Advances in Biological Chemistry
Vol.3 No.5(2013), Article ID:38815,3 pages DOI:10.4236/abc.2013.35057
Molecular cloning and characterization of an ECTO-NOX3 (ENOX3) of Saccharomyces cerevisiae
![]()
1Department of Biology, Valparaiso University, Valparaiso, USA
2MorNuCo, Inc., Purdue Research Park, West Lafayette, USA
Email: *dj_morre@yahoo.com
Copyright © 2013 Sara Dick et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 27 August 2013; revised 27 September 2013; accepted 15 October 2013
Keywords: Age-Related NADH Oxidase (arNOX) ECTO-NOX3; TM9 Superfamily of Transmembrane Proteins; Molecular Cloning; Saccharomyces cerevisiae
ABSTRACT
Exfoliated ECTO-NOX3 (ENOX3) proteins, are members of the human TM9 superfamily of transmembrane proteins that generate superoxide, are present in blood and other body fluids, and increase activity with age beginning about age 30, hence agerelated NOX (arNOX or ENOX3). A yeast deletion library was screened based on NADH fluorescence using a 384 well plate assay to identify a yeast isolate lacking a previously identified cell surface oxidase exhibiting an oscillatory pattern with a period length of 26 min and capable of generating superoxide. The cDNA was cloned from a yeast over expression library using NADH as an impermeant substrate with analysis by Fast Fourier Transform and decomposition fits. The objective was to identify and sequence an ENOX homologue in Saccharomyces cerevisiae with a 26 min rather than a 24 or 25 min period length. The finding identified YER113C as the yeast ENOX3 protein with a 26 min period and capable of generating superoxide. The encoded protein was expressed in bacteria and characterized. Gel slices of expressed proteins revealed a protein of ca. 81,545 kDa with properties paralleling those of human arNOX (periodic NADH oxidation, protein disulfide thiol interchange, inhibited by mammalian arNOX inhibitors and superoxide production inhibited by superoxide dismutase). The YER113C sequence exhibited a 44% similarity and a 26% identity with the mammalian ENOX3 SF4 (arNOX SF4) of the TM9 superfamily of transmembrane proteins1. The YER113C deletion mutant lacked arNOX activity.
1. INTRODUCTION
Our work has identified a group of oscillatory cell surface oxidases in the yeast Saccharomyces cerevisiae [1,2] classified as ECTO-NOX (ENOX) proteins with human homologs [3]. ENOX1 is constitutive [2,4] with oscillations having a period of 24 min. ENOX2 is cancer related [5] with a period length of 22 min. The age-related ENOX (ENOX3) proteins are uniquely superoxide generating members of the human TM9 superfamily of transmembrane proteins that are shed into blood and body fluids and increase in activity with age beginning at about age 30 [6] and have a period length of 26 min.
In this report, we identify an oscillatory, superoxidegenerating NADH oxidase activity with a period length of 26 min of S. cerevisiae. The ENOX3 in yeast was overexpressed by a selection process using a deletion library. One deletion strain had a deletion at the ENOX3 protein sequence and, thus, only showed yeast ENOX1 characteristics. The other deletion strain had a deletion at the yeast ENOX1 sequence and, thus only showed the ENOX3 characteristics. Using this approach, a candidate yeast gene potentially encoding a yeast ENOX3 protein with a 26 min period capable of generating superoxide was identified and characterized.
2. METHODS
Candidates for yeast ENOX3 were selected from a deletion library of nonessential yeast genes which showed altered peak patterns from 7 - 8 to 5 peaks (Figure 1A). The deletion candidates used for ENOX3 showed both a 26 min oscillatory pattern of NADH oxidation (Figure 1B) and/or reduced coenzyme Q10 assays and catalyzed
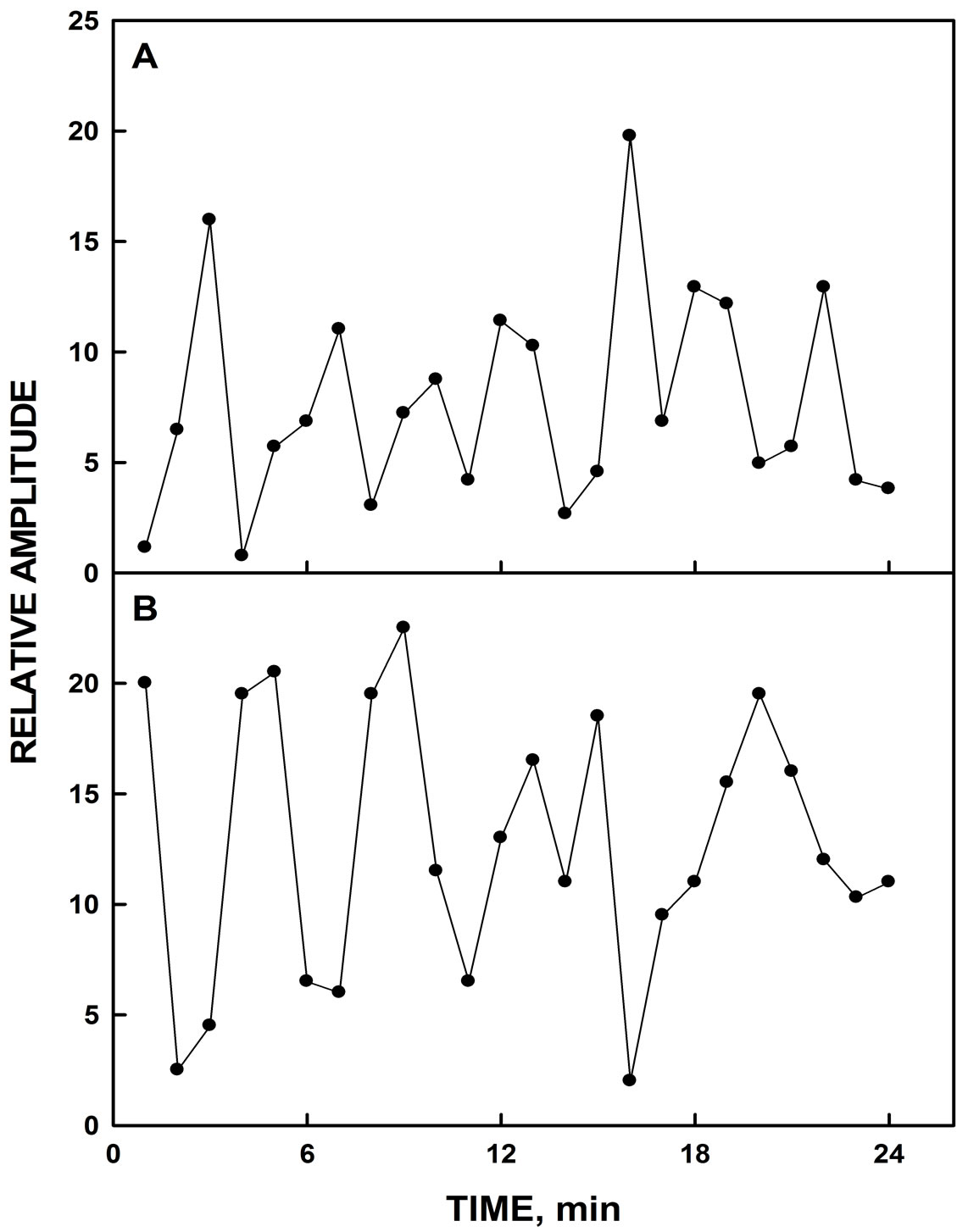
Figure 1. NADH fluorescence decomposition fits. A. Wild type yeast strain showing a pattern of 7 maxima. B. YER113C showing a pattern of 5 maxima.
the formation of superoxide (Results).
