American Journal of Molecular Biology
Vol. 2 No. 4 (2012) , Article ID: 24044 , 8 pages DOI:10.4236/ajmb.2012.24041
GAPDH expression as a measurement of transfection efficiency for p16INK4a gene silencing (siRNA) in senescent human diploid fibroblasts*
![]()
1Department of Biochemistry, Faculty of Medicine, Universiti Kebangsaan Malaysia, Kuala Lumpur, Malaysia
2Department of Physiology, Faculty of Medicine, Universiti Kebangsaan Malaysia, Kuala Lumpur, Malaysia
Email: #suzanamakpol@yahoo.com
Received 19 July 2012; revised 29 August 2012; accepted 19 September 2012
Keywords: GAPDH; Transfection Efficiency; p16INK4a siRNA; HDF Aging model
ABSTRACT
Human diploid fibroblasts (HDFs) undergo a limited number of cell divisions in culture. After certain population doublings, they reach a state of irreversible growth arrest known as replicative senescence. Senescent HDFs showed several molecular and cytological changes such as large flat morphology, expression of senescence-associated b-galactosidase (SA b-gal) activity and altered gene expression. Small interfering RNA (siRNA) has been demonstrated to be a potential research tool to analyse gene function and pathway. Expression of an appropriate housekeeping or reference gene can be used as a measurement of transfection efficiency in siRNA. Therefore this study was designed to determine the suitability of GAPDH expression as a measurement of transfection efficiency for p16INK4a gene silencing in HDFs aging model. GAPDH knockdown with an appropriate transfection reagent was measured by quantitative real time RT-PCR while cellular senescence was characterized based on morphological changes, expression of SA b-gal and p16INK4a expression levels. Our findings showed that GAPDH knockdown represents silencing efficiency and down regulation of p16INK4a in senescent transfected HDFs caused morphological alterations which results in the formation of spindle shaped fibroblasts. This study demonstrated the suitability of GAPDH expression as a measurement of transfection efficiency for p16INK4a gene silencing in HDFs aging model.
1. INTRODUCTION
Cellular senescence is recognized as a general response to a variety of oncogenic and genotoxic stresses. It was originally observed in cultures of primary Human Diploid Fibroblasts (HDFs) as they reached the end of their proliferative life span [1]. Normal HDFs enter the senescence state after about 55 - 60 population doublings [2]. Senescent cells have been shown to accumulate with age in human tissues and thus have been proposed to contribute to organismal aging [3]. Senescent cells are resistant to mitogen-induced proliferation, express senescence-associated β-galactosidase (SA β-gal) activity and have an enlarged and flattened morphology [4-7]. Consequently, senescent cells exhibit a gradual loss of replicative potential that results in reduced cell harvest and saturation densities [8].
A distinctive feature of senescent HDFs is that they express elevated levels of p16INK4a and p21CIP1 CyclinDependent Kinase (CDK) inhibitors [9-13].
Different roles of p21 and p16 in cellular senescence of human diploid fibroblasts have been reported. Up‐ regulation of p16 might be essential for maintaining cell cycle arrest in cellular senescence, whereas p21 might be responsible for the inactivation of both cyclin E and cyclin D1-associated kinase activity at the early stage of replicative senescence. A series of cyclin-dependent kinases (CDKs) such as CDK2, CDK4 and CDK6 has been re- ported to play a critical role in the phosphorylation of pRb [14,15] which results in pRb loses its ability to bind to E2F/DP transcription factor complexes. As a consequence, cells commit to S-phase of the cell cycle. However, in senescent cells the activity of CDKs is inhibited by CDK inhibitors such as p21Cip1 and p16INK4a [10,11,16]. Induction of p16INK4a and p21Cip1 prevents phosphorylation of pRb and results in a stable G1 arrest [17]. Thus, CDK inhibitors have the capacity to arrest cells in the G1- phase of the cell cycle and its probable physiological role is in the implementation of irreversible growth arrest termed cellular senescence [18]. In addition, CDK inhibitors specifically p16INK4a can induce some features of cell senescence [19-23]. Therefore, CDK inhibitor p16 was suggested as an important player in aging and agerelated diseases. Previous study showed that p16 expression was very low or absent in young organisms but was increased with advancing age [24].
