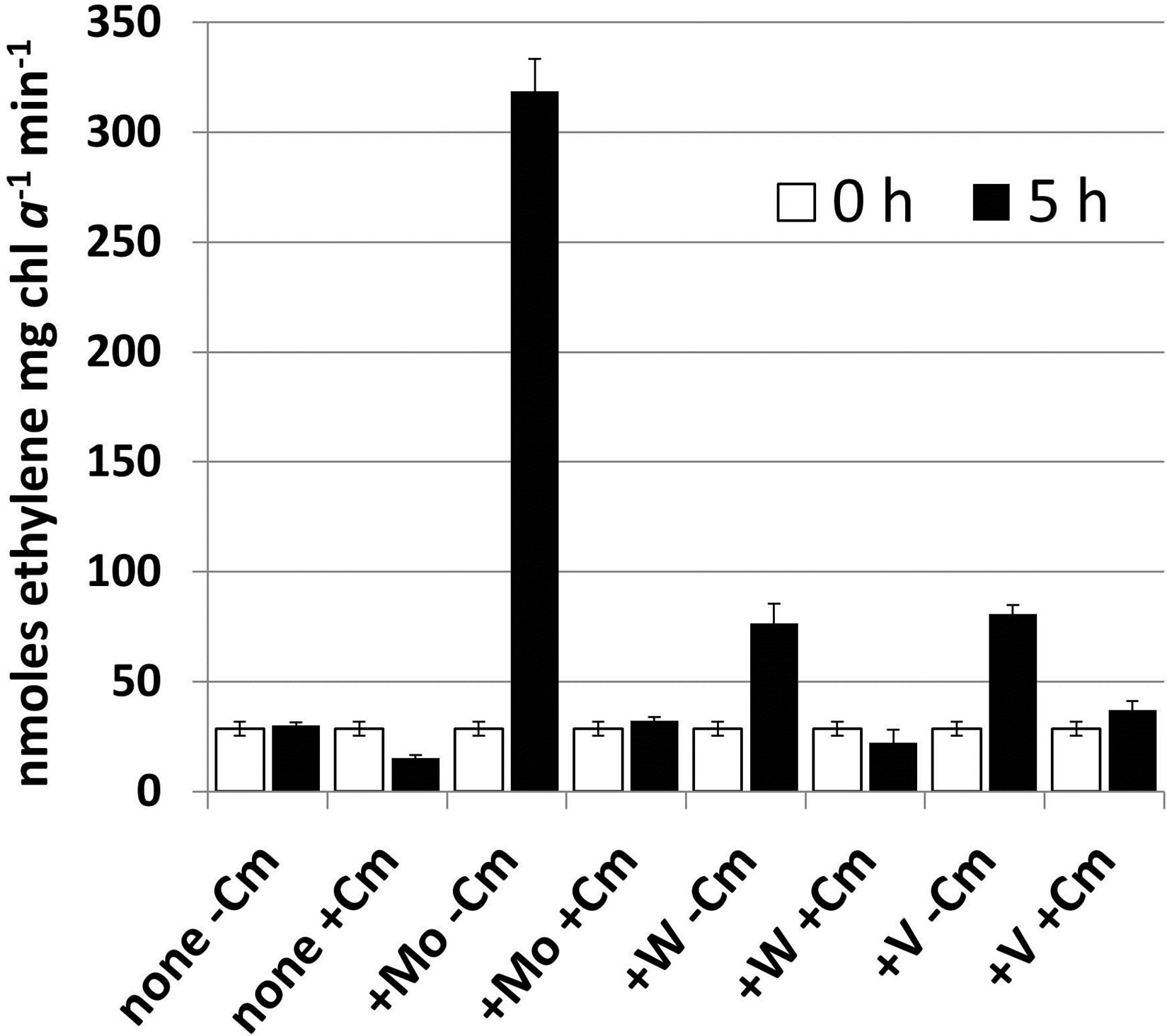
Figure 4. The effect of protein synthesis inhibition on the metal-induced increase in nitrogenase activity in cells starved for molybdate and vanadate. A. variabilis strain FD was grown as described in Figure 3 and divided into two samples, one of which had chloramphenicol (Cm, 50 μg∙ml−1) for 15 min prior to the addition of Na2MoO4, Na3VO4 or Na2WO4 (all at 10−7 M). The cells, with or without Cm, were incubated with each of the metals for 5 hours and then acetylene reduction was determined. The data are the average of triplicate biological samples and the error bars represent the standard deviation in the activity among the triplicate samples. Ethane (about 2% of ethylene) was produced only by the cells with added Na3VO4 without Cm, but not with Cm.