2.1. Saccharomyces Growth and Preparation
Yeast were grown at room temperature with shaking in rich media (YEPD) for 1 - 2 days until saturation. The yeast strains were maintained on YEPD agar plates and stored at 4˚C. Yeast cells were inactivated by heating for 1 h at 70˚C prior to assay.
2.2. Purification of YER113C
The yeast whole cell pellet with overexpression of Histagged YER113C, the putative yeast ENOX3, was resuspended in 20 mM Tris-HCL, pH 8.0 with 0.5 mM benzamidine, 0.5 mM PMSF and 1 mM 6-aminohexanoic acid. Cells were lysed by three passages through a French pressure cell at 20,000 psi. The resultant pellet was extracted sequentially. The supernatant following centrifugation at 10,000 rpm for 15 min at 40˚C was saved. The resultant pellet was then extracted with 20 mM Tris-HCL, pH 8.0, containing 1% Triton X-100, 20 mM Tris-HCL, pH 8.0, with 0.3% sarcosine, and in 20 mM Tris-HCL, pH 8.0 with 0.4% SDS. The combined supernatants were analyzed by SDS-PAGE and western blot and silver stain to determine the location of the yeast ENOX3. Additionally the supernatant was applied to a nickel nitrilotriacetic acid (Ni-NTA) column to bind the histidine tag and eluted with imidazole to further purify the protein. The identity of the YER113C protein was confirmed by sequencing.
2.3. SDS-PAGE Gel Slicing
The SDS-PAGE gels were sliced every 0.5 cm and the slices were eluted with 50 mM Tris-MES, pH 7.0 overnight in 4˚C. The gel slice eluates were assayed for ENOX activity based on NADH oxidation. Proteins were determined by the bicinchoninic acid (BCA) method with bovine serum albumin as a standard [7].
2.4. SDS-PAGE and Western Blot
Yeast samples resolved on 10% SDS polyacrylamide gels were transferred to a nitrocellulose membrane at 90 V for 1 h. A 5% solution of non-fat milk powder was used for blocking and the probe used for the western blot was a 1:2500 anti-histidine antibody (Sigma A5588).
2.5. NAD(P)H Oxidase Activity
The oxidation of NADH was measured from the disappearance of NADH at a wavelength of 340 nm in a reaction mixture containing 50 mM Tris-MES pH 7.0, 2 mM KCN to inhibit mitochondrial oxidase activity, and 150 μM NADH, and YER113C sample at 37˚C with temperature control and stirring [8]. Prior to the assay, 0.1 mM GSH was added to reduce the protein in the presence of its substrate. After 10 min, 0.03% H2O2 was added to reoxidize the protein under renaturing conditions and in the presence of substrate to start the reaction. After an additional 10 min, measurements were recorded continuously over 1 min at intervals of 1.5 min using a SLM Aminco DW 2000 UV-VIS spectrophotometer (Milton Roy, Rochester NY). An extinction coefficient of 6.22 mM−1·cm−1 was used to determine specific activity.
2.6. Measurement of Superoxide Formation
Measurements of superoxide production were based on a standard method where reduction of ferricytochrome c by superoxide was monitored from the increase in absorbance at 550 nm with reference at 540 nm [9]. As a further check for the specificity of the ENOX3 activity, 60 units of superoxide dismutase (SOD) were added near the end of the assay to ascertain that the rate returned to base line. Rates were determined over 1 min at intervals of 1.5 min using a SLM Aminco DW 2000 UV-VIS spectrophotometer (Milton Roy, Rochester NY) in the dual wavelength mode of operation. An extinction coefficient of 19.1 mM−1·cm−1 was used for reduced ferricytochrome c. Superoxide dismutase was added at the end of each assay to ascertain that activity based on superoxide production returned to base line.
2.7. Hydroquinone Oxidation
The oxidation of reduced coenzyme Q10 (CoQ10H2) was determined using a Hitachi U-3110 spectrophotometer from the disappearance of reduced CoQ at both 290 nm and 410 nm [10] in 50 mM Tris-MES pH 7.0 and 40 μL of 0.35% ubiquinol (Tischcon). After 10 min, the yeast sample was added to initiate the reaction. An extinction coefficient of 0.805 mM−1·cm−1 was used to calculate the rate of Q10H2 oxidation.
2.8. DTDP Cleavage
YER113C samples were added to 2.5 ml of reaction buffer (50 mM Tris-Mes, pH 7.0). The reaction was preincubated with 0.5 µmol of 2.2’-dithiodipyridine (DTDP) in 5 µl DMSO. After 10 min of incubation, a further 3.5 µmol of DTDP were added in 35 µl of DMSO to start the reaction. The increase in absorbance due to the cleavage of DTDP was monitored at 340 nm. The specific activity of the cleavage reaction was calculated using a millimolar absorption coefficient of 6.21 [11].
2.9. NADH Fluorescence
As YEPD medium is fluorescent, heat-inactivated yeast cells were washed with PBS by centrifuging the cells in the benchtop mini centrifuge (VWR, Pennsylvaia) for 1 - 2 min, the supernatant was discarded and the pellet was resuspended in PBS. This suspension was used for measurements of NADH fluorescence.
3. RESULT
The inactivated yeast suspension in PBS was added to wells of a black 96-well plate at a dilution of 1:10 in a volume of 20 µl. The PBS for the experiment was supplemented with 2% glucose to support enzyme activity. The plate was loaded into a Fluoroskan fluorescent plate reader and the samples were measured once every min for 2 - 8 h, with excitation at 355 nm and emission at 460 m. Data were analyzed by Fast Fourier Transform (Minitab 15) and decomposition analysis (Minitab 15) [12].