Over expression of any of the CDK inhibitors will induce G1 cell cycle arrest. The INK4 (inhibitors of CDK4) family of CDK inhibitors including p15, p16, p18 and p19, bind to and inhibit CDK4 and CDK6. In contrast, the Cip/Kip (CDK2-interacting protein) family of CDK inhibitors (p21, p27 and p57) exhibits a broad specificity for CDKs [25].
CDK inhibitor p16 has emerged as an important player in aging and age-related diseases. p16INK4a can induce some features of cell senescence [19-23]. p16 expression was very low or absent in young organisms but was increased with advancing age [24].
In recent years, advances in the development of technologies based on short RNAs that silence gene expression and knockdown the production of proteins have been enormous. The initial report on the development of a potent and sequence specific gene silencing by injecting double-stranded RNA (dsRNA) into Caenorhabditis elegans in 1998 sparked the phenomenon known as interference RNA (RNAi) [26,27]. Application of RNAi was restricted to basic research on the mechanisms of gene expression regulation in nematode and flies, until it was found that gene expression in mammalian cells was silenced by transfecting the cells with “short” RNA inhibitors also known as siRNA [28]. Therefore, inhibition of gene expression using the RNAi has become the method of choice for studying gene function in mammalian cells. However, successful knockdown of the target gene requires efficient delivery of short or small interfering RNAs [29].
The efficiency of siRNA delivery can vary between cell types with different delivery methods. Therefore, identifying a suitable housekeeping gene as a positive control for optimizing gene silencing is important to facilitate successful delivery of siRNA without affecting cell viability.
Housekeeping genes or reference genes are essential endogenous regulatory genes that are involved in various processes in the cell such as metabolism, cell structure, gene transcription and homeostasis and are therefore constitutively expressed [30].
The RNA encoding for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is universally expressed. GAPDH catalyzes the oxidative phosphorylation of glyceraldehyde 3-phosphate to 1,3-bisphosphoglycerate during glycolysis as well as the reverse reaction in tissues involved in gluconeogenesis. GAPDH has also been implicated in other ubiquitous processes such as DNA replication and repair as well as in apoptosis [31]. In this study, we determined the transfection efficiency for p16INK4a gene silencing by using GAPDH siRNA as a positive control. The silencing effect of p16INK4a gene in senescent HDFs was also observed.
2. MATERIALS AND METHODS
2.1. Cell Culture and Induction of Cellular Senescence
This research has been approved by the Ethics Committee of Universiti Kebangsaan Malaysia (Approval Project Code: FF-328-2009). Primary HDFs were derived from the circumcision foreskins of 9 to 12 year-old boys. Written informed consents were obtained from parents of all subjects. The samples were aseptically collected and washed several times with 75% alcohol and phosphate buffered saline (PBS) containing 1% antibiotic-antimycotic solution (PAA, Austria). After removing the epidermis, the pure dermis was cut into small pieces and transferred into a falcon tube containing 0.03% collagenase type I solution (Worthington Biochemical Corporation, USA). Pure dermis was digested in the incubator shaker at 37˚C for 6 - 12 h. Then, cells were rinsed with PBS before being cultured in Dulbecco Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) (PAA, Austria) and 1% antibiotic-antimycotic solution at 37˚C in 5% CO2 humidified incubator. After 5 - 6 days, the cultured HDFs were harvested by trypsinization and culture-expand into new T25 culture flasks (Nunc, Denmark) with expansion degree of 1:4. When the subcultures reached 80% - 90% confluence, serial passaging was done by trypsinization and the number of population doublings (PDs) was monitored until HDFs reached senescence. For the subsequent experiments, cells were used at passage 4 (young cells, population doubling; PD < 12), passage 20 (pre-senescent cells, 30 < PD < 40), or passage 30 (senescent cells, PD > 55).
The number of population doubling of the cells was determined by cell count using trypan blue exclusion dye by using the following formula:
Example:
Initial cell seeding—N0: 100, 000.
Total cell count when culture confluent—N1: 800,000.
Therefore 2n = 800,000/100,000 = 8.
n log2= log8.
The number of cell doubling in culture; n = 3.
Figure 1 shows the arrangement of cells treatment for this study.