β-galactosidase cleaves the substrate. Both nifH1 and vnfDG showed heterocyst-specific cell expression (Figure 5, see also [16]). The expression of vnfDG was repressed in heterocysts of cells grown with molybdate. Neither nifH1 nor vnfDG was expressed in cells grown in the presence of ammonium (Figure 5).
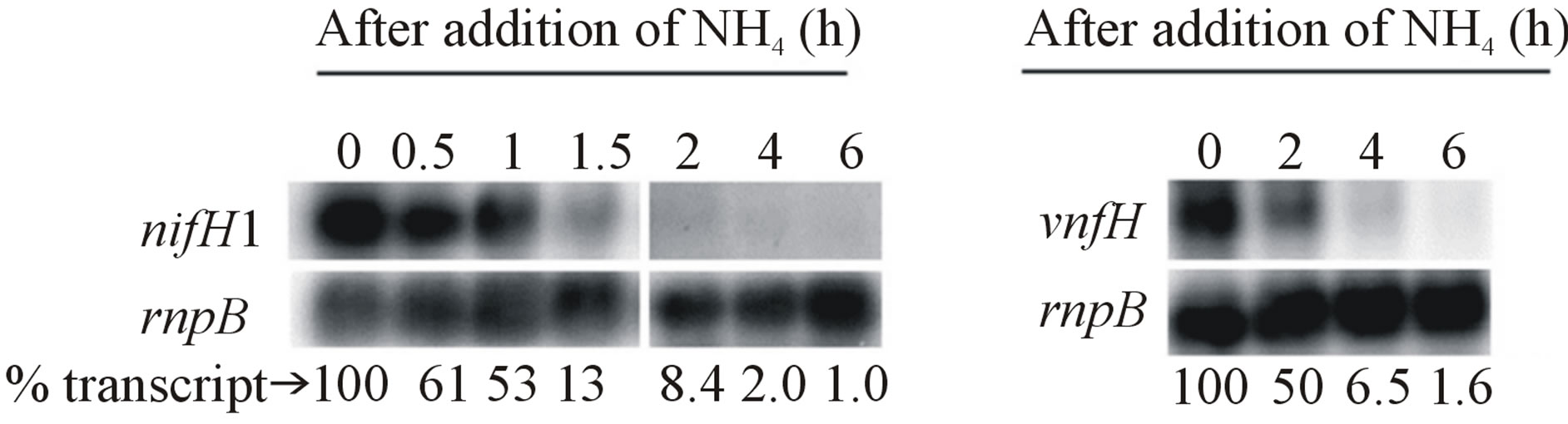
It is known that ammonium inhibits nitrogenase expression because it prevents heterocyst differentiation, which is required for expression of nitrogenase genes. However, we wanted to determine the effect of the addition of ammonium on cells already expressing the nifH1 or the vnfH gene. Ammonium was added to cells expressing the Moor the V-nitrogenase and transcripts of nifH1 or vnfH were determined over a six-hour period. Transcript levels for both genes declined rapidly upon the addition of ammonium indicating that either ammonium or a product of ammonium metabolism quickly represses transcription of both nifH1 and vnfH (Figure 6). Thus, both nitrogenases share some regulatory pathway that quickly turns off gene expression when the enzyme is no longer needed.