The induced yeast lysate when assayed for NADH oxidase activity (Figure 2) exhibited an asymmetric pattern of oscillations with five maxima separated by 6 min. The remaining maxima were separated by an average of 5 min instead of the usual 4 min which, when combined, yielded a total of 26 min to complete the oscillatory ENOX3 cycle. In addition, lysates exhibited a burst of superoxide production coincident with maximum ƒ which is an essential defining characteristic of ENOX3 proteins as measured by reduction of ferricytochrome c (Figure 3). The burst of ferricytochrome c reduction at
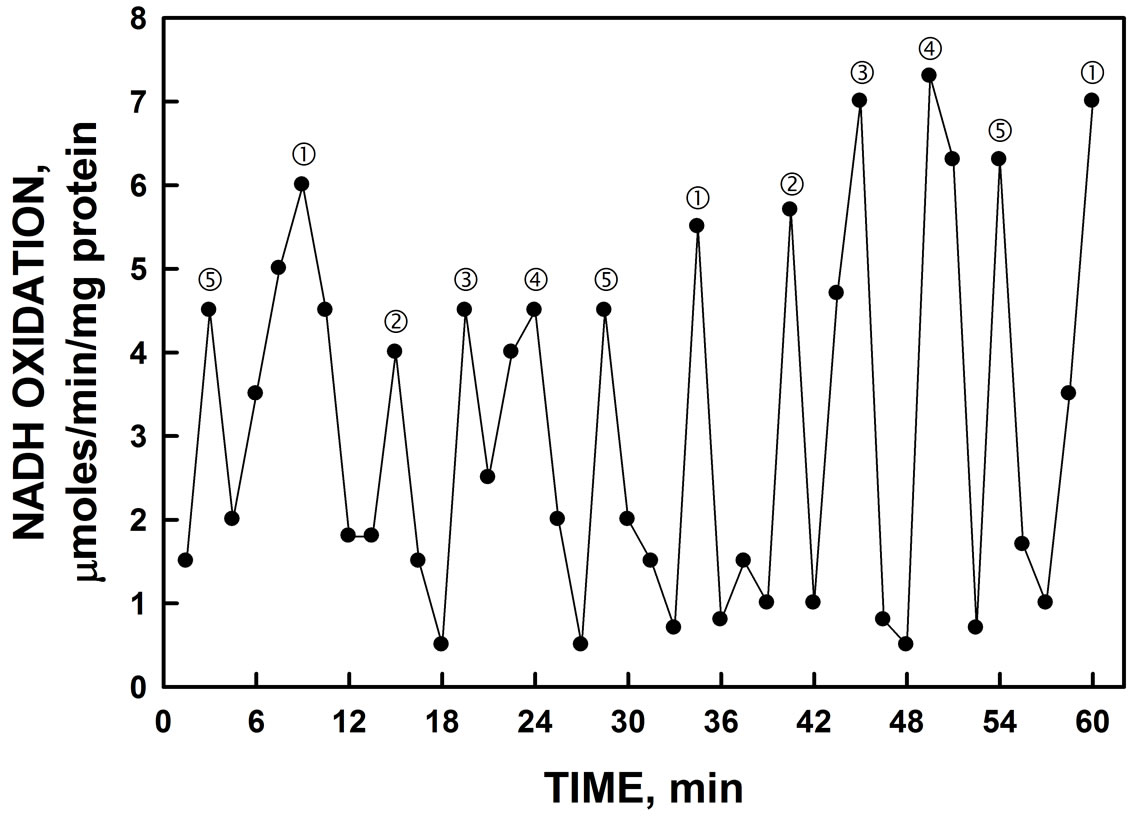
Figure 2. Oxidation of NADH by YER113C yeast lysate. Maxima labeled and ‚ are separated from each other by 6 min whereas maxima labeled ƒ, „ and … are separated from each other and from maxima and ‚ by about 5 min to produce a 26 min period (6 + 4 × 5 min). Activity was unaffected by the addition of the ENOX1 inhibitor 1 µM simalikalactone (not shown).
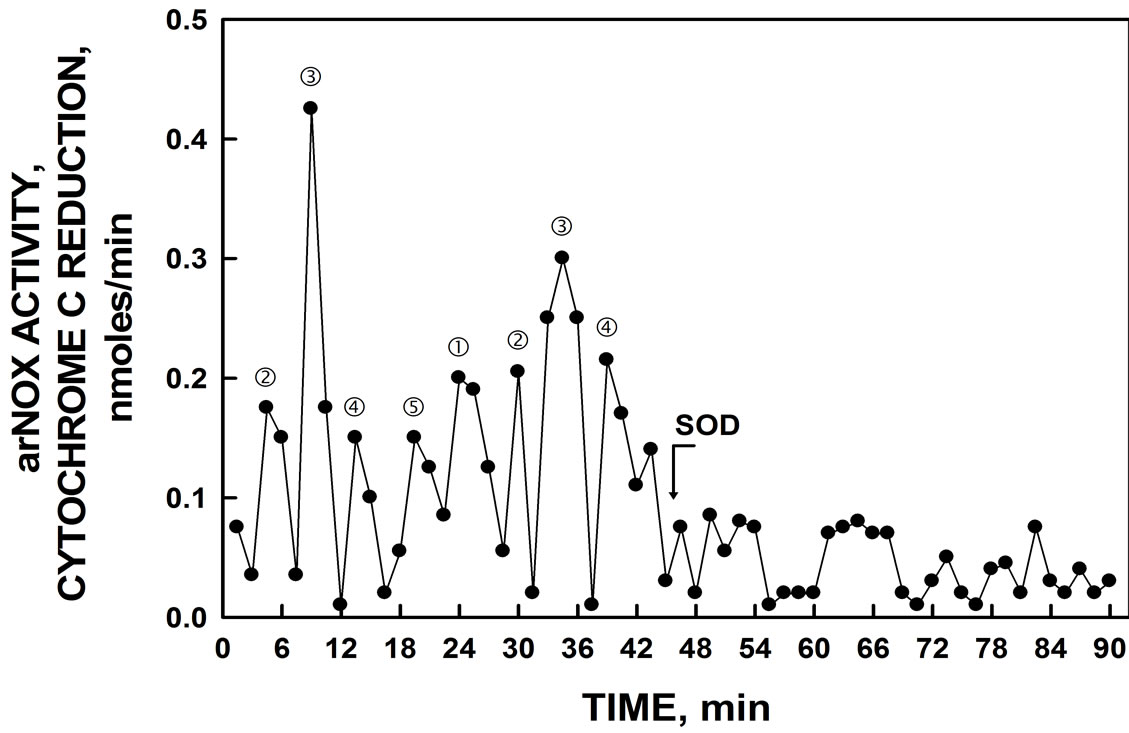
Figure 3. ENOX3 activity by YER113C yeast lysate. Bursts of superoxide generation as measured by reduction of ferricytochrome c were separated by intervals of 26 min and eliminated by the addition of 60 units of superoxide dismutase (SOD) added after 45 min.
peak ƒ was eliminated by the addition of superoxide dismutase to verify its being attributed to superoxide.
YER113C encodes a 706 aa protein of molecular weight 81,545 and isoelectric point pH 7.39 (Figure 4). The amino acid sequence exhibited 26% identity and 44% similarity to the mammalian ENOX3 (ENOX3) SF4 (BK008759), and a 25% identity and 40% similarity to the mammalian arNOX (ENOX3) SF3 (BK008789), 25% identity and 40% similarity to mammalian arNOX (ENOX3) SF2 (BK008790), all of the TM9 superfamily of transmembrane proteins.