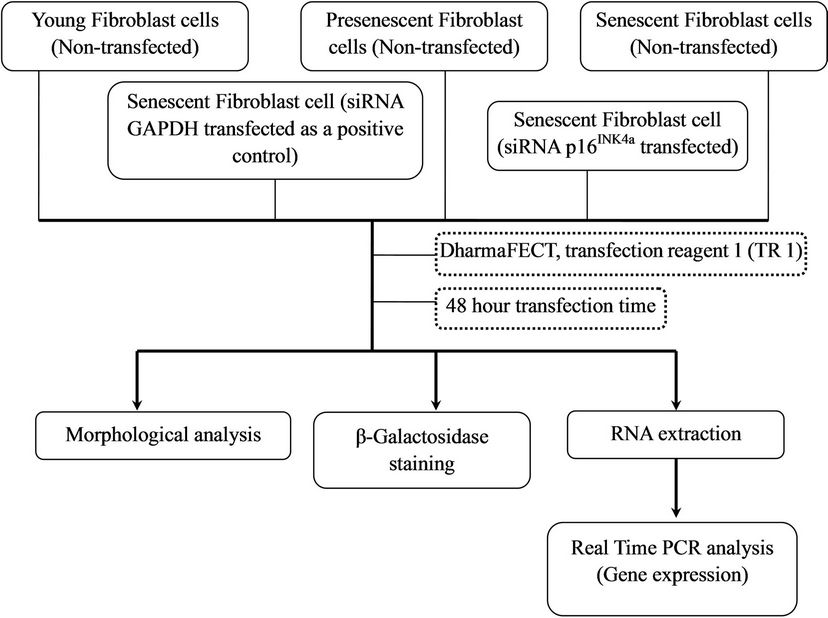
Figure 1. Experimental design.
2.2. Chemical Transfection for 100 nM of siRNA
Senescent HDFs were plated in triplicates with cells density of 2 × 104 per well in 24-well plates. Cells were cultured in antibiotic-free DMEM complete culture medium containing 5% FBS (PAA, Austria) at 37˚C in 5% CO2 humidified incubator. After overnight incubation, 2 μM of ON-TARGETplus GAPDH control pool siRNA and 2 μM ON-TARGETplus SMARTpool p16INK4a siRNA (Dharmacon, USA) solutions were prepared in 1X siRNA buffer (Thermo Scientific, USA). Transfection complexes were prepared in antibiotic-free and serum-free medium by mixing 2 μM of siRNA solution and DharmaFECT tranfection reagents (Thermo Scientific, USA) according to the manufacturer’s recommended protocol. Cells were incubated for 48 h and subsequently the RNA was extracted for real time RT-PCR analysis.
The arrangement of cells treatment for optimization of different transfection reagents is shown in Figure 2.
2.3. Morphology Analysis
Senescent HDFs were transfected with ON-TARGETplus SMARTpool p16INK4a siRNA according to the manufacturer’s instruction. Cells were incubated for 48 h at 37˚C in 5% CO2 humidified incubator. After 48 h transfection, morphology analysis was carried out under inverted microscope.
2.4. Senescence-Associated β-Galactosidase (SA β-Gal) Staining
HDFs positive for SA β-gal activity was determined as described by Dimri et al. 1995 [5]. SA β-gal staining was performed with a senescent cell staining kit (Sigma, USA) according to the manufacturer’s instructions. A total of 1 × 105 cells were seeded in six-well plates and incubated with fixation buffer (2% formaldehyde/0.2% glutaraldehyde) for 6 - 7 min at room temperature. Cells were then rinsed three times with PBS and incubated with 5-bromo- 4-chloro-3-indolyl β-D-galactopyranoside at 1 mg/ml in a buffer containing 40 mM citric acid/phosphate (pH 6.0), 5 mM K3FeCN6, 5 mM K4FeCN6, 150 mM NaCl, and 2 mM MgCl2 for 4 h at 37˚C in the absence of CO2. Blue staining was visible after incubation and the percentage of blue cells observed in 100 cells under a light microscope was calculated.
2.5. RNA Extraction
Total RNA from cultured HDFs in different treatment groups was extracted using TRI Reagent (Molecular Research Center, USA) according to the manufacturer’s instruction. Polyacryl Carrier (Molecular Research Center, USA) was added in each extraction to precipitate the total RNA. Extracted RNA pellet was then washed with 75% ethanol and dried before dissolved in RNase and

Figure 2. Experimental design for different transfection reagents optimization.