4. Discussion
The availability of molybdate or vanadate has a profound effect on the ability of A. variabilis to make and use the Mo-nitrogenase or the V-nitrogenase. Beyond the obvi-
 (a)
(a)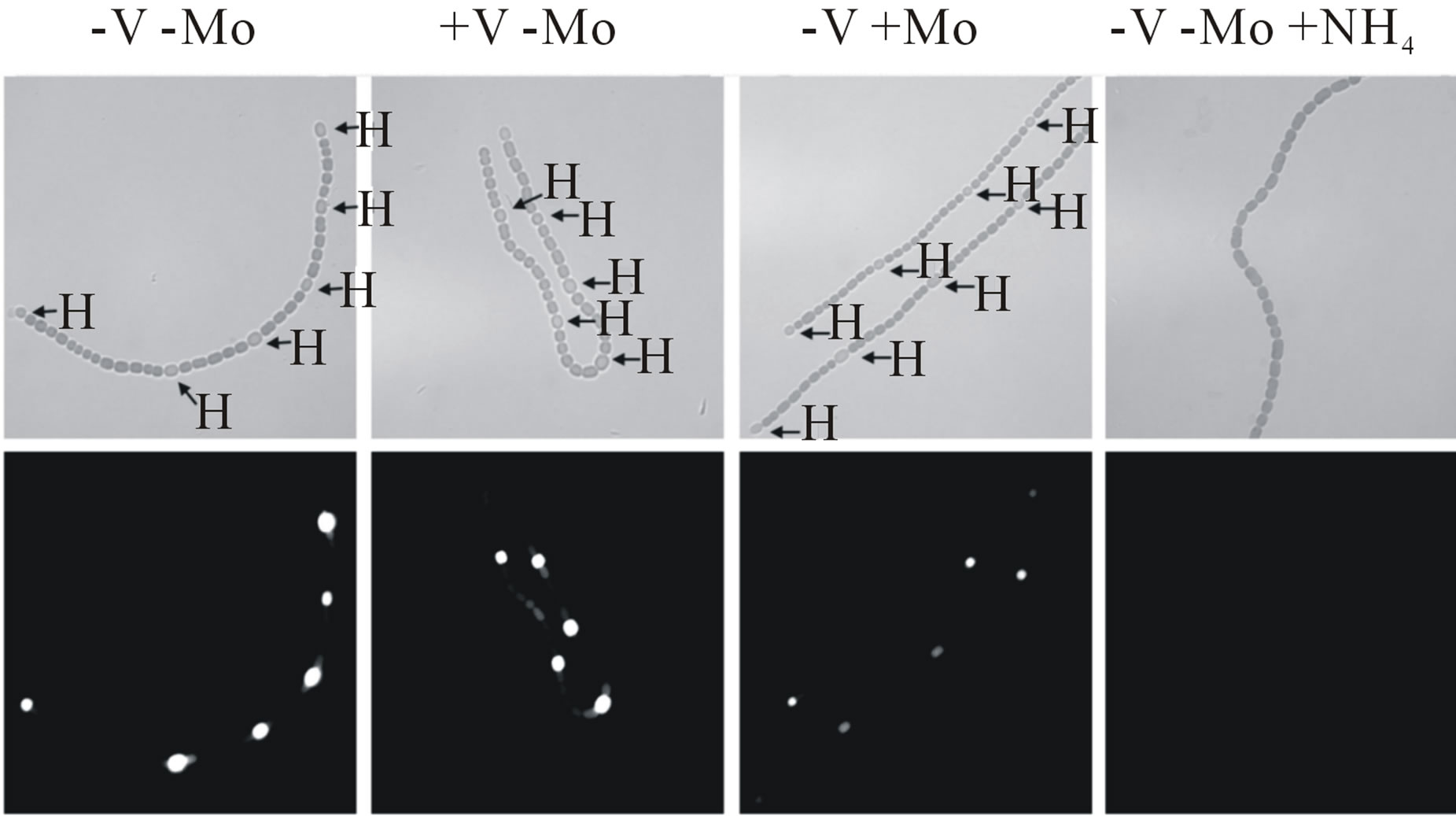 (b)
(b)
Figure 5. In situ localization of (a) nifH1 expression and (b) vnfDG expression. Cells of strain BP221, with lacZ fused to the promoter region of nifH1 (a), or BP193, with lacZ fused to the promoter region of vnfDG, were grown in AA/8 medium, with or without 1.0 μM molybdate or 1.0 μM vanadate or with 5.0 mM NH4Cl and 10 mM TES, pH 7.2. β-galactosidase activity was visualized using fluorescein-β- D-galactopyranoside, as described in Materials and Methods. Top panels are brightfield images showing heterocysts (H). Bottom panels show fluorescein fluorescence.