Functional motifs in common with mammalian arNOX proteins included a 683GLGALS (GXGXXS) adenine binding motif and 576YVY and 590YFY putative copper binding motifs as well as a potential disulfide
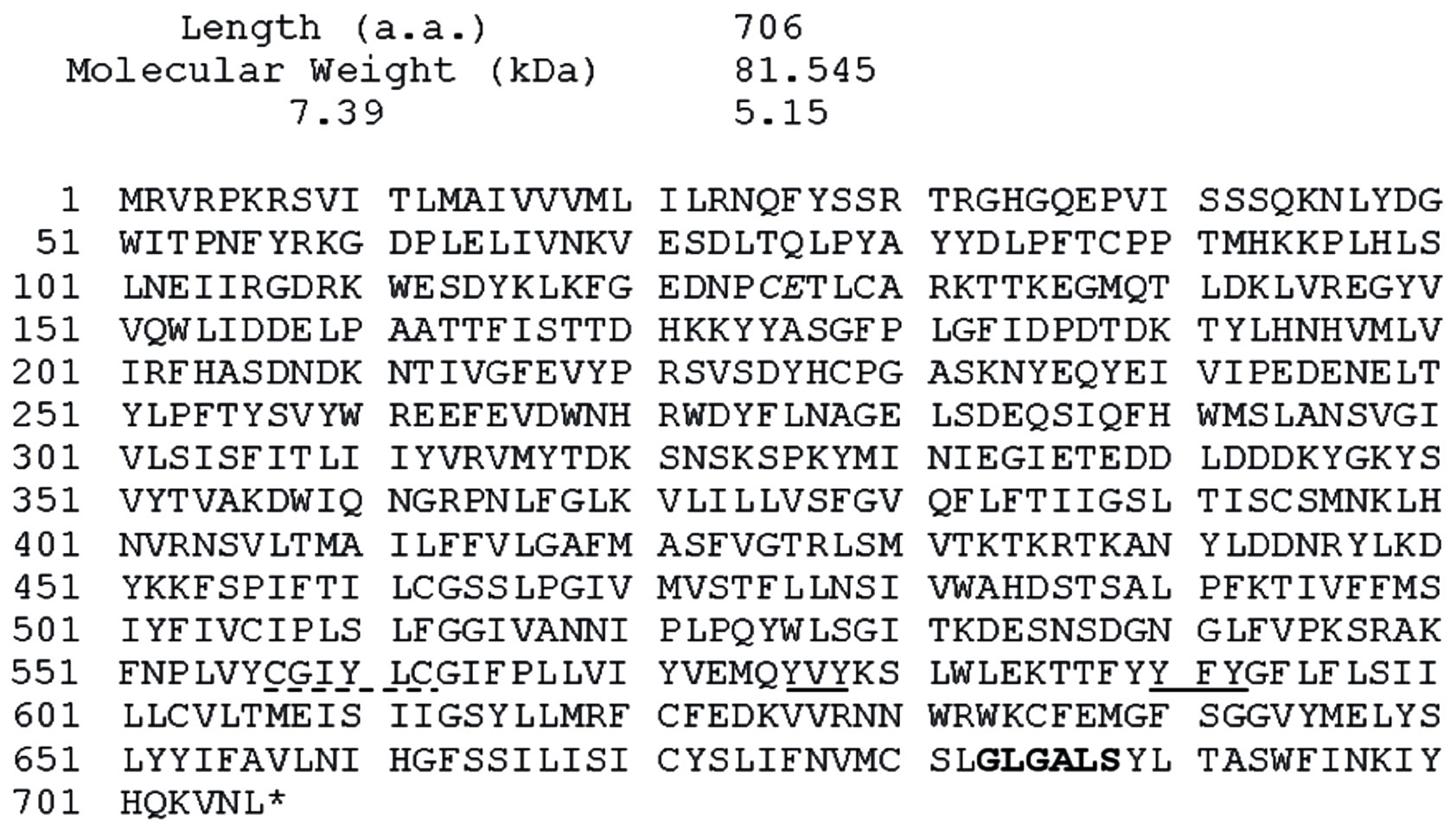
Figure 4. Sequence of YER113C. A putative NADH-binding sequence is bolded, potential protein disulfide-thiol inter-change sites are denoted by dashed underlines and putative copper binding sites are underlined.
interchange site 557CGIYLC (CXXXXC). Also present was a conserved CQ/CE motif common to the mammalian ENOX3 family of proteins at 125CE.
Sliced and pulverized SDS-PAGE gels from which the YER113C proteins were eluted overnight in assay buffer evealed enzymatic activity in the region of the gel capable of superoxide generation based on reduction of ferricytochrome c. The active fraction of resolubilized pelleted material from the yeast lysate corresponded to a molecular weight of about 81.5 kDa (Figure 5A). This slice also contained the protein based on western blot analysis showing the location of the His tag (Lane 2). With soluble material from the yeast lysates, activity corresponding to western blot analyses also was found at molecular weights of about 55 kDa possibly representing a cleavage product (Figure 5B). When hydroquinones were employed as a natural membrane-located substrate, compared to NADH, a single set of 5 maxima were seen with the gel slice elute corresponding to the 55 kDa cleavage product (Figure 6). Measurement, either as an increase in absorbance at 410 nm or as a decrease in absorbance at 290 nm hydroquinone oxidation, yielded the five maxima oscillatory pattern.
The protein disulfide-thiol interchange activity of the gel slice elute containing 55 kDa truncation was determined from the cleavage of dithiodipyridine substrate (Figure 7). The five maxima oscillatory pattern was obtained similar to the NADH oxidase activity. However, in contrast to NADH oxidation where maxima and ‚ often appear to dominate (e.g., Figure 2, the maxima separated by 5 min, labeled ƒ, „ and … were of similar amplitude to those of maxima labeled and ‚ separated by 6 min.