DNase free distilled water [32]. The yield and purity of the extracted total RNA were determined by Nanodrop (Thermo Scientific, USA) in triplicates. Total RNA concentration was then adjusted to 50 - 100 ng/µl before stored at –80˚C immediately after extraction.
2.6. Real Time RT-PCR
Quantitative real-time RT-PCR reaction was carried out to evaluate the expression of GAPDH and p16INK4a using 1 μl total RNA as template, 1 ul of forward and reverse primers and iScript One-Step RT-PCR reagent with SYBR Green (Bio-Rad, USA). All reactions were run in duplicate with reaction profile as follows; cDNA synthesis for 30 min at 50˚C; pre-denaturation for 2 min at 94˚C; PCR amplification for 38 cycles with 10 sec at 94˚C and 30 sec at 61˚C [33] using the Bio-Rad iCycler (Bio-Rad, USA). The primers sequence of GAPDH and p16INK4a are shown in Table 1. Relative expression value of target genes was calculated based on the 2−ΔΔCt method of relative quantification [34] by the following equation:
Relative expression value = 2Ct value of GAPDH – Ct value gene of interest.
2.7. Statistical Analysis
Each experiment was performed in duplicates on three independent cultures. Data were reported as means ± SD. Student’s t-test (two-tailed) was used to compare differences between groups. P < 0.05 was considered statistically significant.
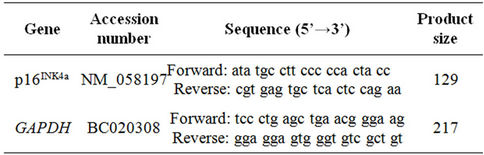
Table 1. Primer sequences for quantitative real-time RT-PCR.
3. RESULTS
3.1. GAPDH as a Positive Control of Gene Silencing
In order to determine the efficiency and successful delivery of siRNA in senescent HDFs, optimization of the different transfection reagents was carried out using GAPDH siRNA as a positive control. The results showed a significant inhibition of GAPDH mRNA expression in transfected senescent HDFs when DharmaFECT transfection reagent 1 was used (p < 0.05) (Figure 3). Similar inhibition of GAPDH mRNA expression was not observed when DharmaFECT transfection reagent 2 (TR 2), 3 (TR 3) and 4 (TR 4) were used. Figure 4 shows a significant inhibition of GAPDH mRNA expression in senescent HDFs transfected with GAPDH siRNA for 48 h (p < 0.05).
3.2. mRNA Expression of p16INK4a in HDFs Aging Model
p16INK4a mRNA expression was increased in senescent
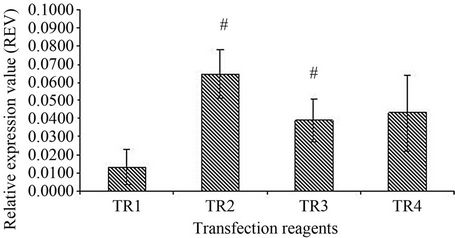
Figure 3. Effect of transfection reagent (TR) on GAPDH expression. Four DharmaFECT transfection reagents were tested for transfection optimization. DharmaFECT transfection reagent 1 (TR1) showed a significant knockdown of GAPDH compared to DharmaFECT 2 (TR2) and DharmaFECT 3 (TR3); (#); p < 0.05. All values were expressed as relative expression value (REV) with mean ± SD, n = 6.

Figure 4. Comparison of GAPDH mRNA expression in transfected and non-transfected senescent HDFs measured 48 h after transfection with 100 nM siRNA targeting GAPDH. GAPDH siRNA was used as a positive control for transfection efficiency. Results showed GAPDH knockdown which represents a relative measurement of transfection efficiency. All values were expressed as relative expression value (REV) with mean ±SD, n = 6.
HDFs as compared to young and pre-senescent cells. Senescent HDFs transfected with p16INK4a siRNA however exhibited down regulation of this gene (Figure 5).