 (a) (b)
(a) (b)
Figure 6. Repression of nifH1 and vnfH by ammonium. FD cells were grown in AA/8 with either 10−6 M Na2MoO4 or 10−6 Na3VO4 prior to the addition of 5 mM NH4Cl + 10 mM TES pH 7.2 at 0 time. RNA was isolated at the indicated times for Northern blots that were hybridized with internal fragments from either (a) nifH1 or (b) vnfH. The control for equal loading of RNA was rnpB. A phosphoimager was used to determine the relative amount of transcript, normalized to rnpB, at each time after ammonium addition.
ous problem that lacks of the appropriate metal blocks synthesis of a functional enzyme, the availability of molybdate also controls the expression of the V-nitrogenase, whereas the availability of vanadate had no effect on expression of either nitrogenase. In Moand V-starved cells, nitrogen fixation continued for many generations in both the wild-type strain and in a mutant lacking the V-nitrogenase; however, a mutant lacking the Mo-nitrogenase showed no growth in the absence of these two metals, suggesting that it is the Mo-nitrogenase which is primarily responsible for the residual growth. Tungstate, which is similar to molybdate and is transported efficiently by the Mo-transport system, acts like Mo in inhibiting the molybdate transport system, but it does not inhibit the transcription of the genes for the V-nitrogenase; hence its effect on regulatory proteins that control the mod genes differs compared to the vnf genes.
In wild-type cells deficient in Mo and V (in which both nitrogenase gene clusters are expressed), the addition of tungstate stimulated acetylene reduction. However, a mutant lacking the V-nitrogenase did not show any increase in acetylene reduction upon addition of tungstate suggesting that, although W was transported into the cells, it could not be assembled into the cofactor for the Mo-nitrogenase. The W-nitrogenase appears to function much better as an acetylene reducing enzyme than as a nitrogen reducing enzyme since growth of the cells with W is poor. The two nitrogenases both function in heterocysts, although not usually at the same time. The Mo-nitrogenase was much better at fixing both acetylene and nitrogen than the V-nitrogenase. Transcription of both the nif1 and the vnf genes was repressed by ammonium, long before heterocysts disappeared, suggesting that the two systems share common mechanisms of activation and repression of expression. The V-nitrogenase is clearly secondary to the Mo-nitrogenase: it requires at least nifB1 nifS1, nifU1 [17] and the V-nitrogenase showed lower activity than the Mo-nitrogenase leading to slower cell growth with this enzyme. In cells starved for Mo and V, the Mo-nitrogenase functioned despite very low intracellular concentrations of Mo to allow cells to survive in an environment deficient in this metal, but the V-nitrogenase was not able to function in cells with very low levels of V.
5. Acknowledgements
Support for this research was provided by National Science Foundation grant MCB-1052241. We thank Jennifer Glass for assaying the Mo content of our Mo-free medium.
REFERENCES
- E. Flores and A. Herrero, “Compartmentalized Function through Cell Differentiation in Filamentous Cyanobacteria,” Nature Reviews Microbiology, Vol. 8, No. 1, 2010, pp. 39-50. http://dx.doi.org/10.1038/nrmicro2242
- T. Thiel, “Nitrogen Fixation in Heterocyst-Forming Cyanobacteria. In: W. Klipp, B. Masepohl, J. R. Gallon and W. E. Newton, Eds., Genetics and Regulation of Nitrogen Fixing Bacteria, Kluwer Academic Publishers, Dordrecht, 2004, pp. 73-110.
- I. Maldener and A. M. Muro-Pastor, “Cyanobacterial Heterocysts,” John Wiley & Sons, Ltd., 2010.
- M. A. Murry and C. P. Wolk, “Evidence That the Barrier to the Penetration of Oxygen into Heterocysts Depends upon Two Layers of the Cell Envelope,” Archives of Microbiology, Vol. 151, No. 6, 1989, pp. 469-474. http://dx.doi.org/10.1007/BF00454860
- M. A. Murry, A. J. Horne and J. R. Benemann, “Physiological Studies of Oxygen Protection Mechanisms in the Heterocysts of Anabaena cylindrical,” Applied and Environmental Microbiology, Vol. 47, 1984, pp. 449-454.