The yeast activity was resistant to simalikalactone D, a
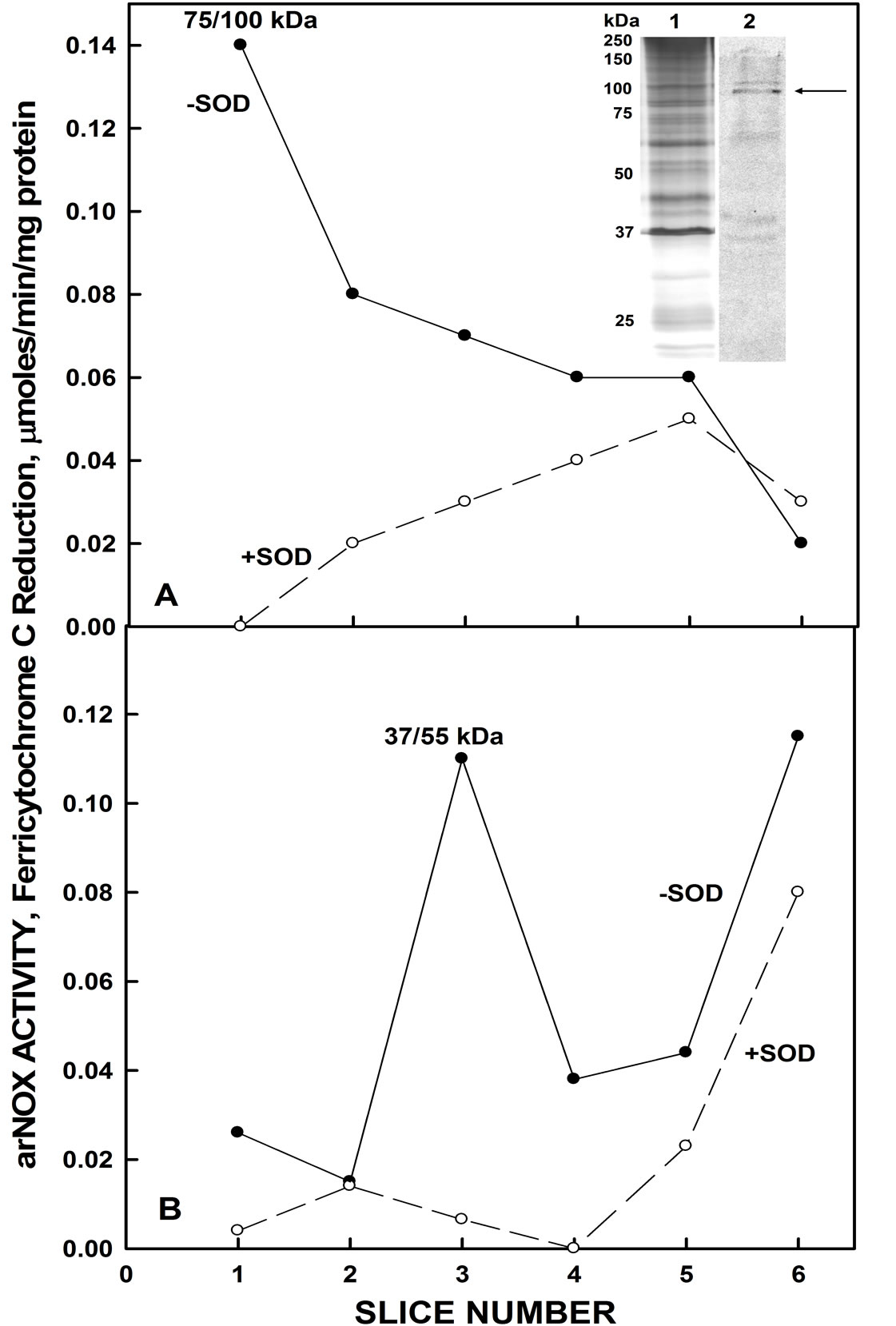
Figure 5. Resolution of recombinant His-tagged YER113C by SDS-Page. A. With the insoluble pellet from yeast lysates, eluted activity correlated with the region of the gel consistent with a molecular weight of about 81.5 kDa (arrow). Inset: Lane 1. Silver stain. Lane 2. Western blot with antihistidine antibody. B. With the soluble fraction from the yeast lysate, activity correlated with a gel slice band of about 37 - 55 kDa.
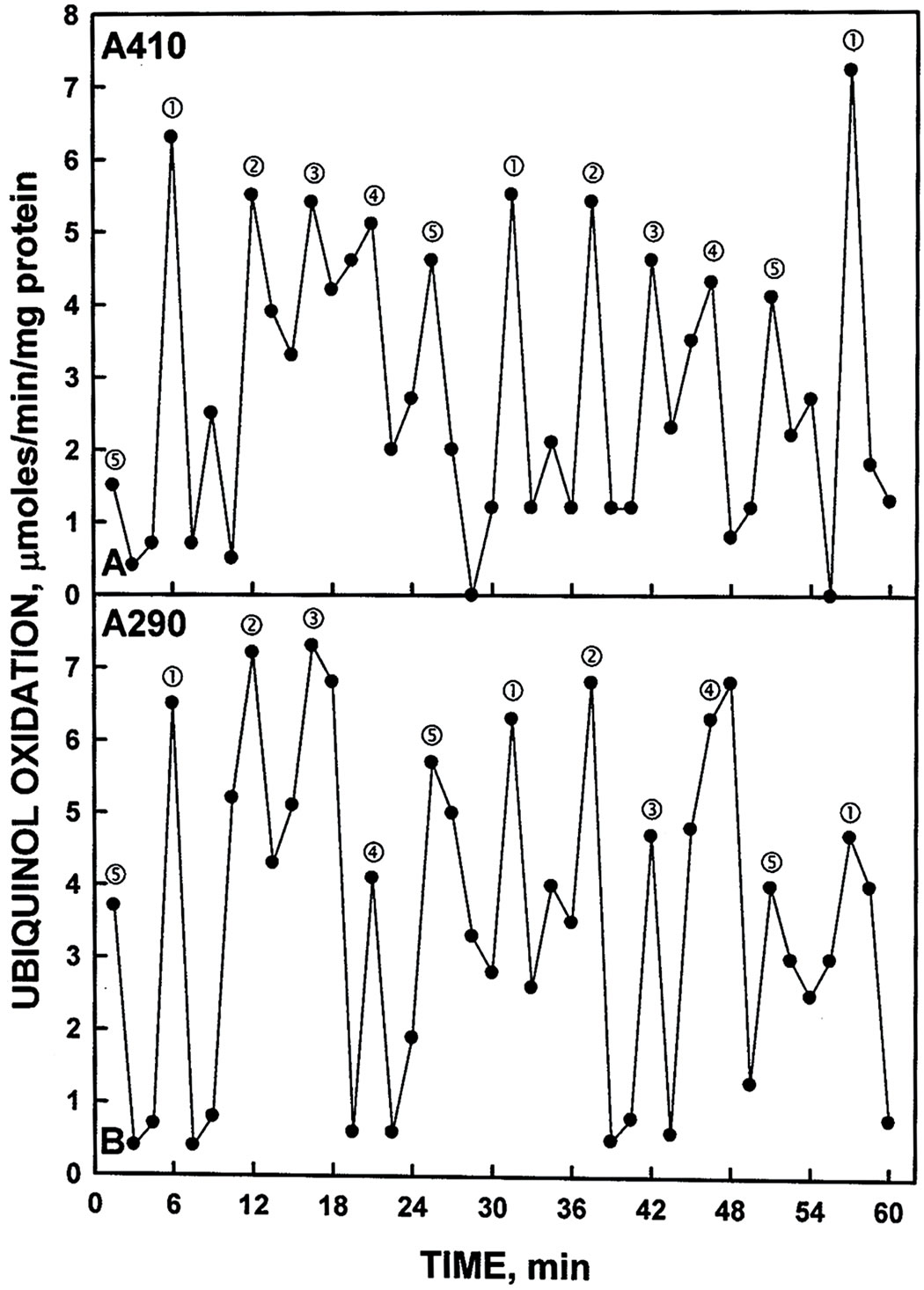
Figure 6. Oxidation of reduced coenzyme Q by a Histagged YER113C measured either by an increase in A410 (A) or by a decrease in A290 (B). As with NADH oxidation of Figure 2, the activity oscillated with prominent maxima separated by 6 min and ‚ followed by 3 additional maxima separated by 5 min to generate the 26 min period.