3.3. Morphology Analysis
Young HDFs exhibit spindle shaped fibroblasts (Figure 6(a)) while senescent HDFs display flattened and enlarged morphology (Figure 6(b)). Senescent HDFs transfected with p16INK4a siRNA showed similar morphology of young cells with the presence of small and spindle shaped fibroblasts (Figure 6(c)).
3.4. Senescence-Associated Marker
An established marker for cellular senescence viz. SA b-gal was present in senescent and p16INK4a siRNA transfected senescent HDFs (Figures 7(b) and (c)). No positive staining for SA b-gal was observed in young HDFs (Figure 7(a)).
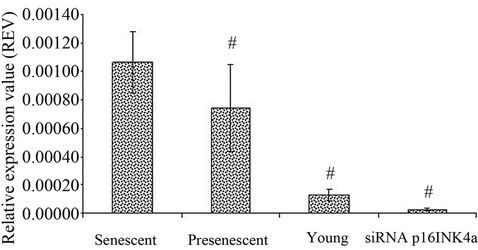
Figure 5. Expression of p16INK4a in HDFs with different experimental treatments. p16INK4a mRNA expression was measured in young, pre-senescent, senescent and transfected senescent HDFs (siRNA p16INK4a). Significant down regulation of p16INK4a expression in pre-senescent, young and transfected senescent HDFs (siRNA p16INK4a) compared to senescent HDFs (#); p < 0.05. All values were normalised to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels. Data was presented as mean ± SD, n = 6.
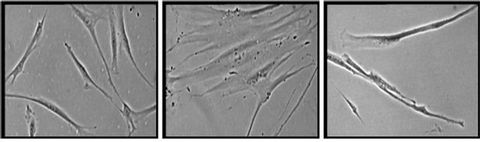 (a) (b) (c)
(a) (b) (c)
Figure 6. Morphology of human diploid fibroblasts with different experimental treatments. HDFs at young age (passage 4) (a), senescent (passage 30) (b) and transfected senescent HDFs (siRNA p16INK4a) (c). Senescent HDFs showed increased cytoplasm volume and vacuoles and loss its original fibroblastic shape by acquiring a flattened feature (b). Senescent cells transfected with siRNA p16INK4a showed similar morphology to young cells (c). Micrographs are shown at ×200 magnification.
 (a) (b) (c)
(a) (b) (c)
Figure 7. Expression of senescence-associated (SA) β-galactosidase with different experimental treatments. HDFs at young age (passage 4) (a), senescent (passage 30) (b) and transfected senescent HDFs (siRNA p16INK4a) (c). Cells with blue staining indicated positive for SA β-galactosidase activity. Micrographs are shown at ×200 magnification.
4. DISCUSSION
Housekeeping gene transcripts such as β-actin and GAPDH have been used in gene expression analysis. These transcripts, due to their stable expression, allow quantification of other gene expression [35]. Housekeeping genes are essential endogenous regulatory genes that are involved in various processes in the cell such as metabolism, cell structure, gene transcription and homeostasis and are therefore constitutively expressed [36]. The expression of housekeeping gene provides simple and precise quantitative assay for quantification of genes of interest.
In RNAi start-up experiments, a positive control siRNA is used to determine the optimal transfection condition which results in complete knockdown of the target gene. siRNAs targeting housekeeping genes are commonly used as positive controls. In this study, the efficiency and delivery of siRNA in senescent HDFs was determined by optimizing four transfection reagents formulated by Dharmacon (DharmaFECT TR 1, TR 2, TR 3 and TR 4) using GAPDH siRNA as a positive control. Results showed that in senescent human diploid fibroblasts (HDFs) model, DharmaFECT 1 transfection reagent exhibited the most efficient delivery of siRNA with significant inhibition of GAPDH mRNA expression indicating the transfection method was optimised for the HDFs aging model used in the study.
When fibroblast cells undergo senescence, distinctive molecular and morphological alterations such as cellular flattening, loss of c-fos induction to serum stimulation and expression of SA β-gal activity were observed [17,37]. Besides the flattened morphology and positive staining for SA β-gal, other characteristics of senescent HDFs are cell growth arrest and up-regulation of cell cycle inhibitors.