- K. Kumar, R. A. Mella-Herrera and J. W. Golden, “Cyanobacterial Heterocysts,” Cold Spring Harbor Perspectives in Biology, Vol. 2, No. 4, 2010, Article ID: a000315. http://dx.doi.org/10.1101/cshperspect.a000315
- T. Thiel, “Characterization of Genes for an Alternative Nitrogenase in the Cyanobacterium Anabaena variabilis,” Journal of Bacteriology, Vol. 175, No. 19, 1993, pp. 6276-6286.
- C. C. Lee, Y. Hu and M. W. Ribbe, “Unique Features of the Nitrogenase VFe Protein from Azotobacter vinelandii,” Proceedings of the National Academy of Sciences, Vol. 106, No. 23, 2009, pp. 9209-9214. http://dx.doi.org/10.1073/pnas.0904408106
- P. E. Bishop and R. Premakumar, “Alternative Nitrogen Fixation Systems,” In: G. Stacey, R. H. Burris and H. J. Evans, Eds., Biological Nitrogen Fixation, Chapman and Hall, Inc., New York, 1992, pp. 736-762.
- C. Ruttimann-Johnson, L. M. Rubio, D. R. Dean and P. W. Ludden, “VnfY Is Required for Full Activity of the Vanadium-Containing Dinitrogenase in Azotobacter vinelandii,” Journal of Bacteriology, Vol. 185, No. 7, 2003, pp. 2383-2386. http://dx.doi.org/10.1128/JB.185.7.2383-2386.2003
- T. Thiel, E. M. Lyons and J. C. Erker, “Characterization of Genes for a Second Mo-Dependent Nitrogenase in the Cyanobacterium Anabaena variabilis,” Journal of Bacteriology, Vol. 179, No. 16, 1997, pp. 5222-5225.
- T. Thiel, E. M. Lyons, J. C. Erker and A. Ernst, “A Second Nitrogenase in Vegetative Cells of a HeterocystForming Cyanobacterium,” Proceedings of the National Academy of Sciences, Vol. 92, No. 20, 1995, pp. 9358- 9362. http://dx.doi.org/10.1073/pnas.92.20.9358
- T. Thiel, “Isolation and Characterization of the vnfEN Genes of the Cyanobacterium Anabaena variabilis,” Journal of Bacteriology, Vol. 178, No. 15, 1996, pp. 4493-4499.
- B. S. Pratte, K. Eplin and T. Thiel, “Cross-Functionality of Nitrogenase Components NifH1 and VnfH in Anabaena variabilis,” Journal of Bacteriology, Vol. 188, No. 16, 2006, pp. 5806-5811. http://dx.doi.org/10.1128/JB.00618-06
- B. S. Pratte and T. Thiel, “High-Affinity Vanadate Transport System in the Cyanobacterium Anabaena variabilis ATCC 29413,” Journal of Bacteriology, Vol. 188, No. 2, 2006, pp. 464-468. http://dx.doi.org/10.1128/JB.188.2.464-468.2006
- B. S. Pratte, R. Sheridan, J. A. James and T. Thiel, “Regulation of V-Nitrogenase Genes in Anabaena Variabilis by RNA Processing and by Dual Repressors,” Molecular Microbiology, Vol. 88, No. 2, 2013, pp. 413-424. http://dx.doi.org/10.1111/mmi.12197
- E. M. Lyons and T. Thiel, “Characterization of nifB, nifS, and nifU Genes in the Cyanobacterium Anabaena variabilis: NifB Is Required for the Vanadium-Dependent Nitrogenase,” Journal of Bacteriology, Vol. 177, No. 6, 1995, pp. 1570-1575.