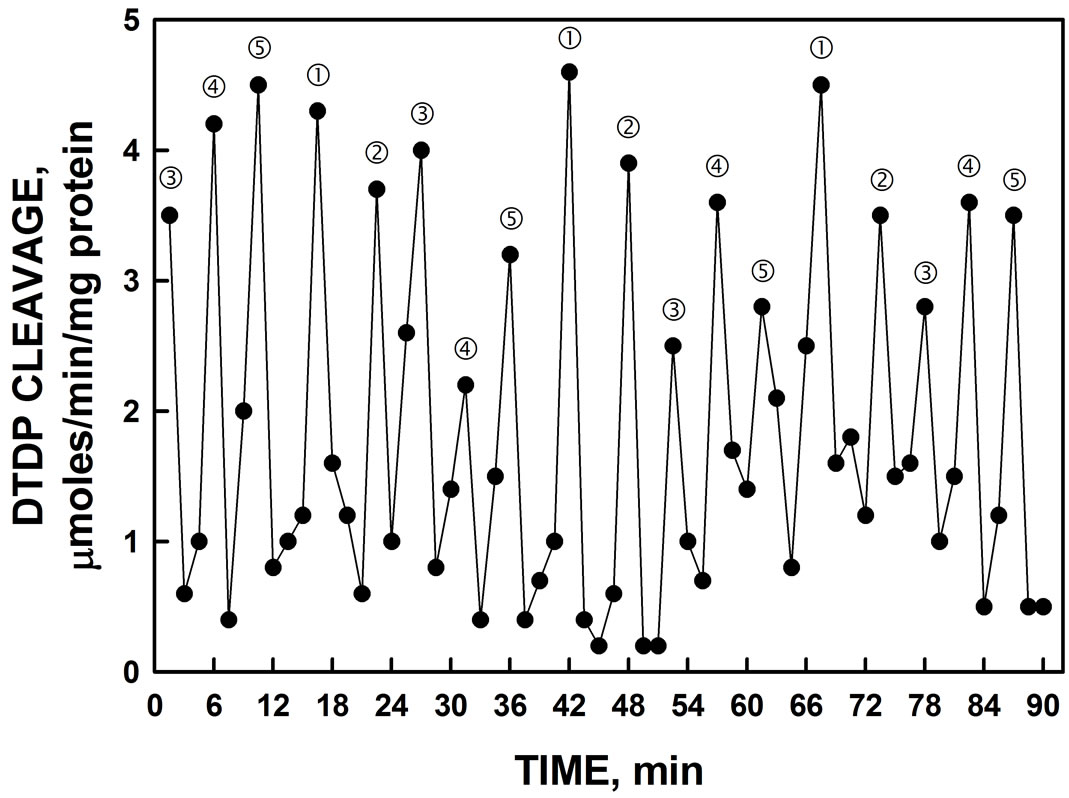
Figure 7. Protein disulfide-thiol interchange activity of Histagged YER113C determined by cleavage of a dithiodipyridine (DTDP) substrate. The spacing of labeled maxima is approximately the same as that of Figure 2.
specific inhibitor of ENOX1 nor were the activity patterns phased by the addition of melatonin. Phasing by melatonin is an ENOX1 characteristic [2,4]. The period length, however, was increased to about 32 min by assay in D2O in place of water (Figure 8) as is characteristic of ENOX proteins generally. In contrast to melatonin, the activity was phased by exposure to low frequency electromagnetic fields (20 sec, 50 µT) (Figure 9).
A specific ENOX3 inhibitor mixture of dormin + Schizandra + salicin [to 2.5 ml of assay volume were added 60 µl of an aqueous mixture of 4 mg/ml Schizandra (Schizandra chinensis extract, 9% schizandrins, Draco, San Jose, CA) plus 1 mg/ml salicin (Sigma, St. Louis, MO) and 20 µl of IBR Dormin (Israli Biotechnology Research, Ramat-Gan, Israel) = AgeLoc (NuSkin Enterprises, Provo, UT)] inhibited the activity by >90%. Also inhibitory were a herbal hot water infusion of dried savory at a concentration of 125 mg/ml (>90%), gallic acid (200 µM), and coenzyme Q10. The ENOX1-specific
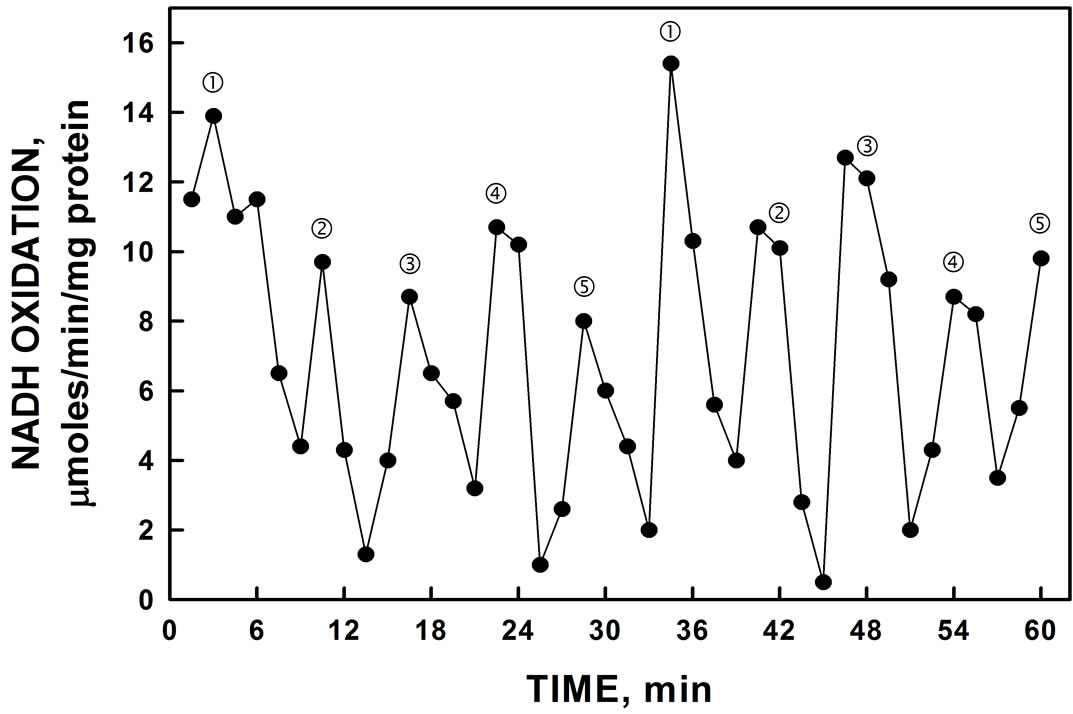
Figure 8. His-tagged YER113C activity assayed in D2O rather than H2O. In the presence of D2O, the period length of the oscillation was lengthened from 26 min to approximately 32 min. The effect of heavy water to increase period length is one of the hallmarks of the ENOX family of proteins.