Accumulation of p16INK4a in senescent cells inhibits retinoblastoma (Rb) phosphorylation and this is regarded as the most important tumour suppressing function of p16INK4a. Hypophosphorylated Rb family proteins, enhancement of actin stress fibers and the redistribution of focal adhesion proteins were reported to contribute to the morphological changes observed in senescent HDFs [38]. p16INK4a maintains Rb in the active state independent of E2F binding [39] and impairment of this mechanism in senescent cells retains Rb in the hypophosphorylated inhibitory form [40].
In this study, transfected senescent HDFs exhibited down regulation of p16INK4a gene which results in more cells showed spindle shaped morphology which resem- bles that of young cells even though SA b-gal activity is observed. The alteration in HDFs morphology which resembles that of young cells with p16INK4a knock down may be explained by inhibition of extra cellular matrix degradation as shown by down regulation of MMP1 gene and up-regulation of COL1A1 gene via Rb regulation [36] indicating p16INK4a is involved in the morphogenesis of HDFs during cellular senescence. Kato et al. (1998) reported that expression of p16INK4a in glioma cells, diploid fibroblasts and squamous carcinoma cells was associated with cell flattening and expression of senescence-associated marker, SA β-gal at pH 6.0. The increased expression of p16INK4a subsequently was reported to cause senescence of cell [41].
Senescent fibroblasts contain p16INK4a levels of at least 40-fold greater than cells at early passage. An increase in p16INK4a expression caused cell growth arrest and induced cellular senescence [9] while inhibition of the p16INK4a expression leads to the extension of proliferative lifespan and finally delay the onset of cellular senescence [2].
5. CONCLUSION
The findings of the present study suggest that GAPDH expression can be used as a measurement of transfection efficiency for p16INK4a gene silencing particularly in senescent HDFs and it is suitable as a positive control for siRNA optimization. Silencing of p16INK4a gene caused alteration in the morphology of senescent HDFs which resembles that of young cells.
6. ACKNOWLEDGEMENTS
This study was funded by the Ministry of Higher Learning under the Fundamental Research Grant Scheme UKM-FF-03-FRGS0043-2009 and National University of Malaysia grant FF-328-2009.
REFERENCES
- Hayflick, L. and Moorhead, P.S. (1961) The serial cultivation of human diploid cell strains. Experimental Cell Research, 25, 585-621. doi:10.1016/0014-4827(61)90192-6
- Zheng, W., Wang, H., Xue, L., Zhang, Z. and Tong, T. (2004) Regulation of cellular senescence and p16INK4a expression by Id1 and E47 proteins in human diploid fibroblast. The Journal of Biological Chemistry, 279, 31524-31532. doi:10.1074/jbc.M400365200
- Campisi, J. (2000) Cancer, aging and cellular senescence. In Vitro, 14, 183-188.
- Park, W.Y., Park, J.S., Cho, K.A., Kim, D.I., Ko, Y.G., Seo, J.S. and Park, S.C. (2000) Up-regulation of caveolin attenuates epidermal growth factor signaling in senescent cells. Journal of Biological Chemistry, 275, 20847-20852. doi:10.1074/jbc.M908162199
- Dimri, G.P., Lee, X., Basile, G., Acosta, M., Scott, G., Roskelley, C., Medrano, E.E., Linskens, M., Rubelj, I., Pereira-Smith, O., Peacocke, M. and Campisi, J. (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proceedings of the National Academy of Sciences of the United States of America, 92, 9363-9367. doi:10.1073/pnas.92.20.9363
- Wagner, M., Hampel, B., Bernhard, D., Hala, M., Zwerschke, W. and Jansen-Durr, P. (2001) Replicative senescence of human endothelial cells in vitro involves G1 arrest, polyploidization and senescence-associated apoptosis. Experimental Gerontology, 36, 1327-1347. doi:10.1016/S0531-5565(01)00105-X
- Cho, K.A., Ryu, S.J., Oh, Y.S., Park, J.H., Lee, J.W., Hwang-Phill, K., Kim, K.T., Jang, I.S. and Park, S.C. (2004) Morphological adjustment of senescent cells by modulating caveolin-1 status. The Journal of Biological Chemistry, 279, 42270-42278. doi:10.1074/jbc.M402352200
- Cristofalo, V.J. (1988) Cellular biomarkers of aging. Experimental Gerontology, 23, 297-305. doi:10.1016/0531-5565(88)90032-0
- Alcorta, D.A., Xiong, Y., Phelps, D., Hannon, G., Beach, D. and Barrett, J.C. (1996) Involvement of the cyclindependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proceedings of the National Academy of Sciences of the United States of America, 93, 13742-13747. doi:10.1073/pnas.93.24.13742
- Hara, E., Smith, R., Parry, D., Tahara, H., Stone, S. and Peters, G. (1996) Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Molecular Cell Biology, 16, 859-867.