- L. M. Rubio and P. W. Ludden, “Biosynthesis of the Iron-Molybdenum Cofactor of Nitrogenase,” Annual Review of Microbiology, Vol. 62, No. 1, 2008, pp. 93-111. http://dx.doi.org/10.1146/annurev.micro.62.081307.162737
- J. W. Golden and D. R. Wiest, “Genome Rearrangement and Nitrogen Fixation in Anabaena Blocked by Inactivation of xisA Gene,” Science, Vol. 242, No. 4884, 1988, pp. 1421-1423. http://dx.doi.org/10.1126/science.3144039
- J. C. Meeks, E. L. Campbell and P. S. Bisen, “Elements Interrupting Nitrogen Fixation Genes in Cyanobacteria: Presence and Absence of a nifD Element in Clones of Nostoc sp. Strain Mac,” Microbiology, Vol. 140, No. 12, 1994, pp. 3225-3232. http://dx.doi.org/10.1099/13500872-140-12-3225
- J. S. Brusca, M. A. Hale, C. D. Carrasco and J. W. Golden, “Excision of an 11-Kilobase-Pair DNA Element from within the nifD Gene in Anabaena variabilis Heterocysts,” Journal of Bacteriology, Vol. 171, No. 8, 1989, pp. 4138-4145.
- O. Stricker, B. Masepohl, W. Klipp and H. Böhme, “Identification and Characterization of the nifV-nifZ-nifT Gene Region from the Filamentous Cyanobacterium Anabaena sp. Strain PCC 7120,” Journal of Bacteriology, Vol. 179, No. 9, 1997, pp. 2930-2937.
- B. Masepohl, K. Scholisch, K. Gorlitz, C. Kutzki and H. Bohme, “The Heterocyst-Specific fdxH Gene Product of the Cyanobacterium Anabaena sp. PCC 7120 Is Important But Not Essential for Nitrogen Fixation,” Molecular and General Genetics, Vol. 253, No. 6, 1997, pp. 770- 776. http://dx.doi.org/10.1007/s004380050383
- J. A. Maupin-Furlow, J. K. Rosentel, J. H. Lee, U. Deppenmeier, R. P. Gunsalus and K. T. Shanmugam, “Genetic Analysis of the modABCD (Molybdate Transport) Operon of Escherichia coli,” Journal of Bacteriology, Vol. 177, No. 17, 1995, pp. 4851-4856.
- W. T. Self, A. M. Grunden, A. Hasona and K. T. Shanmugam, “Molybdate Transport,” Research in Microbiology, Vol. 152, No. 3-4, 2001, pp. 311-321. http://dx.doi.org/10.1016/S0923-2508(01)01202-5
- M. Zahalak, B. Pratte, K. J. Werth and T. Thiel, “Molybdate Transport and Its Effect on Nitrogen Utilization in the Cyanobacterium Anabaena variabilis ATCC 29413,” Molecular Microbiology, Vol. 51, No. 2, 2004, pp. 539- 549. http://dx.doi.org/10.1046/j.1365-2958.2003.03851.x
- T. Thiel, B. Pratte and M. Zahalak, “Transport of Molybdate in the Cyanobacterium Anabaena variabilis ATCC 29413,” Archives of Microbiology, Vol. 179, No. 1, 2002, pp. 50-56. http://dx.doi.org/10.1007/s00203-002-0499-y
- I. Martinez-Argudo, R. Little, N. Shearer, P. Johnson and R. Dixon, “Nitrogen Fixation: Key Genetic Regulatory Mechanisms,” Biochemical Society Transactions, Vol. 33, Pt. 1, 2005, pp. 152-156.
- M. E. Mulligan and R. Haselkorn, “Nitrogen Fixation (nif) Genes of the Cyanobacterium Anabaena Species Strain PCC 7120. The nifB-fdxN-nifS-nifU Operon,” The Journal of Biological Chemistry, Vol. 264, No. 32, 1989, pp. 19200-19207.
- J. W. W. L. L. Golden and D. R. Wiest, “Independent Regulation of nifHDK Operon Transcription and DNA Rearrangement during Heterocyst Differentiation in the Cyanobacterium Anabaena sp. Strain PCC 7120,” Journal of Bacteriology, Vol. 173, 1991, pp. 7098-7105.
- R. Haselkorn, D. Rice, S. E. Curtis and S. J. Robinson, “Organization and Transcription of Genes Important in Anabaena Heterocyst Differentiation,” Annals of Microbiology, Vol. 134b, 1983, pp. 181-193.