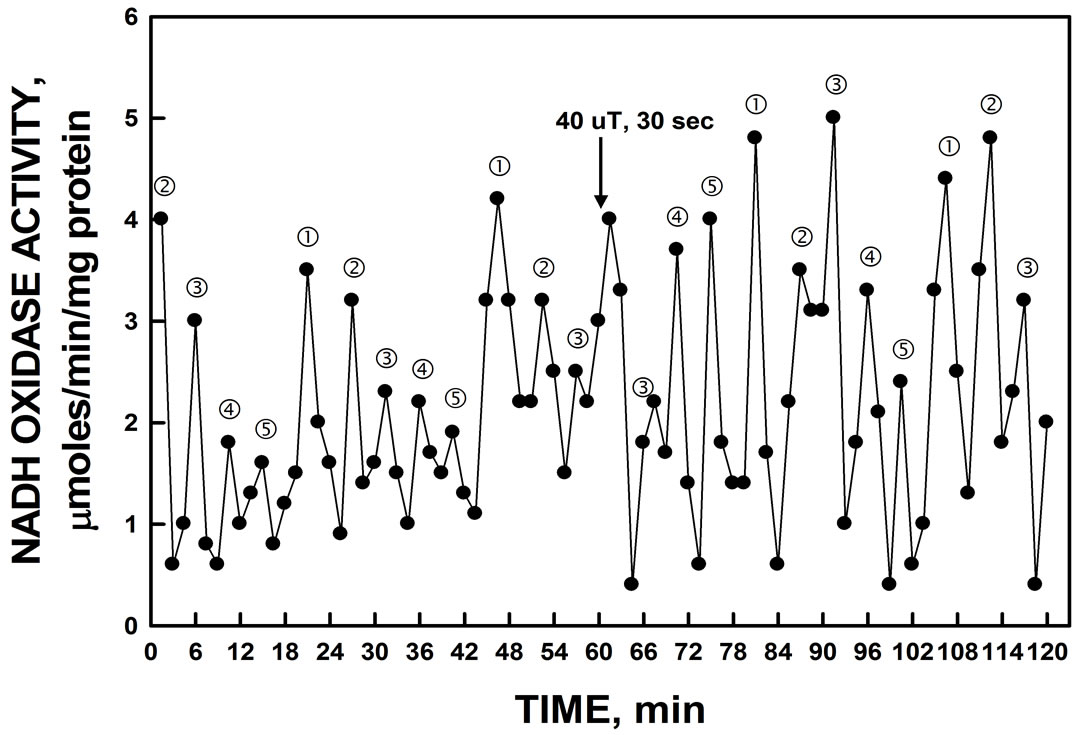
Figure 9. The activity period of NADH oxidation by recombinant YER113C was phased by the application of low-frequency EMF applied at 60 min. A control cuvette, protected by a Faraday cage, was assayed in parallel in a second spectrophotometer. All maxima were shifted by approximately 7.5 min by the 20 sec EMF exposure to 50 µT relative to the control preparation which was unchanged (not shown).
NOX inhibitor simalikalactone D, the mammalian ENOX3 inhibitor tyrosol (Figure 10) and a peptide antibody to mammalian TM9SF4 (not shown) were without effects.
4. DISCUSSION
Based on DNA sequence, YER113C, the human TM9SF4 homolog was expressed in E. coli and shown to fulfill the requirements for an age-related ENOX (arNOX or ENOX3) protein. The pattern of activity oscillations consisted of 5 unequally spaced maxima with the requisite 26 min period length and the generation of superoxide. Superoxide generation was evidenced by the superoxide dismutase inhibited reduction of ferricytochrome c. Superoxide is not a reaction product of the two other ENOX proteins: ENOX1 with a 24 min period length [2] and YNOX with a 25 min period length [13].
The human ENOX3 cDNAs all encode polypeptides having a highly hydrophobic C-terminal portion were organized into 9 transmembrane regions [14,15]. The transmembrane domains all have a similar structure and sequence to form a novel family of multispanning domain proteins designated “TM9SF” (transmembrane protein 9 superfamily) by the Human Gene Nomenclature Committee. The leader member of the TM9SF family is the S. cerevisiae EMP70 gene product, a 70 kDa precursor that is processed into a 24 kDa protein (p24a) located in the endosomes [14]. Full length members of the TM9 protein superfamily are all characterized as cell surface proteins having the characteristic series of membrane spanning hydrophobic helices that criss-cross the plasma membrane and are also present on endosomes [14,16,17]. YER113C also localizes to the Golgi apparatus in yeast [18].
There are 5 TM9 super family members known in the human genone (1 with two transcript variants, 2, 3 and 4).
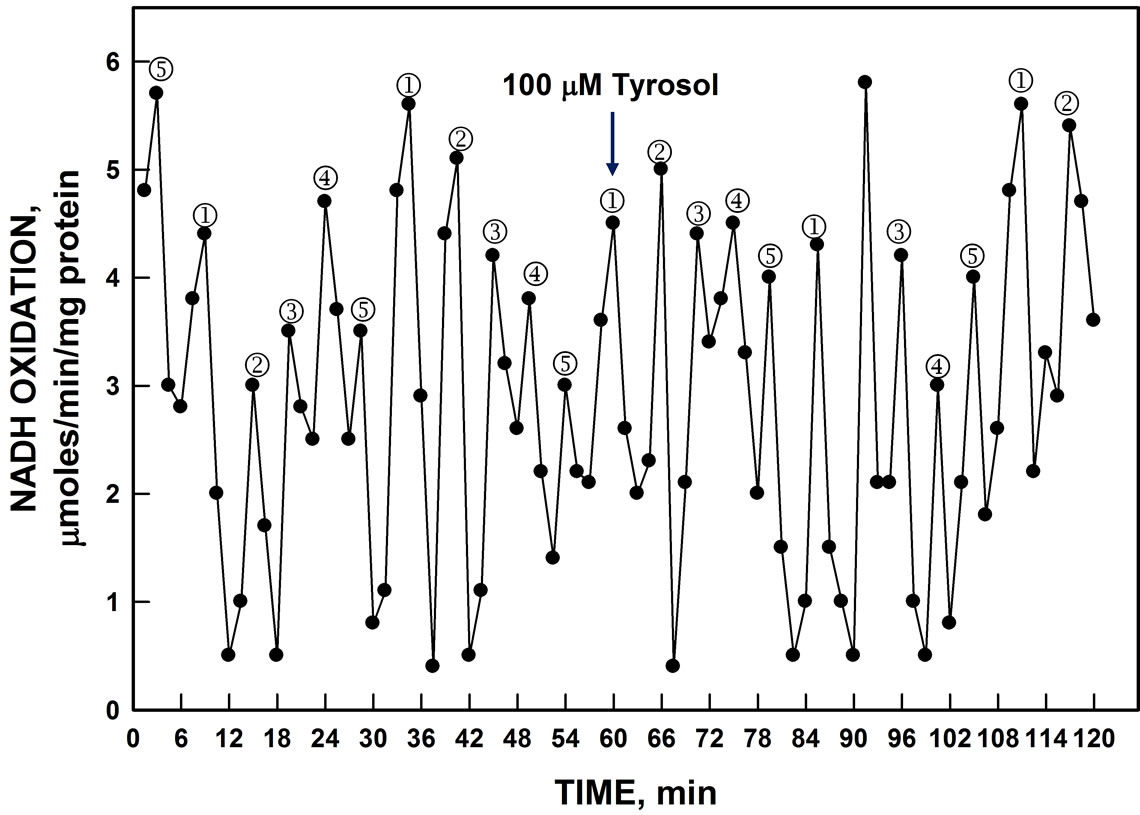
Figure 10. Oxidation of NADH by YER113C lysate was unaffected by addition of 100 µM tyrosol (arrow).
The two transcript variants of human family member 1 are similar with the exception that number 1a transcript variant contains additional C-terminal residues absent from transcription variant 1b. The yeast homolog most closely resembles family member 4. Interestingly, the human TM9SF4 is highly expressed in human melanoma cells [17].