- Noda, A., Ning, Y., Venable, S.F., Pereira-Smith, O.M. and Smith, J.R. (1994) Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Experimental Cell Research, 211, 90-98. doi:10.1006/excr.1994.1063
- Stein, G.H., Drullinger, L.F., Soulard, A. and Dulic, V. (1999) Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Molecular and Cellular Biology, 19, 2109-2117.
- Wong, H. and Riabowol, K. (1996) Differential CDK inhibitor gene expression in aging human diploid fibroblasts. Experimental Gerontology, 31, 311-325. doi:10.1016/0531-5565(95)00025-9
- Berthet, C., Klarmann, K.D., Hilton, M.B., Suh, H.C., Keller, J.R., Kiyokawa, H. and Kaldis, P. (2006) Combined loss of Cdk2 and Cdk4 results in embryonic lethality and Rb hypophosphorylation. Developmental Cell, 10, 563-573. doi:10.1016/j.devcel.2006.03.004
- Malumbres, M. and Barbacid, M. (2007) Cell cycle kinases in cancer. Current Opinion in Genetic Development, 17, 60-65. doi:10.1016/j.gde.2006.12.008
- Gil, J. and Peters, G. (2006) Regulation of the INK4bARF-INK4a tumour suppressor locus: All for one or one for all. National Review of Molecular and Cellular Biology, 7, 667-677. doi:10.1038/nrm1987
- Campisi, J. (2001) Cellular senescence as a tumor-suppressor mechanism. Trends in Cell Biology, 11, S27-S31.
- Ohtani, N., Yamakoshi, K., Takahashi, A. and Hara, E. (2004) The p16INK4a-RB pathway: Molecular link between cellular senescence and tumor suppression. The Journal of Medical Investigation, 51, 146-153. doi:10.2152/jmi.51.146
- Campisi, J., Dimiri, G. and Hara, E. (1996) Control of replicative senescence. In: Schneider, E.L., Rowe, J.W., Johnson, T.G., Holbrook, N.J. and Morrison, J.H., Eds., Handbook of the Biology of Aging, Academic Press, New York.
- Brenner, A.J., Stampfer, M.R. and Aldaz, C.M. (1998) Increased p16 expression with first senescence arrest in human mammary epithelial cells and extended growth capacity with p16 inactivation. Oncogene, 17, 199-205. doi:10.1038/sj.onc.1201919
- Jarrad, D.F., Sarkar, S., Shi, Y., Yeager, T.R., Magrane, G., Kinoshita, H., Nassif, N., Meisner, L., Newton, M.A., Waldman, F.M. and Reznikoff, C.A. (1999) p16/pRb pathway alterations are required for bypassing senescence in human prostate epithelial cells. Cancer Research, 59, 2957-2964.
- You, Y.O., Lee, G. and Min, B.M. (2000) Retinoic acid extends the in vitro life span of normal human oral keratinocytes by decreasing p16 (INK4A) expression and maintaining telomerase activity. Biochemical and Biophysical Research Communications, 268, 268-274. doi:10.1006/bbrc.2000.2101
- Sandhu, C., Peehl, D.M. and Slingerland, J. (2000) p16INK4a mediates cyclin dependent kinase 4 and 6 inhibition in senescent prostatic epithelial cells. Cancer Research, 60, 2616-2622.