- A. Herrero, A. M. Muro-Pastor and E. Flores, “Nitrogen control in cyanobacteria,” Journal of Bacteriology, Vol. 183, No. 2, 2001, pp. 411-425. http://dx.doi.org/10.1128/JB.183.2.411-425.2001
- J. L. Ungerer, B. S. Pratte and T. Thiel, “RNA Processing of Nitrogenase Transcripts in the Cyanobacterium Anabaena variabilis,” Journal of Bacteriology, Vol. 192, No. 13, 2010, pp. 3311-3320. http://dx.doi.org/10.1128/JB.00278-10
- D. M. Jackman and M. E. Mulligan, “Characterization of a Nitrogen-Fixation (nif) Gene Cluster from Anabaena azollae 1a Shows That Closely Related Cyanobacteria Have Highly Variable But Structured Intergenic Regions,” Microbiology, Vol. 141, No. 9, 1995, pp. 2235- 2244. http://dx.doi.org/10.1099/13500872-141-9-2235
- T. C. Currier and C. P. Wolk, “Characteristics of Anabaena variabilis Influencing Plaque Formation by Cyanophage N-1,” Journal of Bacteriology, Vol. 139, No. 1, 1979, pp. 88-92.
- M. B. Allen and D. I. Arnon, “Studies on Nitrogen-Fixing Blue-Green Algae. I. Growth and Nitrogen Fixation by Anabaena cylindrica Lemm,” Plant Physiology, Vol. 30, 1955, pp. 366-372. http://dx.doi.org/10.1104/pp.30.4.366
- K. Schneider, A. Müller, K. U. Johannes, E. Diemann and J. Kottmann, “Selective Removal of Molybdenum Traces from Growth Media of N2-Fixing Bacteria,” Analytical Biochemistry, Vol. 193, No. 2, 1991, pp. 292-298. http://dx.doi.org/10.1016/0003-2697(91)90024-N
- J. Elhai and C. P. Wolk, “Conjugal Transfer of DNA to Cyanobacteria,” Methods in Enzymology, Vol. 167, 1988, pp. 747-754. http://dx.doi.org/10.1016/0076-6879(88)67086-8
- J. L. Ungerer, B. S. Pratte and T. Thiel, “Regulation of Fructose Transport and Its Effect on Fructose Toxicity in Anabaena spp.,” Journal of Bacteriology, Vol. 190, No. 24, 2008, pp. 8115-8125. http://dx.doi.org/10.1128/JB.00886-08
- P. B. Hebbar and S. E. Curtis, “Characterization of devH, a Gene Encoding a Putative DNA Binding Protein Required for Heterocyst Function in Anabaena sp. Strain PCC 7120,” Journal of Bacteriology, Vol. 182, No. 12, 2000, pp. 3572-3581. http://dx.doi.org/10.1128/JB.182.12.3572-3581.2000
- R. H. Burris, “Nitrogen Fixation—Assay Methods and Techniques,” Methods in Enzymology, Vol. 24, 1972, pp. 415-431. http://dx.doi.org/10.1016/0076-6879(72)24088-5
- A. Vioque, “The RNase P RNA from Cyanobacteria: Short Tandemly Repeated Repetitive (STRR) Sequences Are Present within the RNase P RNA Gene in Heterocyst-Forming Cyanobacteria,” Nucleic Acids Research, Vol. 25, No. 17, 1997, pp. 3471-3477. http://dx.doi.org/10.1093/nar/25.17.3471
- M. J. Dilworth, R. R. Eady and M. E. Eldridge, “The Vanadium Nitrogenase of Azotobacter chroococcum. Reduction of Acetylene and Ethylene to Ethane,” Biochemical Journal, Vol. 249, No. 3, 1988, pp. 745-751.
- L. E. Bevers, P.-L. Hagedoorn and W. R. Hagen, “The Bioinorganic Chemistry of Tungsten,” Coordination Chemistry Reviews, Vol. 253, No. 3-4, 2009, pp. 269-290. http://dx.doi.org/10.1016/j.ccr.2008.01.017
- W. R. Hagen, “Cellular Uptake of Molybdenum and Tungsten,” Coordination Chemistry Reviews, Vol. 255, No. 9- 10, 2011, pp. 1117-1128. http://dx.doi.org/10.1016/j.ccr.2011.02.009
NOTES
*Corresponding author.