5. ACKNOWLEDGEMENTS
We thank Debby Parisi for assistance and Peggy Runck for manuscript preparation.
REFERENCES
- Dick, S.S., Ryuzoji, A., Morré, D.M. and Morré, D.J. (2013) Ultradian oscillators of the circadian clock in Saccharomyces cerevisiae. Advances in Biological Chemistry, 3, 59-69. http://dx.doi.org/10.4236/abc.2013.31008
- Dick, S.S., Ryuzoji, A., Morré, D.M. and Morré, D.J. (2013) Identification of the constitutive ultradian oscillator of the circadian clock (ENOX1) in Saccharomyces cerevisiae. Advances in Biological Chemistry, in press.
- Morré, D.J. and Morré, D.M. (2013) ECTO-NOX Proteins. Springer, New York, 507 p. http://dx.doi.org/10.1007/978-1-4614-3958-5
- Jiang, Z., Goldstein, N. M., Morré, D.M. and Morré, D.J. (2008) Molecular cloning and characterization of a candidate human growth-relaqted ant time-keeping constitutive cell surface hydroquinone (NADH) oxidase. Biochemistry, 47, 14028-14038. http://dx.doi.org/10.1021/bi801073p
- Chueh, P.-J., Kim, C., Cho, N., Morré, D.M. and Morré, D.J. (2002) Molecular cloning and characterization of a tumor-associated, growth-related and time-keeping hydroquinone (NADH) oxidase (NOX) of the HeLa cell surface. Biochemistry, 41, 3732-3741. http://dx.doi.org/10.1021/bi012041t
- Tang, X., Parisi, D., Spicer, B. Morré, D.M. and Morré, D.J. (2013) Molecular cloning and characterization of human age-related NADH oxidase (ENOX3) proteins as members of the TM9 superfamily of transmembrane proteins. Advances in Biological Chemistry, 3, 187-197. http://dx.doi.org/10.4236/abc.2013.32024
- Smith, P.K., Krohn, R.I., Hermanson, G.T., Mailia, A.K., Gartner, F.H., Provenzano, M.D., Fujimoto, E.K., Goeke, N.M., Olson, F.J. and Klenk, D.C. (1985) Measurement of protein using bicinchoninic acid. Analytical Biochemistry, 150, 76-85. http://dx.doi.org/10.1016/0003-2697(85)90442-7
- Brightman, A.O., Wang, J., Miu, R.K., Sun, L.L., Barr, R., Crane, F.L. and Morré, D.J. (1992) A growth factorand hormone-stimulated NADH oxidase from rat liver plasma membrane. Biochimica et Biophysica Acta, 1105, 109-117. http://dx.doi.org/10.1016/0005-2736(92)90168-L
- Butler, J., Koppenol, W.H. and Margoliash, E. (1982) Kinetics and mechanism of the reduction of ferricytochrome c by the superoxide anion. The Journal of Biological Chemistry, 257, 10747-10750.
- Kishi, T., Morré, D.M. and Morré, D.J. (1999) The plasma membrane NADH oxidase of HeLa cells has hydroquinone oxidase activity. Biochimica et Biophysica Acta, 1412, 66-77. http://dx.doi.org/10.1016/S0005-2728(99)00049-3
- Morré, D.J., Gomez-Rey, M.L., Schramke, C., Em, O., Lawler, J., Hobeck, J. and Morré, D.M. (1999) Use of dipyridyl-dithio substrates to measure directly the protein disulfide-thiol interchange activity of the auxin stimulated NADH: Protein disulfide reductases (NADH oxidase) of soybean plasma membranes. Molecular and Cellular Biochemistry, 207, 7-13. http://dx.doi.org/10.1023/A:1006916116297
- Foster, K., Anward, N., Pogue, R., Morré, D.M., Keenan, T.W. and Morré, D.J. (2003) Decomposition analyses applied to a complex ultradian biorhythm: The oscillating NADH oxidase activity of plasma membraneshaving a potential time-keeping (clock) function. Nonlinearity in Biology, Toxicology and Medicine, 1, 51-70.
- Ryuzoji, A.F., Parisi, D.H., Dick, S.S., Kim, J., Dick, S.S., Ryuzoji, A., Morré, D.M. and Morré, D.J. (Submitted) Molecular cloning and characterization of a candidate ENOX protein of Saccharomyces cerevisiae with a 25 min period insensitive to simalikalactone D inhibition and melatonin. Advances in Biological Chemistry.
- Schimmöller, F., Diaz, E. Mühlbauer, B. and Pfeffer, S.R. (1998) Characterization of a 76 kDa endosomal, multispanning membrane protein that is highly conserved throughout evolution. Gene, 216, 311-318. http://dx.doi.org/10.1016/S0378-1119(98)00349-7
- Sugasawa, T., Lenzen, G., Simon, S., Hidaka, J., Cahen, A.W., Guillaume, J.-L., Camoin, L., Strosberg, A. and Nahmias, C. (2001) The iodocyanopindolol and SM- 11044 binding protein belongs to the TM9SF multispanning membrane protein superfamily. Gene, 273, 283-289. http://dx.doi.org/10.1016/S0378-1119(01)00587-X
- Singer-Krüger, B., Frank, R., Crausaz, E. and Riezman, H. (1993) Partial purification and characterization of early and late endosomes from yeast. Identification of four novel proteins. The Journal of Biological Chemistry, 268, 14376-14386.
- Lozupone, F., Perdicchio, M., Brambilla, D., Borghi, M., Meschini, S., Barca, S., Marino, M.L., Logozzi, M., Federici, C., Iessi, E., de Milito, A. and Fais, S. (2009) The human homologue of Dictyostelium discoideum phg1A is expressed by human metastatic melanoma cells. EMBO Reports, 10, 1348-1354. http://dx.doi.org/10.1038/embor.2009.236
- Huh, W.K., Falvo, J.V., Gerke, L.C., Carroll, A.S., Howson, R.W., Weissman, J.S. and O’Shea, E.K. (2003) Global analysis of protein localization in budding yeast. Nature, 425, 686-691. http://dx.doi.org/10.1038/nature02026
ABBREVIATIONS
BCA: bicinchoninic acid;
ENOX3: age-related NADH oxidase;
CAPS: 3(cyclohexylamino)-1-propane sulfonic acid;
DTDP: dithiodipyridine;
ELISA: enzyme-linked immunosorbent assay;
ENOX: ECTO-NOX;
GSH: reduced glutathione;
Ni-NTA: nickel nitrilotriacetic acid;
IPTG: isopropyl-beta-D-thiogalactopyranoside;
PMSF: phenylmethylsulfonyl fluoride;
SDS-PAGE: sodium dodecyl sulfate polyacrylamide gel electrophoresis;
SF: superfamily;
SOD: superoxide dismutase;
TFA: trifluoroacetic acid;
TM: transmembrane.
NOTES
*Corresponding author.
1Mammalian nucleotide sequence data for comparison are available in The Third Party Annotation section of the DDBJ/EMBL/GenBank databases under the accession numbers TPA:BKi008759, BK008789 nd BK008790.