- Zindy, F., Quelle, D.E., Roussel, M.F. and Sherr, C.J. (1997) Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene, 15, 203-211. doi:10.1038/sj.onc.1201178
- Gao, C.Y. and Zelenka, P.S. (1997). Cyclins, cyclindependent kinases and differentiation. BioEssays, 19, 307. doi:10.1002/bies.950190408
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E. and Mello, C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenor-habditis elegans. Nature, 391, 806-811. doi:10.1038/35888
- Lee, S.H. and Sinko, P.J. (2006) SiRNA-Getting the message out. European Journal of Pharmaceutical Sciences, 27, 401-410. doi:10.1016/j.ejps.2005.12.002
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. and Tuschl, T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494-498. doi:10.1038/35078107
- Rozema, D.B. and Lewis, D.L. (2003) SiRNA delivery technologies for mammalian systems. Targets, 2, 253- 260. doi:10.1016/S1477-3627(03)02381-X
- Touchberry, C.D., Wacker, M.J., Richmond, S.R., Whitman, S.A. and Godard, M.P. (2006) Age-Related Changes in Relative Expression of Real-Time PCR Housekeeping Genes in Human Skeletal Muscle. Journal of Biomolecular Techniques, 17, 157-162.
- Corbin, I.R., Gong, Y., Zhang, M. and Minuk, G.Y. (2002) Proliferative and nutritional dependent regulation of glycer-aldehyde-3-phosphate dehydrogenase expression in the rat liver. Cell Proliferation, 35, 173-182. doi:10.1046/j.1365-2184.2002.00236.x
- Chua, K.H., Aminuddin, B.S., Fuzina, N.H. and Ruszymah, B.H.I. (2005) Insulin-transferrin-selenium prevent human chondrocyte dedifferentiation and promote the formation of high quality tissue engineered human hyaline cartilage. European Cells and Materials, 9, 58-67.
- Zainuddin, A., Chua, K.H., Abdul Rahim, N. and Makpol, S. (2010) Effects of experimental treatment on GAPDH mRNA expression as a housekeeping gene in human diploid fibroblasts. BMC Molecular Biology, 11, 59. doi:10.1186/1471-2199-11-59
- Carthagena, L., Bergamaschi, A., Luna, J.M., David, A., Uchil, P.D., Margottin-Goguet, F., Mothes, W., Hazan, U., Transy, C., Pancino, G. and Nisole, S. (2009) Human TRIM gene expression in response to interferons. PloS One, 4, e4894. doi:10.1371/journal.pone.0004894
- Thellin, O., Zorzi, W., Lakaye, B., De Borman, B., Coumans, B., Hennen, G., Grisar, T., Igout, A. and Heinen, E. (1999) Housekeeping genes as internal standards: Use and limits. Journal of Biotechnology, 75, 291-295. doi:10.1016/S0168-1656(99)00163-7
- Chad, D.T., Michael, J.W., Scott, R.R., Samantha, A.W. and Michael, P.G. (2006) Age-related changes in relative expression of real time RT PCR housekeeping genes in human skeletal muscle. Journal of Biomolecular Techniques, 17, 157-162.
- Sharpless, N.E. (2003) The persistence of senescence. Science of Aging Knowledge Environment, PE24. doi:10.1126/sageke.2003.33.pe24
- Chen, Q.M., Tu, V.C., Catania, J., Burton, M., Toussaint, O. and Dilley, T. (2000) Involvement of Rb family proteins, focal adhesion proteins and protein synthesis in senescent morphogenesis induced by hydrogen peroxide. Journal of Cell Science, 113, 4087-4097.
- Sellers, W.R., Novitch, B.G., Miyake, S., Heith, A., Otterson, G.A., Kaye, F.J., Lassar, A.B. and Kaelin, W.G. (1998) Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation and suppress tumor cell growth. Genes & Development, 12, 95-106. doi:10.1101/gad.12.1.95
- Makpol, S., Zainuddin, A., Chua, K.H., Yusof, Y.A., Wan, N.W.Z. (2012) Gamma-tocotrienol modulation of senescence-associated gene expression prevents cellular aging in human diploid fibroblasts. Clinics, 67, 135-143. doi:10.6061/clinics/2012(02)08
- Kato, D., Miyazawa, K., Starborg, M., Wada, I., Oka, T., Sakai, T., Peters, G. and Hara, E. (1998) Features of replicative senescence induced by direct addition of antennapedia-p16INK4a fusion protein to human diploid fibroblasts. FEBS Letters, 427, 203-208. doi:10.1016/S0014-5793(98)00426-8
NOTES
*The authors of these manuscripts have no competing financial interests.
#Corresponding author.

